Summary
Deubiquitinating enzymes (DUBs) control vital processes in eukaryotes by hydrolyzing ubiquitin adducts. Their activities are tightly regulated, but the mechanisms remain elusive. In particular, the DUB UCH-L5 can be either activated or inhibited by conserved regulatory proteins RPN13 and INO80G, respectively. Here we show how the DEUBAD domain in RPN13 activates UCH-L5 by positioning its C-terminal ULD domain and crossover loop to promote substrate binding and catalysis. The related DEUBAD domain in INO80G inhibits UCH-L5 by exploiting similar structural elements in UCH-L5 to promote a radically different conformation, and employs molecular mimicry to block ubiquitin docking. In this process, large conformational changes create small but highly specific interfaces that mediate activity modulation of UCH-L5 by altering the affinity for substrates. Our results establish how related domains can exploit enzyme conformational plasticity to allosterically regulate DUB activity. These allosteric sites may present novel insights for pharmaceutical intervention in DUB activity.
Graphical Abstract
Highlights
-
•
The RPN13 DEUBAD domain activates UCH-L5 by positioning its CL and ULD domain
-
•
The INO80G DEUBAD domain inhibits UCH-L5 by blocking ubiquitin binding
-
•
The FRF hairpin in the DEUBAD domain of INO80G drives UCH-L5 inhibition
-
•
DEUBAD domains regulate UCH-L5 activity by tuning UCH-L5 substrate affinity
Deubiquitinating enzyme UCH-L5 (UCH37) is involved in DNA repair and the proteasome system. Sahtoe et al. uncover how two evolutionarily related DEUBAD domains in RPN13 (ADRM1) and INO80G (NFRKB) can either activate or inhibit UCH-L5. These remarkable regulatory modes exploit flexible structural elements in UCH-L5 to modulate activity.
Introduction
The ubiquitin conjugation machinery regulates almost every process in the eukaryotic cell. Deubiquitinating enzymes (DUBs) are a critical component of the machinery since they can remove ubiquitin adducts and thereby control the level of ubiquitin signals (Komander et al., 2009). In accordance with their important roles, DUBs are frequently deregulated in human pathologies including cancer and neurological disease (Clague et al., 2013), making DUBs potential prime targets for therapeutic intervention.
The level of the intrinsic DUB activity is important and requires precise control. For a subset of DUBs, there is emerging evidence that the catalytic activity can be modulated by regulatory proteins or by internal domains (Sowa et al., 2009). Notable examples include USP7 activation by its HUBL domain and GMPS (Faesen et al., 2011), USP1 activation by UAF1 (Cohn et al., 2007), and Ubp8 activation in the SAGA complex (Köhler et al., 2008; Lee et al., 2005). The most striking example is UCH-L5, for which both activation and inhibition have been observed (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006, 2008) by two different proteins, RPN13 (ADRM1) and INO80G (NFRKB), respectively.
Understanding the mechanisms of DUB activation is important for interpreting their roles in specific cellular contexts. Mechanistic insight into regulatory mechanisms also can provide vital information for the development of inhibitors or activators. So far, the only available crystal structure of a DUB-activator complex is that of the SAGA DUB module (Köhler et al., 2010; Samara et al., 2010), but no structure is available for its inactive state. Due to this lack of structural data, detailed mechanisms of DUB regulation are still poorly understood.
UCH-L5 (UCH37) is a cysteine protease of the ubiquitin C-terminal hydrolase (UCH) family of DUBs, which also includes UCH-L1, UCH-L3, and BAP1. UCH-L5 is overexpressed in several carcinomas (Chen et al., 2012; Fang et al., 2012, 2013) and knockout of the gene is embryonically lethal in mice (Al-Shami et al., 2010). Functionally, it has been linked to TGF-β signaling, Alzheimer’s disease, and longevity (Kikuchi et al., 2013; Matilainen et al., 2013; Wicks et al., 2005, 2006). UCH-L5 constitutes a component of proteasomes and INO80 chromatin remodeling complexes, where it is activated and inhibited, respectively.
As a non-essential component of the proteasome 19S regulatory particle, UCH-L5 catalyzes K48-linked polyubiquitin hydrolysis. This activity requires the RPN13 subunit whose C-terminal domain binds UCH-L5 (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006). In vitro, RPN13 is able to directly promote UCH-L5 activity against a minimal substrate (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006).
UCH-L5 has a less well-defined role in metazoan INO80 chromatin remodeling complexes. INO80 is an essential determinant of embryonic stem cell identity (Chia et al., 2010; Wang et al., 2014) and participates in the DNA damage response (Smeenk and van Attikum, 2013), but the function of the metazoan-specific subunits, such as INO80G, is poorly defined. A recent report has implicated UCH-L5 and INO80G as key factors of the DNA double-strand-break response (Nishi et al., 2014). Interestingly, in the context of the INO80 complex, the DUB activity of UCH-L5 is inhibited by the INO80G subunit (Yao et al., 2008). Intriguingly, an artificial shorter version of INO80G was found to activate UCH-L5 in vitro (Yao et al., 2008).
The UCH enzymes have a small highly conserved papain-like catalytic domain (CD) characterized by a flexible active site cross-over loop (CL). The CL is thought to select substrates according to leaving group size (Popp et al., 2009; Zhou et al., 2012). In UCH-L5 and UCH family member BAP1 the CL is relatively large, enabling them to process larger substrates (Zhou et al., 2012).
Within the UCH family UCH-L5 and BAP1 are close relatives. BAP1 is a critical tumor suppressor whose regulation is important for proper gene regulation (Carbone et al., 2013; Goldstein, 2011; White and Harper, 2012). UCH-L5 and BAP1 share an unusual C-terminal helical extension, called ULD (Misaghi et al., 2009). The ULD domain could mediate protein-protein interactions, including higher-order homo-oligomerization (Burgie et al., 2012; Jiao et al., 2014), and was proposed to act as an auto-inhibitory module (Yao et al., 2006).
Like UCH-L5, BAP1 can be activated by a regulatory protein, in this case ASX, to promote H2A deubiquitination (Scheuermann et al., 2010). Phylogenetic analyses have uncovered a conserved domain within the UCH regulatory proteins RPN13, INO80G, and ASX, which was named the DEUBAD domain (Sanchez-Pulido et al., 2012). As all three proteins affect UCH activity, it was proposed that the DEUBAD domain is responsible for this modulation. The conservation suggests a common mechanism of regulation, but where ASX and RPN13 activate their cognate DUB, INO80G inhibits it. Thus, the DEUBAD domain has shifted from activator to inhibitor mode. The mechanistic details of this dual mode of action of the DEUBAD domains are unclear.
Here we present structural and functional analyses that explain how DