ABSTRACT
The type III protein secretion system (T3SS) encoded by the locus of enterocyte effacement (LEE) is essential for the pathogenesis of attaching/effacing bacterial pathogens, including enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), and Citrobacter rodentium. These pathogens use the T3SS to sequentially secrete three categories of proteins: the T3SS needle and inner rod protein components; the EspA, EspB, and EspD translocators; and many LEE- and non-LEE-encoded effectors. SepD and SepL are essential for translocator secretion, and mutations in either lead to hypersecretion of effectors. However, how SepD and SepL control translocator secretion and secretion hierarchy between translocators and effectors is poorly understood. In this report, we show that the secreted T3SS components, the translocators, and both LEE- and non-LEE-encoded effectors all carry N-terminal type III secretion and translocation signals. These signals all behave like those of the effectors and are sufficient for mediating type III secretion and translocation by wild-type EPEC and hypersecretion by the sepD and sepL mutants. Our results extended previous observations and suggest that the secretion hierarchy of the different substrates is determined by a signal other than the N-terminal secretion signal. We identified a domain located immediately downstream of the N-terminal secretion signal in the translocator EspB that is required for SepD/SepL-dependent secretion. We further demonstrated that this EspB domain confers SepD/SepL- and CesAB-dependent secretion on the secretion signal of effector EspZ. Our results thus suggest that SepD and SepL control and regulate secretion hierarchy between translocators and effectors by recognizing translocator-specific export signals.
IMPORTANCE Many bacterial pathogens use a syringe-like protein secretion apparatus, termed the type III protein secretion system (T3SS), to secrete and inject numerous proteins directly into the host cells to cause disease. The secreted proteins perform different functions at various stages during infection and are classified into three substrate categories (T3SS components, translocators, and effectors). They all contain secretion signals at their N termini, but how their secretion hierarchy is determined is poorly understood. Here, we show that the N-terminal secretion signals from different substrate categories all behave the same and do not confer substrate specificity. We further characterize the secretion signals of the translocators and identify a translocator-specific signal, demonstrating that substrate-specific secretion signals are required in regulating T3SS substrate hierarchy.
INTRODUCTION
The type III secretion system (T3SS) is one of the best-studied bacterial protein secretion systems and plays a central role in the virulence and pathogenesis of a large number of medically and agriculturally important bacterial pathogens, including Yersinia, Salmonella, Pseudomonas, Shigella, and Escherichia spp. (1–3). The T3SS consists of a series of multicomponent, ring-shaped protein structures spanning the bacterial envelope, the extracellular space, and the host cellular membrane, enabling direct delivery of bacterial virulence proteins, called effectors, into the host cell, where the effectors modulate and subvert host cellular functions and immune responses, leading to diseases (1–3).
The T3SS sequentially secretes a large number of diverse proteins, and these secreted proteins can be divided into at least three categories based on their functions, including the early substrates (T3SS needle and inner rod components), the intermediate substrates (translocators), and the late substrates (effectors), although some regulatory proteins in certain T3SSs are also secreted (1–3). The needle and inner rod proteins are secreted first via a basal body of the T3SS to complete the assembly of a fully functional protein export apparatus and are required for type III secretion (T3S) of the intermediate and late substrates. The translocators are presumably secreted after the needle and inner rod components but before the effectors, since they are needed for translocating the effectors into host cells. In addition, some effectors have opposing functions, and their secretion may be required at different infection stages. Therefore, these different substrates need to be secreted in a programmed temporal hierarchy.
There are at least two distinct substrate specificity switching events operating in the T3SSs (2), the first switching from secreting the early substrates (needle and inner rod proteins) to secreting the intermediate substrates (translocators), and the second from translocators to effectors. Several proteins controlling these switches and regulating T3S hierarchy have been identified. The YscU/FlhB/EscU/SpaS family of proteins regulates the first switch, whereas the YopN/SepL/MxiC/InvE family of proteins controls the second switch (reviewed in reference 2). Customized chaperones are often required for the stability and secretion of different substrates, and it has been proposed that the SpaO-OrgA-OrgB complex serves as a cytoplasmic sorting platform for the complexes of different substrates and their T3S chaperones, and the affinity between the chaperone-substrate complexes and the sorting platform may determine the secretion hierarchy (4, 5).
Proteins secreted from bacteria often contain a tag or epitope that serves as the targeting signal for a specific secretion pathway (6). The proteins secreted via the T3SS also carry targeting signals usually located at the N-terminal 20 or so amino acid residues (7) (reviewed in reference 2), although the T3S targeting signals are located in the mRNA of some effectors (8, 9). Since most T3SS substrates appear to possess secretion signals in their amino acid sequences, several machine-learning computational approaches based on the N-terminal sequences of effectors have been used to predict T3S signals (10). Biochemical and structural analyses of the T3S signals of several effectors have failed to reveal a common structural motif and found these signals to be intrinsically disordered (11). The N-terminal T3S signals do not seem to determine the substrate hierarchy (reviewed in reference 2). It has recently been shown that the translocator PopD of Pseudomonas aeruginosa contains two additional signals, besides the N-terminal secretion signal, that are required for its proper hierarchical secretion (12). There is some evidence that the N-terminal T3S signal of the translocator YopD of Yersinia pseudotuberculosis appears to be distinct from secretion signals of other T3SS substrates, such as effectors, but this distinction in sequence and/or structure is yet to be defined (13). How the N-terminal T3S signal is recognized and connected to the T3SS is poorly understood.
Enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), and the mouse pathogen Citrobacter rodentium are the prototypical members of the family of attaching/effacing (A/E) bacterial pathogens. EPEC and EHEC are diarrheagenic in humans and cause significant morbidity and mortality worldwide, as well as severe economic loss resulting from food and water contamination (14). The A/E pathogens all possess a highly conserved T3SS encoded by a chromosomally located pathogenicity island, the locus of enterocyte effacement (LEE), and this T3SS is crucial to the virulence and pathogenesis of these pathogens (14–16). The LEE-encoded T3SS is used by the A/E pathogens to secrete a large number of proteins, including the needle protein EscF and the inner rod component EscI of the T3SS, the three translocators EspA, EspB, and EspD, and a varying number of both LEE- and non-LEE-encoded effectors that can range from as few as 22 in EPEC to as many as 41 in certain EHEC strains (17–23).
The regulation of the secretion hierarchy of the LEE-encoded T3SS in the A/E pathogens is not well understood, with the molecular details still lacking. The secretion hierarchy appears to be regulated at the posttranslational level in the A/E pathogens (24, 25). Several proteins, namely, EscU, SepD, and SepL, have been implicated in regulating the secretion hierarchy in these pathogens. The autocleavage of EscU, a homolog of YscU and FlhB, is implicated in regulating the secretion hierarchy between the early substrates and the later substrates (2). We have shown that EscU and its autocleavage are essential for type III secretion of the translocators and effectors (26). SepD and SepL are required for the secretion of the translocators EspA, EspB, and EspD, and mutation of either sepD or sepL abolishes secretion of the translocators and leads to hypersecretion of the effectors, suggesting that SepD and SepL control the secretion hierarchy between translocators and effectors (17, 24, 27–29). SepL is a member of the Yersinia YopN-TyeA family of gatekeeper proteins, including InvE and SsaL in Salmonella and MxiC in Shigella, while SepD is a homolog and functional equivalent of SpiC in Salmonella (2, 24, 28, 30, 31). The homologs of SepD and SepL in other T3SSs are also implicated in regulating the secretion hierarchy between translocators and effectors in response to certain environmental cues (2, 31–34). It is hypothesized that SepD and SepL form a secretion switch (17, 24). SepL has been shown to bind to Tir, suggesting that SepL may prevent Tir secretion before the translocators are secreted (29). However, how SepD and SepL control the secretion of translocators is not understood. It has been shown that the N-terminal 20 amino acid residues of translocators and effectors contain sufficient information for type III secretion and translocation and that the N-terminal signals of the translocators behave like those of the effectors by conferring hypersecretion in the sepD and sepL mutants in the A/E pathogens (25, 35, 36).
In this report, we confirm and extend these observations. We found that, similar to the translocators and effectors, the N termini of the secreted T3SS components EscF and EscI are also sufficient for type III secretion and translocation. We further demonstrated that these N-terminal type III secretion and translocation signals do not confer substrate specificity and hierarchy. By using a domain swapping strategy, we were able to identify a domain in the translocator EspB that is required for SepD/SepL-dependent secretion. Placing this EspB domain immediately downstream of the N-terminal secretion signal of effector EspZ rendered the hybrid protein to be preferentially secreted in an SepD/SepL-dependent fashion. In addition, we showed that the C terminus of another translocator EspD is critical for its type III secretion and may contain an accessory secretion signal, as truncations and epitope tagging at the C terminus of EspD prevent its type III secretion. Our results suggest that type III secretion hierarchy of certain substrates is determined by signals other than the N-terminal secretion and translocation signal.
MATERIALS AND METHODS
Bacterial strains, cell lines, and growth conditions.
A streptomycin-resistant derivative of EPEC O127:H6 strain E2348/69 and its escN, sepD, and sepL gene deletion mutants, as described previously (24, 37), were used in this study. E. coli strain DH10B was used as the transformation host for cloning. Bacteria were grown in Luria-Bertani (LB) broth or agar plates at 37°C supplemented with the appropriate antibiotics at the following concentrations (μg/ml): streptomycin, 50; kanamycin, 50; and tetracycline, 10. Human epithelial cell line HeLa (CCL2; American Type Culture Collection) was used for the type III translocation assay. HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) from HyClone (Logan, UT) supplemented with 10% (vol/vol) fetal calf serum (FCS), 4,500 mg/liter glucose, and 4 mM l-glutamine at 37°C in a tissue culture incubator with an atmosphere containing 5% (vol/vol) CO2.
Construction of β-lactamase TEM-1 fusions.
Plasmid pCX341, a derivative of pCX340 with its origin of replication replaced with that of pBR322, was used to generate N-terminal translational fusions to the mature form of TEM-1 β-lactamase (36, 38). The coding regions encoding various fragments of EPEC T3SS-secreted substrates, including the needle and inner rod components EscF and EscI/rOrf8, translocators EspA, EspB, and EspD, LEE-encoded effectors EspZ and Tir, and non-LEE-encoded effectors NleA and NleD, were amplified by PCR and cloned first into pCR2.1-TOPO (Invitrogen) before being subcloned into pCX341. This cloning step was carried out in E. coli strain DH10B. All the constructs were verified by DNA sequencing before transformation into EPEC strains by electroporation. For plasmid constructs that were recalcitrant to be transformed directly into our EPEC strains, they were first passed through restriction-minus EPEC E2348/69 ΔhsdR strain NH4 before electroporation (39).
(i) N-terminal fragments of type III secretion substrates fused to TEM.
It has been shown in A/E pathogens that the N-terminal ca. 20 amino acid residues of the translocators and some effectors are sufficient for serving as signals for type III secretion and translocation (25, 36). The coding regions of the N-terminal 20 amino acid residues of EPEC EscF, EspA, EspB, EspD, Tir, and NleD, 21 amino acid residues of EspZ and NleA, and 23 amino acid residues of EscI were cloned immediately upstream of the blaM gene (encoding TEM-1 β-lactamase) in pCX341. The same pair of restriction sites, NdeI and EcoRI, was incorporated into the PCR primers and used for cloning for all these TEM-1 fusion proteins, and the variation in amino acid residue numbers from 20 to 23 was the result of this constraint of restriction sites, in order to avoid the introduction of any foreign amino acid residue between the type III secretion substrates and TEM-1.
(ii) Generation of TEM fusions to nested truncations of EspB.
To delineate the region required for SepD/SepL-dependent secretion, the coding regions of the full-length EspB as well as its N-terminal 20, 50, 60, 70, 80, and 100 amino acid residues were amplified by PCR and cloned in front of blaM in pCX341 by using the restriction enzymes NdeI and EcoRI. Similarly, fusions of the 21 to 70 and 21 to 96 amino acid residues of EspB to TEM-1 were also generated.
(iii) Construction of TEM fusions to different regions of EspA and EspD.
A series of constructs expressing TEM fusions of different parts of EspA, including the full-length protein as well as the N-terminal 20, 50, 100, 140, and 175 amino acid residues, were generated. Likewise, the coding regions for the full-length EspD and its N-terminal 20, 81, 144, 179, 233, 336, and 368 amino acid residues were amplified by PCR and cloned upstream of the blaM gene in pCX341. All these constructs used restriction enzymes NdeI and EcoRI for cloning.
(iv) Generation of TEM fusions containing chimeric type III secretion signals of effector EspZ and translocator EspB.
As described earlier, a plasmid construct (pEPespZ21-TEM) was generated to express an EspZ21-TEM fusion protein by cloning the coding region of the N-terminal 21 amino acid residues of EspZ into the NdeI and EcoRI restriction sites of pCX341. To facilitate the generation of hybrid secretion signal constructs between EspZ and EspB, a KpnI site was introduced at the end of the EspZ21 coding region immediately upstream of the EcoRI site in pEPespZ21-TEM by PCR, creating pEPespZ21-TEM/KpnI. The coding regions of EspB amino acid residues 51 to 96, 21 to 70, and 21 to 96 were then amplified by PCR with primers containing KpnI (in forward primers) and EcoRI (in reverse primers) sites and cloned into pEPespZ21-TEM/KpnI digested by KpnI/EcoRI to generate 3 TEM fusions containing EspZ and EspB chimeric secretion signals (EspZ21-EspB51–96, EspZ21-EspB21–96, and EspZ21-EspB21–70) at the N terminus.
Construction of untagged and C-terminally FLAG-tagged full-length EspD, EspD1–336, and EspD1–368.
The coding regions with stop codons for full-length EspD (380 amino acid residues) and its N-terminal 336 and 368 amino acid residues were amplified by PCR and cloned upstream of the blaM gene in pCX341 using the restriction sites NdeI and EcoRI to express untagged EspD and its C-terminal truncations, EspD1–336 and EspD1–368. To add a C-terminal FLAG tag to these three EspD constructs, oligonucleotides (5′-GATTACAAGGATGACGACGATAAG-3′) encoding the FLAG epitope (DYKDDDDK) were added in front of the stop codons in the reverse PCR primers for the coding regions of full-length EspD, EspD1–336, and EspD1–368 to amplify the respective espD regions. These PCR products were then cloned upstream of the blaM gene in the restriction sites NdeI and EcoRI of pCX341 to express C-terminally FLAG-tagged EspD, EspD1–336, and EspD1–368.
Generation of nonpolar ΔespD, ΔescN ΔespD, ΔsepD ΔespD, ΔcesAB, and ΔsepD ΔcesAB deletion mutants in EPEC.
In-frame, nonpolar espD deletion mutants were generated in EPEC strain E2348/69 using the allelic exchange method as described before (24). Two PCR products were generated using primer pairs EPespD-1/EPespD-DR (EPespD-1, 5′-GCGGTACCGGGCGCTATCGTTCGATCTGTC-3′ [KpnI]; EPespD-DR, 5′-GCGCTAGCCACAGACTGGATATCGTTATTTAC-3′ [NheI]) and EPespD-DF/EPespD-2 (EPespD-DF, 5′-GCGCTAGCATCCGTATTGTCAGCGGTCGAG-3′ [NheI]; EPespD-2, 5′-GCGAGCTCGCCCTGTTTGGTTACGTGCTTC-3′ [SacI]) (the restriction sites indicated in brackets are underlined). The two PCR products were digested with KpnI/NheI and NheI/SacI, respectively, and cloned together into KpnI/SacI-digested pRE112. The resulting suicide plasmid construct was introduced into the EPEC wild-type (WT), ΔescN, and ΔsepD strains by conjugation to generate EPEC ΔespD, ΔescN ΔespD, and ΔsepDΔespD mutants via allelic exchange using sucrose selection. The mutants were verified by PCR. The region of espD from codons 12 to 372 was deleted.
In-frame cesAB deletion mutants were constructed in EPEC strain E2348/69 in a similar manner. Two DNA fragments were amplified by PCR using primer pairs EPorf3-1/DEPorf3-R and DEPorf3-F/EPorf3-2. The sequences for the primers are as follows: EPorf3-1, 5′-CGGTACCGAGATCTATCTTATAAAGAGAAACGC-3′ (KpnI); DEPorf3-R, 5′-CGCTAGCTTGGCTCACAATACTCATCCTC-3′ (NheI); DEPorf3-F, 5′-CGCTAGCAGAAATAATAGAAAAATAGTATGAC-3′ (NheI); and EPorf3-2 5′-CGAGCTCATTATGCGGTGTGTTTGCATC-3′ (SacI) (the restriction sites indicated in parentheses are underlined). The PCR products were digested with KpnI/NheI and NheI/SacI and cloned into KpnI/SacI-digested pRE112 in a 3-way ligation. The resulting plasmid was introduced into EPEC WT and the ΔsepD mutant by conjugation to generate EPEC ΔcesAB and ΔsepD ΔcesAB mutants via allelic exchange. The region of cesAB from codons 7 to 100 was deleted.
Type III secretion assay.
EPEC wild-type (WT), ΔescN, ΔsepD, and/or ΔsepL strains containing pCX341 or derivatives were grown in LB broth containing streptomycin and tetracycline overnight in a shaker at 225 rpm. The cultures were then diluted 1:40 into 3 ml of prewarmed DMEM (HyClone) supplemented with 4,500 mg/liter glucose, 4 mM l-glutamine, and 110 mg/liter sodium pyruvate without any antibiotics in a 6-well plate and grown statically at 37°C for 6 h in a tissue culture incubator containing 5% CO2 (vol/vol) to induce type III secretion. The cultures were centrifuged at 16,100 × g for 10 min to pellet the bacteria, and the bacterial pellet was resuspended in SDS-PAGE sample buffer to generate whole-cell lysates. The supernatant was collected and filtered through a 0.22-μm-pore-size filter unit (Millipore), and the proteins in the supernatant were precipitated with 10% (vol/vol) trichloroacetic acid (TCA). After centrifugation at 16,100 × g for 20 min, the precipitated protein pellet was dried in air and dissolved in SDS-PAGE sample buffer. The volume of the sample buffer used to resuspend the bacterial pellet or dissolve the precipitated proteins was normalized relative to the optical density at 600 nm of the cultures to ensure equal loading of samples.
Protein analysis and immunoblotting.
Total bacterial lysates and secreted proteins of EPEC were run on an SDS-12% PAGE gel according to standard protocols, and protein gels were stained with Coomassie brilliant blue G (Sigma). For detection of TEM-1 and its fusion proteins by immunoblotting, proteins resolved by SDS-PAGE were transferred onto pure nitrocellulose membranes (Bio-Rad) and detected by using mouse monoclonal antibodies against N-terminally His-tagged TEM-1 β-lactamase (QED Bioscience Inc.) at 1:2,000 dilutions and enhanced chemiluminescence (ECL) reagents (GE Healthcare) according to the manufacturers' instructions. Mouse monoclonal anti-FLAG M2 antibodies were used to detect FLAG-tagged EPEC proteins at 1:2,000 dilutions. For detection of native, untagged EPEC EspD, polyclonal antibodies previously raised in rats against purified EspD from EPEC culture supernatant were used at 1:2,000 dilutions. The secondary antibodies, horseradish peroxidase (HRP)-conjugated goat anti-mouse and anti-rat antibodies (Jackson ImmunoResearch), were used at 1:5,000 dilutions.
Type III translocation assay.
Translocation of TEM-1 fusions into HeLa cells was assayed using the TEM-1 β-lactamase translocation assay as described before (18, 36, 38). Briefly, EPEC WT and ΔescN strains carrying pCX341 or derivatives expressing TEM-1 fusion proteins of EPEC type III-secreted protein substrates were grown overnight statically at 37°C in LB and preinduced in DMEM for 3 h before being used to infect HeLa cells. The cells were incubated with the fluorescent substrate CCF2-AM (Invitrogen) for 90 min at room temperature, and fluorescence was quantified using an Infinite M200 microplate reader (TECAN) with excitation at 409 nm. Relative TEM-1 translocation efficiency was expressed as the emission ratio of 460/530 nm (between blue fluorescence at 460 nm and green fluorescence at 530 nm) as previously described (18, 36).
RESULTS AND DISCUSSION
TEM-1 fusions of full-length translocator EspB and effector EspZ retain the translocator and effector substrate specificity, respectively, for type III secretion.
Type III secretion of translocators and effectors is regulated differentially in the A/E pathogens. While the translocators require SepD and SepL for their secretion, the effectors do not and are hypersecreted by the sepD and sepL mutants instead (17, 24, 25, 27–29). It has been shown that deleting the genes for all the translocators does not result in hypersecretion of effectors in the presence of SepD and SepL (24), suggesting that competition alone among the secretion substrates cannot explain the secretion hierarchy and that SepD and SepL must play an essential role in facilitating translocator secretion and preventing effector secretion prior to the completion of translocator secretion or host cell contact. The exact molecular mechanism for this reciprocal regulation of translocator secretion and effector secretion mediated by SepD/SepL has not been elucidated. We reasoned that the differential secretion patterns of translocators and effectors are mediated by different types of secretion signals.
We sought to determine whether translocators and effectors contain distinct, substrate class-specific type III secretion signals in EPEC. To do this, we generated fusions of a translocator (EspB) and an effector (EspZ) to a reporter protein (TEM-1) and then made truncations and deletions of the translocators and effectors to delineate the secretion signals. EspZ was chosen as a representative for effectors, as it is one of the first secreted/translocated and most abundant effectors and has the smallest size among all the effectors identified thus far (18–20, 38, 40), making it favorable for deletion and truncation analysis. Mature TEM-1 β-lactamase without its N-terminal secretion signal can be secreted via the T3SS when fused downstream of a type III secretion signal and has been used extensively as a reporter in type III secretion and translocation studies (13, 20, 25, 36, 38).
We cloned the full-length coding regions of EspB and EspZ into the vector pCX341 to make N-terminal fusions to TEM-1. These constructs were introduced into EPEC WT, ΔescN, ΔsepD, and ΔsepL strains and assayed for type III secretion of the TEM-1 fusion proteins. As shown in Fig. 1, we observed that these fusion proteins were expressed equally well in all 4 strains (Fig. 1A) and were not secreted by the escN mutant (defective for the T3SS ATPase), as expected (Fig. 1B). Full-length EspB-TEM was secreted by WT EPEC but not by the sepD and sepL mutants. In contrast, the full-length EspZ-TEM fusion was poorly secreted by WT EPEC but hypersecreted by the sepD and sepL mutants (Fig. 1B). We then determined if type III translocation into host cells is affected in these TEM-1 fusion proteins. While EspZ-TEM was efficiently translocated into cultured HeLa cells in a type III secretion-dependent manner, EspB-TEM was not translocated (Fig. 2B). This is consistent with previously reported secretion and translocation profiles of translocators and effectors in EPEC WT and the sepD and sepL mutants (17, 24, 29, 38). Collectively, these results demonstrated that EspB-TEM and EspZ-TEM fusions recapitulate the type III secretion and translocation patterns of EspB and EspZ, respectively, and we therefore used these fusions to further delineate their type III secretion and translocation signals.
FIG 1.
Expression and type III secretion profiles of TEM-1 fusions to full-length translocator EspB and effector EspZ or to the N-terminal secretion signals of EspB and EspZ. EPEC WT, ΔescN, ΔsepD, and ΔsepL strains expressing TEM (from vector pCX341), EspB-TEM, EspB1–20-TEM, EspZ-TEM, or EspZ1–21-TEM were grown in DMEM under type III secretion-inducing conditions. Whole-cell lysates (A) or secreted proteins (B) from these bacterial strains were separated on an SDS-13% PAGE gel and analyzed by Western blotting using mouse monoclonal antibodies against TEM-1 β-lactamase. WCL, whole-cell lysate; SP, secreted proteins concentrated from bacterial culture supernatant. Molecular mass markers in kilodaltons (kDa) are indicated on the left.
FIG 2.
The N-terminal 20 to 23 amino acid residues of translocators (EspA, EspB, and EspD), secreted T3SS components (EscF and EscI), and both LEE-encoded (Tir and EspZ) and non-LEE-encoded (NleA and NleD) effectors all confer type III-specific secretion and translocation when fused to TEM-1. (A) Type III secretion assay (using the same conditions as described for Fig. 1) of TEM-1 fusions to the N-terminal secretion signals of EspA, EspB, EspD, EscF, EscI, Tir, EspZ, NleA, and NleD in EPEC WT, ΔescN, and ΔsepD strains. Whole-cell lysates (WCL) and secreted proteins (SP) were resolved by SDS-PAGE and analyzed by Western blotting using mouse antibodies against TEM-1. (B) Type III translocation assay of the same set of strains as described for Fig. 2A. EPEC WT and ΔescN strains carrying different TEM-1 fusion constructs were used to infect cultured HeLa cells. The infected cells were loaded with CCF2/AM and measured for fluorescence using a microplate reader. Fluorescence quantification data are presented as the emission ratio between blue fluorescence at 460 nm and green fluorescence at 530 nm. Positive type III translocation is indicated by higher 460/530 (nm/nm) ratios by WT EPEC but not the ΔescN mutant. The results shown are mean values with standard deviations from triplicates in one representative experiment out of 3 experiments.
The N-terminal 20 and 21 amino acid residues of EspB and EspZ contain type III secretion and translocation signals.
It has been shown that the N terminus, usually the first 20 amino acid residues, of type III-secreted proteins in EPEC and other pathogens contains sufficient information to mediate type III secretion and translocation when fused to a secretion-competent reporter protein, such as CyaA and TEM-1 (10, 25, 35, 36, 40). To confirm this under our experimental conditions, we cloned the coding regions of the N-terminal 20 (EspB20) and 21(EspZ21) amino acid residues of EspB and EspZ into pCX341 to make N-terminal fusions to TEM-1. When the N-terminal 20 and 21 amino acid residues of EspB and EspZ were fused to TEM-1, both TEM-1 fusions showed identical secretion profiles in the EPEC strains (Fig. 1). Both EspZ21-TEM and EspB20-TEM were marginally secreted by WT EPEC and hypersecreted by the sepD and sepL mutants but were not secreted by the escN mutant (Fig. 1B). In addition, both these fusion proteins were translocated into HeLa cells in a T3SS-dependent fashion (Fig. 2B). These results confirm previous reports that the N-terminal 20 and 21 amino acid residues of EspB and EspZ are sufficient for type III secretion and translocation (25, 40) and that the EspB secretion signal behaves like the secretion signal of effectors and confers hypersecretion in the sepD and sepL mutants (25).
Since the secretion profiles of all these constructs showed very similar patterns in both the sepD mutant and the sepL mutant (Fig. 1) (17, 24, 25), and since all the observations thus far indicated that SepD and SepL appear to be part of the same secretion switch mechanism (17, 24, 29), type III secretion assays in all subsequent experiments were done only in the sepD mutant.
A previous study by Chiu et al. on the secretion signal of EspB from EHEC O157:H7 was inconclusive (41). They showed that the N-terminal 117 residues of EHEC EspB played a major role in secretion, but the N-terminal 21 residues were not sufficient (41). The reporter, an antigen of hepatitis D virus (HDAg), used by these authors to tag the proteins may have contributed to this discrepancy, as EspB-HDAg fusion proteins were often unstable and may not be competent for secretion (41). Our result that the N-terminal 20 and 21 residues of EPEC EspB and EspZ are sufficient for type III secretion and translocation is consistent with recent findings in characterizing type III secretion and translocation signals using CyaA and TEM-1 reporters (10, 25, 35, 36, 40).
The N termini of the translocators, the secreted T3SS components, and both LEE- and non-LEE-encoded effectors all contain type III secretion and translocation signals but do not confer substrate specificity and secretion hierarchy.
To determine whether the observation that the N-terminal amino acid residues of EspB and EspZ contain both type III secretion and translocation signals applies to other substrate classes of type III-secreted proteins, we extended our analysis to the two secreted T3SS components, EscF and EscI, the other two translocators, EspA and EspD, another LEE-encoded effector, Tir, and two non-LEE-encoded effectors, NleA and NleD. We cloned the coding regions of the N-terminal 20 to 23 amino acid residues of EPEC EscF, EscI, EspA, EspD, Tir, NleA, and NleD into pCX341 to generate TEM-1 fusions. These different sequences were cloned into the same restriction enzyme sites (NdeI and EcoRI). However, the design of the vector pCX341 requires the last nucleotide of the cloned EPEC sequences to be a G (part of the EcoRI restriction site, GAATTC) (36, 38), and this cloning limitation necessitated the total number of the N-terminal amino acid residues of the EPEC proteins in the TEM-1 fusions to vary from 20 to 23, in order to avoid adding any foreign amino acid residue that is not encoded by the LEE sequences in front of TEM-1.
All these constructs were expressed equally well when introduced into the EPEC WT, ΔescN, and ΔsepD strains (Fig. 2A). All the TEM-1 fusions, with their expression under the control of the same Ptrc promoter and identical regulatory elements to facilitate fair and standard comparison, showed very similar secretion patterns, i.e., low levels of secretion by WT EPEC, nondetectable secretion by the ΔescN mutant, and hypersecretion by the ΔsepD mutant (Fig. 2A), regardless of their function and secretion substrate categories. In addition, all these TEM-1 fusions were translocated into cultured HeLa cells requiring a functional T3SS (Fig. 2B). The translocation efficiency varied among the different TEM fusions, but there was no direct correlation between type III secretion and translocation efficiencies for these TEM-1 fusions, although the TEM-1 fusions to the N-terminal secretion signals of effectors (Tir, NleA, EspZ, and NleD) were more efficiently translocated. As expected, the TEM-1 fusions to full-length translocators, the needle and inner rod components, were not translocated (Fig. 2B). This demonstrated that the N-terminal type III secretion signals of all the tested type III secretion substrates behave like that of the effectors. This was particularly surprising for EscF (needle) and EscI (inner rod), two secreted proteins that are structural components of the T3SS and should not be translocated into host cells, yet they both contain effector-like secretion and translocation signals.
These results confirm and extend previous observations on the N-terminal type III secretion signals of EPEC translocators and a few selected effectors (25, 35, 36, 40) and further demonstrate that the N-terminal secretion signals of the three different categories of secretion substrates, i.e., secreted T3SS components, translocators, and both LEE- and non-LEE-encoded effectors, all behave like those of the effectors. These results suggest that the extreme N-terminal secretion and translocation signal does not determine the secretion hierarchy and substrate specificity. Indeed, it has been demonstrated that exchange or swapping of the N-terminal secretion and translocation signals between translocators and effectors does not influence the secretion and function of the chimeric translocators and effectors (25). These results indicate that an additional signal(s) must be needed in the type III-secreted proteins to define their substrate category and secretion hierarchy.
EspB encodes a signal that confers SepD/SepL-dependent secretion.
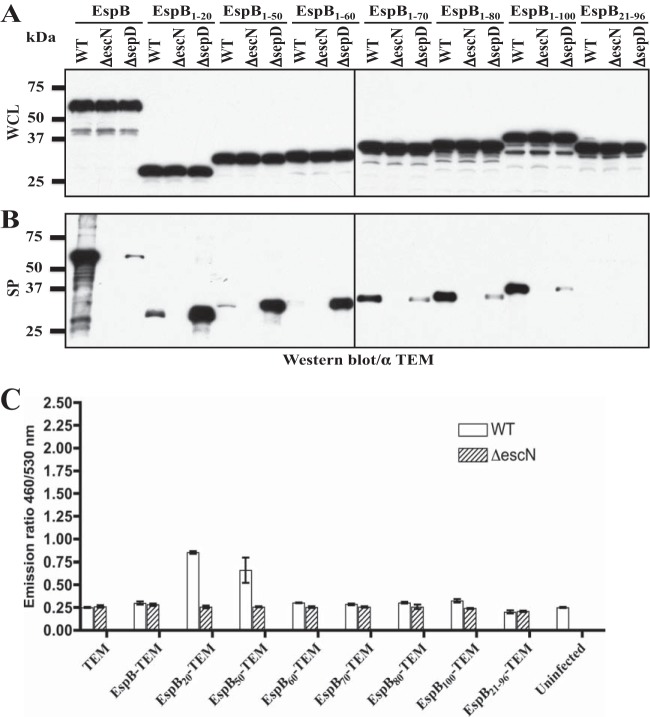
Whether the different classes of type III secretion substrates (needle protein and inner rod protein, translocators, and effectors) contain substrate-specific secretion signals, in addition to the N-terminal secretion signals, is not well understood. There is evidence that the N-terminal secretion signal may determine the substrate categories of translocators and effectors in Yersinia enterocolitica (13, 42). However, it has also been shown that the N-terminal secretion signals of translocators and effectors in EPEC and P. aeruginosa are exchangeable without affecting the secretion and function of the chimeric translocators and effectors (25, 43). Translocator-specific export signals have recently been identified in PopD and PopB in P. aeruginosa (12). Since full-length EspB-TEM fusion acted like a translocator and was secreted by EPEC WT at low levels but not the ΔsepD mutant, while EspB1–20-TEM behaved like an effector and was secreted by WT EPEC and hypersecreted by the ΔsepD mutant, we reasoned that EspB contains a signal or domain, in addition to its N-terminal type III secretion and translocation signal, that defines EspB as a translocator and confers SepD/SepL-dependent secretion.
To identify the potential domain/signal in EspB that is required for SepD/SepL-dependent secretion, we first generated two TEM fusions using the N-terminal 50 and 100 amino acid residues of EspB, EspB1–50, and EspB1–100. While EspB50-TEM behaved similarly to EspB20-TEM and its secretion did not require SepD (secreted by WT EPEC and hypersecreted by the sepD mutant), EspB100-TEM showed a secretion profile similar to that of EspB-TEM (Fig. 3A and B), suggesting that the SepD/SepL-dependent secretion signal in EspB is located between amino acid residues 21 and 100 of EspB.
FIG 3.
A domain/signal located immediately downstream of the N-terminal type III secretion/translocation signal in EspB is required for efficient SepD/SepL-dependent secretion. (A) Western blotting against TEM-1 of whole bacterial cell lysates of EPEC WT, ΔescN, and ΔsepD strains expressing EspB-TEM and a series of TEM-1 fusions containing different N-terminal regions of EspB (EspB1–20, EspB1–50, EspB1–60, EspB1–70, EspB1–80, EspB1–100, and EspB21–96) grown in DMEM under type III secretion-inducing conditions. (B) Secreted proteins of the same set of strains were analyzed by Western blotting. (C) Type III translocation assay in HeLa cells using EPEC WT and ΔescN strains carrying the same set of constructs as described in Fig. 3A. The infected cells were loaded with CCF2/AM and measured for fluorescence using a microplate reader. Fluorescence quantification data are presented as the emission ratio between blue fluorescence at 460 nm and green fluorescence at 530 nm. Positive type III translocation is indicated by higher 460/530 (nm/nm) ratios by WT EPEC than by the ΔescN mutant. The results shown are mean values with standard deviations.
There is a predicted transmembrane domain from amino acid residues 97 to 117 in EspB (44, 45) (Fig. 4A), and we reasoned that the SepD/SepL-dependent secretion signal of EspB may precede the transmembrane domain of EspB. We next generated a series of EspB-TEM fusions using different numbers of the N-terminal 96 amino acid residues of EspB. As shown in Fig. 3A and B, the secretion pattern of EspB1–70, EspB1–80, and EspB1–100 TEM fusions resembled that of EspB-TEM (high levels of secretion by WT EPEC and low or no secretion by the sepD mutant), while shorter EspB fusions (from amino acid residues 1 to 60) were secreted like effectors (low or no secretion by WT EPEC and hypersecretion by the sepD mutant). Since the N-terminal 20 amino acid residues of EspB contain the type III secretion signal, the removal of amino acid residues 2 to 20 (EspB21–96-TEM) abolished the type III secretion of the fusion protein, as expected (Fig. 3B). These data indicated that the sequence of EspB between amino acid residues 21 and 70 is essential for SepD/SepL-dependent secretion of EspB. Our results also suggest that, while not sufficient by itself to confer type III secretion, this sequence (EspB21–70) serves as an accessory secretion signal specific for the translocators.
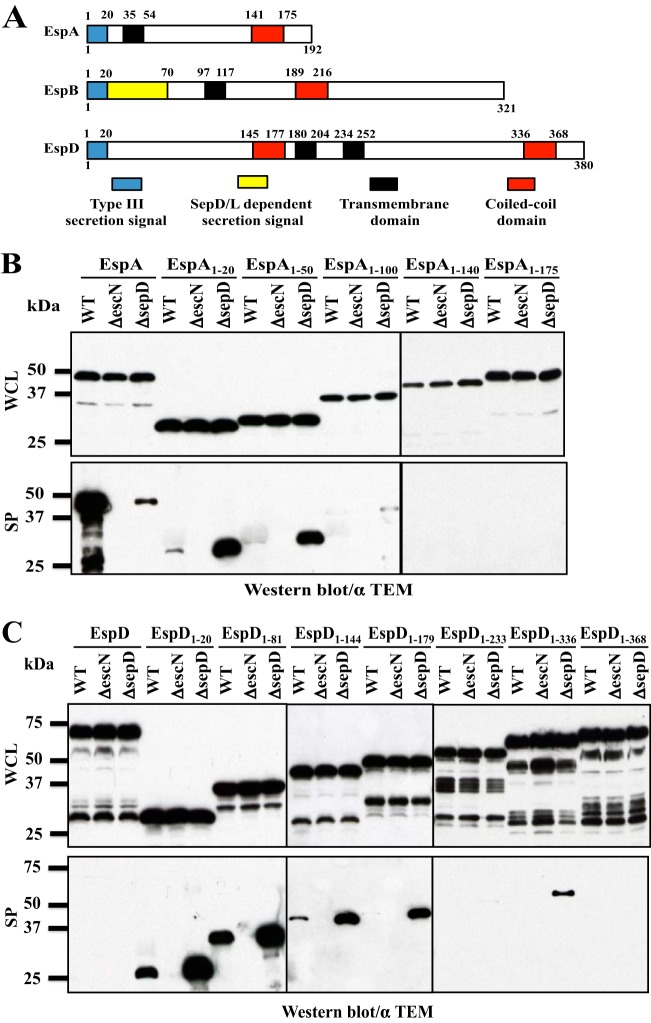
FIG 4.
SepD/SepL-dependent secretion signals in translocators EspA and EspD. (A) Schematic diagrams of EspA, EspB, and EspD. (B) Type III secretion assay of TEM-1 fusions to different EspA truncations. Whole bacterial lysates (WCL) and secreted proteins (SP) were analyzed by Western blotting using antibodies against TEM-1. (C) Type III secretion assay of TEM-1 fusions to different EspD truncations. Whole bacterial lysates (WCL) and secreted proteins (SP) were analyzed by Western blotting using antibodies against TEM-1.
This EspB21–70 domain containing the SepD/SepL-dependent secretion signal is not located in any of the strongly predicted structural domains (transmembrane and coiled-coil domains) in EspB (Fig. 4A). While it is not predicted to form any obvious secondary structures, this region does overlap a weak coiled-coil domain predicted by Luo and Donnenberg (46). Interestingly, this EspB domain is particularly sensitive to insertion mutagenesis of a five-codon linker, and insertions in this region often interfere with the proper translocator function of EspB (46, 47), indicating that this EspB21–70 domain is essential for EspB function.
These EspB-TEM fusions were also tested in the type III translocation assay. As shown in Fig. 3C, among all the EspB-TEM fusions, only EspB20-TEM and EspB50-TEM were translocated into HeLa cells at a significant level. This suggested that the EspB domain spanning amino acid residues 21 to 70 not only confers SepD/SepL-dependent secretion of EspB but also inhibits type III translocation efficiency of EspB.
The translocators EspA and EspD also contain SepD/SepL-dependent secretion signals, but these signals are organized differently from that of EspB.
The three translocator proteins EspA, EspB, and EspD behave similarly in their type III secretion patterns, and they all require SepD and SepL for their secretion (17, 24). EspA20-TEM and EspD20-TEM were secreted in the same fashion as EspB20-TEM, exhibiting low levels of secretion by WT EPEC and hypersecretion by the sepD and sepL mutants (25) (Fig. 2A). This suggests that EspA and EspD may also contain SepD/SepL-dependent secretion signals. Hence, we generated a series of TEM-1 fusions containing different regions and domains of EspA or EspD at the N terminus of TEM-1. These EspA and EspD regions were chosen based on the bioinformatic predictions of secondary structures, such as transmembrane and coiled-coiled domains, in EspA and EspD (44, 45) (Fig. 4A). These constructs were introduced into EPEC WT, ΔescN, and ΔsepD strains and assayed for type III secretion.
While the full-length EspA-TEM fusion was secreted similarly to native EspA (high levels of secretion by WT EPEC and low or no secretion by the sepD mutant), EspA20-TEM and EspA50-TEM showed secretion profiles of effectors (low or no secretion by WT EPEC and hypersecretion by the sepD mutant). This suggests that EspA contains an SepD/SepL-dependent secretion signal immediately downstream of the N-terminal type III secretion and translocation signal. However, this could not be definitively determined by deletion analyses, as longer EspA (EspA100, EspA140, and EspA175)-TEM fusions were all less stable, either in bacteria (EspA100 and EspA140) or when secreted into the culture media, since there was very little EspA100-TEM, EspA140-TEM, and EspA175-TEM detected in the secreted proteins enriched from the culture supernatant (Fig. 4B). The high instability of secreted mutant EspA has been reported previously (48).
Attempts to delineate the secretion signals of EspD were inconclusive. EspD81-TEM, EspD144-TEM, and EspD179-TEM had the same secretion pattern as EspD20-TEM, indicating that EspD does not contain an SepD/SepL-dependent secretion signal immediately downstream of the N-terminal type III secretion and translocation signal similar to that in EspB. However, longer EspD (EspD233, EspD336, and EspD368) fusions to TEM resulted in unstable fusion proteins as assessed by the degradation products visible in Fig. 4C, and these EspD-TEM fusions were poorly secreted (Fig. 4C), similar to the full-length EspD-TEM fusion. It is possible that the instability of these fusion proteins affects their type III secretion. In addition, all the fusions containing EspD longer than the N-terminal 179 amino acid residues exhibited dominant negative effects and inhibited overall type III secretion of other proteins in WT EPEC and the sepD mutant (data not shown). This suggests that the C terminus of EspD may contain an accessory signal required for EspD secretion, with TEM-1 fusions at the C terminus thus blocking the type III secretion of the fusion proteins.
To exclude the possibility that the bulky size of TEM-1 (29 kDa) is responsible for blocking secretion of TEM-1 fusions to EspD233, EspD336, EspD368, and full-length EspD, we tagged EspD336, EspD368, and full-length EspD at the C terminus with the much smaller FLAG tag (1 kDa) and analyzed their secretion in the EPEC WT, ΔescN, and ΔsepD strains. As shown in Fig. 5A, EspD-FLAG, EspD336-FLAG, and EspD368-FLAG were all expressed well and appeared to be stable, but none of them was secreted by either WT EPEC or the ΔsepD mutant, indicating that C-terminal epitope tagging, regardless of the size of the tag, blocks EspD secretion. Similar to their TEM-1 fusions (Fig. 4C), the FLAG-tagged EspD, EspD336, and EspD368 also displayed a dominant negative phenotype and inhibited type III secretion of other proteins in WT EPEC and the sepD mutant (data not shown). This suggests that the C terminus of EspD is critical for secretion and that EspD may contain an accessory secretion signal in the C terminus that is required for the secretion of full-length, native EspD, in addition to the N-terminal secretion signal.
FIG 5.
The C terminus of EspD is required for type III secretion of EspD, and C-terminal epitope tagging inhibits EspD secretion. (A) EPEC WT, ΔescN, and ΔsepD strains containing the vector pCX341 or pCX341 expressing C-terminally FLAG-tagged full-length EspD, EspD1–336, and EspD1–368 were grown in DMEM under type III secretion-inducing conditions. Whole-cell lysates (WCL) and secreted proteins (SP) were analyzed in SDS-PAGE by Western blotting using mouse monoclonal antibodies against FLAG. (B) EPEC ΔespD, ΔescN ΔespD, and ΔsepD ΔespD strains carrying the vector pCX341 or pCX341 expressing untagged full-length EspD, EspD1–336, and EspD1–368 were subjected to type III secretion assays. Whole-cell lysates and secreted proteins were analyzed by Western blotting using rat polyclonal antibodies against EPEC EspD. EPEC WT was included as a positive control on the left. Note that a faint band cross-reacting with the antibodies is located underneath the full-length EspD band.
To test explicitly whether the C terminus of EspD contains any additional secretion signal, we made constructs expressing untagged full-length EspD and EspD derivatives EspD1–336 and EspD1–368, with the C-terminal 44 and 12 amino acid residues truncated, respectively. We then generated ΔespD, ΔescN ΔespD, and ΔsepD ΔespD deletion mutants in EPEC to remove native EspD, introduced the plasmids expressing EspD, EspD1–336, and EspD1–368 into these mutant strains, and carried out type III secretion assays. As shown in Fig. 5B, EspD, EspD1–336, and EspD1–368 were all expressed well from the constructs and stable in the EPEC mutants, but only the full-length EspD was secreted by the ΔespD mutant. EspD1–336 and EspD1–368 were not secreted by either the ΔespD or the ΔsepD ΔespD mutant, although they carry the N-terminal secretion signal and both the ΔespD and ΔsepD ΔespD mutants have a functional T3SS and can secrete other proteins (data not shown). These results suggest that the C-terminal 12 amino acid residues of EspD may represent a C-terminal secretion signal. However, this signal is different in nature from the SepD/SepL-dependent, translocator-specific secretion signal identified in EspB, as deletion of the EspD C-terminal signal does not promote EspD secretion in the ΔsepD mutant, although it is required for EspD secretion in WT EPEC. An alternative possibility is that the C terminus of EspD does not constitute a secretion signal per se but is important for proper folding of EspD. It is hypothesized that proteins need to be unfolded in order to be secreted through the T3SS channel (1–3). Without the C terminus, EspD may misfold or fold prematurely, preventing its secretion. We showed that TEM-1 fusions that contain the N-terminal 20, 81, 144, and 179 amino acid residues of EspD are type III secreted, and only the longer EspD fusions (EspD233, EspD336, EspD368, and full-length EspD), all of which contain one or both of the predicted transmembrane domains in EspD (Fig. 4A), fail to be secreted (Fig. 4C). It is possible that the C terminus of EspD is important for stabilizing these transmembrane domains and maintaining EspD in a secretion-competent state, thus promoting EspD secretion.
It has recently been shown that the translocator protein PopD (a homolog of EspB) in the Pseudomonas aeruginosa T3SS contains discrete translocator-specific secretion signals, and there is some evidence that the translocator protein PopB (a homolog of EspD) may also contain a similar signal(s) (12). However, there seems to be at least two such signals in PopD, one located adjacent to the chaperone binding site and the other in the C terminus (12). Interestingly, PopD is also sensitive to tagging at the C terminus, as full-length PopD fusions to TEM-1 and the smaller vesicular stomatitis virus glycoprotein (VSV-G) epitope display severe secretion defects (12). Our results showed that EspB has only one translocator-specific secretion signal located immediately downstream of the N-terminal secretion signal and that such signals, if any, would be organized differently in the other two translocator proteins (EspA and EspD) in EPEC. EspD may require a signal in the C terminus for secretion, in addition to its N-terminal secretion signal, but both signals in EspD do not confer translocator specificity. It is possible that the secretion hierarchy of EspA and EspD is regulated by a different mechanism. This suggests that the secretion hierarchy of translocators may not be controlled by a universal mechanism.
The SepD/SepL-dependent secretion signal of EspB confers SepD/SepL-dependent secretion on the N-terminal secretion signal of effector EspZ.
SepD and SepL are required for secretion of translocators EspA, EspB, and EspD but are dispensable for secretion of effectors, such as Tir and EspZ (17, 24). To investigate whether the SepD/SepL-dependent secretion signal of EspB can confer SepD/SepL-dependent secretion to other proteins, we engineered chimeric proteins by placing different portions of EspB (EspB21–96, EspB21–70, and EspB51–96) immediately downstream of the N-terminal secretion signal of the effector EspZ (EspZ21) and then fused it to the N terminus of TEM-1. These constructs were introduced into EPEC WT, ΔescN, and ΔsepD strains and assayed for type III secretion. While EspZ1–21-TEM showed the typical secretion profile of effectors, i.e., secreted at low levels by WT EPEC and hypersecreted by the ΔsepD mutant, EspZ1–21-EspB21–70 and EspZ1–21-EspB21–96 TEM fusions were secreted predominantly by WT EPEC and only weakly by the ΔsepD mutant, mimicking the type III secretion profile of EspB and other translocators (Fig. 6B). Although the steady protein level of EspZ1–21-EspB51–96-TEM was much lower than that of EspZ1–21-EspB21–70 and EspZ1–21-EspB21–96 TEM fusions in the whole-cell lysate (Fig. 6A), and it was probably less stable, EspZ1–21-EspB51–96-TEM evidently displayed the secretion profile of effectors. It should be noted that the Western blots shown in Fig. 6A and B were from relatively long exposures of the blots during detection, in order to show the weak signals for EspZ1–21-EspB51–96-TEM. Our results indicated that the EspB domain conferring SepD/SepL-dependent secretion lies between amino acid residues 21 and 70, and this domain is sufficient to change the secretion pattern of the type III secretion signal of EspZ from effector-like to translocator-like. This EspB domain also significantly reduced the translocation efficiency of the EspZ1–21 secretion and translocation signal (Fig. 6C).
FIG 6.
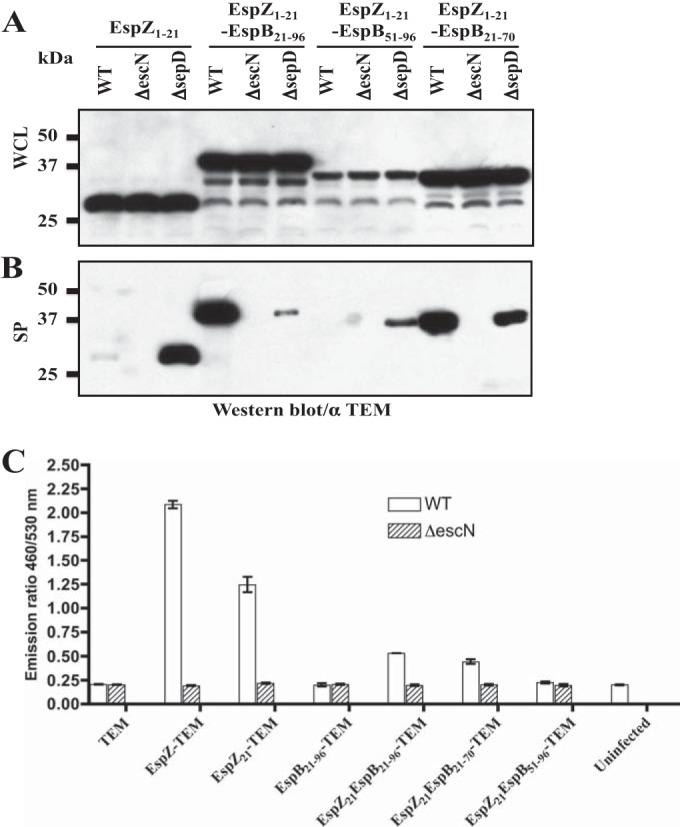
The SepD/SepL-dependent secretion signal of EspB confers SepD/SepL-dependent secretion when placed downstream of the type III secretion/translocation signal of effector EspZ. (A and B) Type III secretion assay of TEM-1 fusions to different EspZ, EspB, and EspZ-EspB chimeric secretion signals in EPEC WT, ΔescN, and ΔsepD strains. Shown are proteins from whole bacterial cell lysates (WCL) (A) and secreted proteins (SP) (B) analyzed by Western blotting using antibodies against TEM-1. (C) Type III translocation assay of the same set of constructs as described for Fig. 6A. EPEC WT and ΔescN strains carrying different TEM-1 fusion constructs were used to infect HeLa cells. The infected cells were loaded with CCF2/AM and measured for fluorescence using a microplate reader. Fluorescence quantification data are presented as the emission ratio between blue fluorescence at 460 nm and green fluorescence at 530 nm. Positive type III translocation is indicated by higher 460/530 (nm/nm) ratios by WT EPEC than by the ΔescN mutant. The results shown are mean values with standard deviations.
The SepD/SepL-dependent secretion signal of EspB confers CesAB-dependent secretion when fused downstream of the secretion signal of effector EspZ.
It has been shown that type III secretion chaperones recognize specific domains or motifs, often located immediately downstream of the N-terminal secretion signals, in the cognate effector proteins and target the effectors to the type III secretion apparatus (49, 50). CesAB is a chaperone for EspA and EspB, and it has been shown to interact with EspA and EspB (51, 52). While required for EspB secretion, CesAB plays no role in the intracellular stability of EspB (52). We therefore examined whether CesAB is required for SepD/SepL-dependent secretion of EspB and whether the SepD/SepL-dependent secretion signal of EspB confers CesAB-dependent secretion when placed downstream of the secretion signal of an effector.
As shown in Fig. 7, both WT EPEC and the cesAB mutant secreted no or very low levels of EspZ-TEM, EspZ1–21-TEM, and EspB1–20-TEM, in agreement with our previous results that these TEM fusions behave like effectors and are not highly secreted under conditions favoring translocator secretion (Fig. 1B and 2A). As expected, deletion of cesAB abolished the type III secretion of the EspB-TEM fusion (Fig. 7B), demonstrating that CesAB is essential for secretion of EspB-TEM, which behaves like a translocator. This is consistent with previously published secretion data of EspB and EspA in the cesAB mutant (52). While CesAB was not essential for the secretion of EspB1–20-, EspZ-, and EspZ1–21-TEM fusions in the ΔsepD mutant, the secretion of the EspZ1–21-EspB21–70-TEM fusion protein depended on the presence of CesAB (Fig. 7B), indicating that EspB20–70 contains a secretion signal conferring dependence on both SepD/SepL and CesAB. It should be noted that deleting cesAB in the ΔsepD mutant background seemed to also result in reduced secretion of EspZ-TEM, EspZ1–21-TEM, and EspB1–20-TEM, with a greater effect on EspB1–20-TEM secretion (Fig. 7B). The reason for this observation was not known, but it is possible that in the absence of both CesAB and SepD, the intracellular pool of unsecreted translocators accumulates, competes with, and slows down the secretion of these TEM-1 fusions, especially EspB1–20-TEM. Nevertheless, our results suggest that under translocator-secreting conditions, CesAB facilitates the secretion of translocators probably by interacting with the SepD/SepL-dependent secretion signal, but it does not play a significant role in secreting substrates bearing only effector-like secretion signals.
FIG 7.
The chaperone CesAB for translocators EspA and EspB is required for secretion of translocator secretion signal-TEM fusions but not for effector secretion signal-TEM fusions. (A and B) Type III secretion assay of TEM-1 fusions to different EspZ, EspB, and EspZ-EspB chimeric secretion signals in EPEC WT, ΔescN, ΔsepD, ΔcesAB, and ΔsepD ΔcesAB strains. Shown are proteins from whole bacterial cell lysates (WCL) (A) and secreted proteins (SP) (B) analyzed by Western blotting using antibodies against TEM-1 β-lactamase.
SepD and SepL are required for the secretion of translocators EspA, EspB, and EspD (17, 24, 29). Various biochemical and genetic analyses have established the direct interaction between SepD and SepL (24, 28, 51), and it has also been shown that SepL binds to Tir, perhaps to delay the secretion of Tir (29). However, direct interactions between translocators (or their chaperones) and SepD or SepL have not been demonstrated. Although it has been shown that CesAB interacts with EspB (51, 52), our attempts using biochemical and proteomic methods to show direct binding of CesAB to the SepD/SepL-dependent secretion signal in EspB (EspB21–70) were not successful, suggesting that the yet-to-be-mapped chaperone-binding domain in EspB may be distinct from the SepD/SepL-dependent secretion signal. We could not detect direct interaction between CesAB and SepD or SepL using coexpression and pulldown assays either (data not shown). It is possible that such interactions occur only immediately before the substrates are secreted and are transient in nature, which makes it difficult to simulate or capture using conventional biochemical and proteomic techniques. An alternative possibility is that the interactions between CesAB and SepD or SepL depend upon the presence of other proteins. Indeed, InvE, a Salmonella homolog of SepL and YopN, does not interact with either the translocators or their chaperone individually but can form a complex when all these proteins are present (53). It has recently been shown by using nuclear magnetic resonance (NMR) spectroscopy that the interaction between EscN and CesAB occurs only after binding of CesAB to EspA, which results in conformational changes in CesAB exposing a targeting signal to EscN (54).
Concluding remarks.
The type III-secreted T3SS components (needle and inner rod components), translocators, and effectors function at distinct stages of the T3SS and can therefore be classified into different groups of type III-secreted substrates. In this report, we have shown that the N-terminal 20 to 23 amino acid residues from translocators, both LEE- and non-LEE-encoded effectors, and the T3SS needle and inner rod components are all sufficient for conferring type III secretion and can serve as type III secretion and translocation signals. However, these N-terminal signals do not confer substrate specificity, as they all behave like the secretion signals of effectors. We propose that translocators and T3SS components contain additional signals specifying their substrate groups. We show that at least in the translocator EspB, an SepD/SepL-dependent secretion signal exists immediately following the N-terminal type III secretion signal. Such translocator-specific secretion signals have recently been identified in another translocator protein, PopD, in the P. aeruginosa T3SS (12). PopD appears to require two translocator-specific secretion signals, one located downstream of the N-terminal secretion signal and upstream of the chaperone binding domain, and the other at the C terminus, and both signals are needed for PopD to be exported before the effectors (12). However, it seems that EPEC EspB requires only one translocator-specific signal located immediately downstream of the N-terminal secretion signal (Fig. 3 and 6). This suggests that the organization of such substrate-specific secretion signals may vary with each substrate. Indeed, it seems that EspD secretion may also need a second secretion signal located in the C terminus, and that the translocator-specific secretion signals in EspA and EspD of EPEC, if any, are organized differently from those of EspB. Nonetheless, these results raise the possibility that other type III secretion substrates, such as effectors and the needle and inner rod components, also possess distinct signals that determine their secretion hierarchy. The challenge now is to elucidate the mechanism as to how such signals are differentially recognized by the T3SS in controlling type III secretion hierarchy.
ACKNOWLEDGMENTS
This work was funded by operating grants from the Canadian Institutes of Health Research (CIHR) to B.B.F. B.B.F. is the UBC Peter Wall Distinguished Professor. H.B.Y. was supported by a CIHR postdoctoral fellowship.
We are grateful to Stefanie Vogt and Tracy Raivio for providing restriction-minus EPEC E2348/69 ΔhsdR strain NH4.
REFERENCES
- 1.Cornelis GR. 2006. The type III secretion injectisome. Nat Rev Microbiol 4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Büttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev 76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stamm LM, Goldberg MB. 2011. Establishing the secretion hierarchy. Science 331:1147–1149. doi: 10.1126/science.1203195. [DOI] [PubMed] [Google Scholar]
- 6.Filloux A. 2010. Secretion signal and protein targeting in bacteria: a biological puzzle. J Bacteriol 192:3847–3849. doi: 10.1128/JB.00565-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci U S A 92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DM, Schneewind O. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 9.Niemann GS, Brown RN, Mushamiri IT, Nguyen NT, Taiwo R, Stufkens A, Smith RD, Adkins JN, McDermott JE, Heffron F. 2013. RNA type III secretion signals that require Hfq. J Bacteriol 195:2119–2125. doi: 10.1128/JB.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDermott JE, Corrigan S, Peterson E, Oehmen C, Niemann G, Cambronne ED, Sharp D, Adkins JN, Samudrala R, Heffron F. 2011. Computational prediction of type III and IV secreted effectors in Gram-negative bacteria. Infect Immun 79:23–32. doi: 10.1128/IAI.00537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchko GW, Niemann G, Baker ES, Belov ME, Smith RD, Heffron F, Adkins JN, McDermott JE. 2010. A multi-pronged search for a common structural motif in the secretion signal of Salmonella enterica serovar Typhimurium type III effector proteins. Mol Biosyst 6:2448–2458. doi: 10.1039/c0mb00097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomalka AG, Stopford CM, Lee P-C, Rietsch A. 2012. A translocator-specific export signal establishes the translocator-effector secretion hierarchy that is important for type III secretion system function. Mol Microbiol 86:1464–1481. doi: 10.1111/mmi.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amer AAA, Åhlund MK, Bröms JE, Forsberg Å Francis MS. 2011. Impact of the N-terminal secretor domain on YopD translocator function in Yersinia pseudotuberculosis type III secretion. J Bacteriol 193:6683–6700. doi: 10.1128/JB.00210-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 15.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 16.Wong ARC, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol 80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 17.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vázquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A 101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng W, de Hoog CL, Yu HB, Li Y, Croxen MA, Thomas NA, Puente JL, Foster LJ, Finlay BB. 2010. A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. J Biol Chem 285:6790–6800. doi: 10.1074/jbc.M109.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W, Yu HB, de Hoog CL, Stoynov N, Li Y, Foster LJ, Finlay BB. 2012. Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol Cell Proteomics 11:692–709. doi: 10.1074/mcp.M111.013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobe T, Beatson S, Taniguchi H, Abe H, Bailey C, Fivian A, Younis R, Matthews S, Marches O, Frankel G, Hayashi T, Pallen M. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A 103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, Henderson IR, Harris D, Asadulghani M, Kurokawa K, Dean P, Kenny B, Quail MA, Thurston S, Dougan G, Hayashi T, Parkhill J, Frankel G. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J Bacteriol 191:347–354. doi: 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petty NK, Bulgin R, Crepin VF, Cerdeño-Tárraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. 2010. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J Bacteriol 192:525–538. doi: 10.1128/JB.01144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA, Lee S, Gruenheid S, Strynadka NCJ, Puente JL, Finlay BB. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun 73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munera D, Crepin VF, Marches O, Frankel G. 2010. N-terminal type III secretion signal of enteropathogenic Escherichia coli translocator proteins. J Bacteriol 192:3534–3539. doi: 10.1128/JB.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NCJ. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- 27.Kresse AU, Beltrametti F, Muller A, Ebel F, Guzman CA. 2000. Characterization of SepL of enterohemorrhagic Escherichia coli. J Bacteriol 182:6490–6498. doi: 10.1128/JB.182.22.6490-6498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ, Luo W, Albert MJ, Kaper JB, Frankel G, Donnenberg MS. 2004. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol 52:1613–1625. doi: 10.1111/j.1365-2958.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DJ. 2008. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol 69:1499–1512. doi: 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
- 30.Younis R, Bingle LEH, Rollauer S, Munera D, Busby SJ, Johnson S, Deane JE, Lea SM, Frankel G, Pallen MJ. 2010. SepL resembles an aberrant effector in binding to a class 1 type III secretion chaperone and carrying an N-terminal secretion signal. J Bacteriol 192:6093–6098. doi: 10.1128/JB.00760-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu XJ, Liu M, Holden DW. 2004. SsaM and SpiC interact and regulate secretion of Salmonella pathogenicity island 2 type III secretion system effectors and translocators. Mol Microbiol 54:604–619. doi: 10.1111/j.1365-2958.2004.04297.x. [DOI] [PubMed] [Google Scholar]
- 32.Ramamurthi KS, Schneewind O. 2002. Type III protein secretion in Yersinia species. Annu Rev Cell Dev Biol 18:107–133. doi: 10.1146/annurev.cellbio.18.012502.105912. [DOI] [PubMed] [Google Scholar]
- 33.Botteaux A, Sory MP, Biskri L, Parsot C, Allaoui A. 2009. MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol Microbiol 71:449–460. doi: 10.1111/j.1365-2958.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- 34.Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science 328:1040–1043. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford JA, Kaper JB. 2002. The N terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol Microbiol 46:855–868. doi: 10.1046/j.1365-2958.2002.03214.x. [DOI] [PubMed] [Google Scholar]
- 36.Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based vector. J Bacteriol 186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gauthier A, Puente JL, Finlay BB. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect Immun 71:3310–3319. doi: 10.1128/IAI.71.6.3310-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills E, Baruch K, Charpentier X, Kobi S, Rosenshine I. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3:104–113. doi: 10.1016/j.chom.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Hobson N, Price NL, Ward JD, Raivio TL. 2008. Generation of a restriction minus enteropathogenic Escherichia coli E2348/69 strain that is efficiently transformed with large, low copy plasmids. BMC Microbiol 8:134. doi: 10.1186/1471-2180-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanack KJ, Crawford JA, Tatsuno I, Karmali MA, Kaper JB. 2005. SepZ/EspZ is secreted and translocated into HeLa cells by the enteropathogenic Escherichia coli type III secretion system. J Bacteriol 73:4327–4337. doi: 10.1128/IAI.73.7.4327-4337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu H-J, Lin W-S, Syu W-J. 2003. Type III secretion of EspB in enterohemorrhagic Escherichia coli O157:H7. Arch Microbiol 180:218–226. doi: 10.1007/s00203-003-0579-7. [DOI] [PubMed] [Google Scholar]
- 42.Sorg I, Wagner S, Amstutz M, Müller SA, Broz P, Lussi Y, Engel A, Cornelis GR. 2007. YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J 26:3015–3024. doi: 10.1038/sj.emboj.7601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee PC, Stopford CM, Svenson AG, Rietsch A. 2010. Control of effector export by the P. aeruginosa type III secretion proteins PcrG and PcrV. Mol Microbiol 75:924–941. doi: 10.1111/j.1365-2958.2009.07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daniell SJ, Delahay RM, Shaw RK, Hartland EL, Pallen MJ, Booy F, Ebel F, Knutton S, Frankel G. 2001. Coiled-coil domain of enteropathogenic Escherichia coli type III secreted protein EspD is involved in EspA filament-mediated cell attachment and hemolysis. Infect Immun 69:4055–4064. doi: 10.1128/IAI.69.6.4055-4064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delahay RM, Frankel G. 2002. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol Microbiol 45:905–916. doi: 10.1046/j.1365-2958.2002.03083.x. [DOI] [PubMed] [Google Scholar]
- 46.Luo W, Donnenberg MS. 2006. Analysis of the function of enteropathogenic Escherichia coli EspB by random mutagenesis. Infect Immun 74:810–820. doi: 10.1128/IAI.74.2.810-820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo W, Donnenberg MS. 2011. Interactions and predicted host membrane topology of the enteropathogenic Escherichia coli translocator protein EspB. J Bacteriol 193:2972–2980. doi: 10.1128/JB.00153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh MP, Shaw RK, Knutton S, Pallen MJ, Crepin VF, Frankel G. 2008. Identification of amino acid residues within the N-terminal domain of EspA that play a role in EspA filament biogenesis and function. J Bacteriol 190:2221–2226. doi: 10.1128/JB.01753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa SCP, Schmitz AM, Jahufar FF, Boyd JD, Cho MY, Glicksman MA, Lesser CF. 2012. A new means to identify type 3 secreted effectors: functionally interchangeable class IB chaperones recognize a conserved sequence. mBio 3(1):e00243–11. doi: 10.1128/mBio.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lilic M, Vujanac M, Stebbins CE. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol Cell 21:653–664. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 51.Creasey EA, Delahay RM, Daniell SJ, Frankel G. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149:2093–2106. doi: 10.1099/mic.0.26355-0. [DOI] [PubMed] [Google Scholar]
- 52.Creasey EA, Friedberg D, Shaw RK, Umanski T, Knutton S, Rosenshine I, Frankel G. 2003. CesAB is an enteropathogenic Escherichia coli chaperone for the type-III translocator proteins EspA and EspB. Microbiology 149:3639–3647. doi: 10.1099/mic.0.26735-0. [DOI] [PubMed] [Google Scholar]
- 53.Kubori T, Galan JE. 2002. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J Bacteriol 184:4699–4708. doi: 10.1128/JB.184.17.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Ai X, Portaliou AG, Minetti CASA, Remeta DP, Economou A, Kalodimos CG. 2013. Substrate-activated conformational switch on chaperones encodes a targeting signal in type III secretion. Cell Rep 3:709–715. doi: 10.1016/j.celrep.2013.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]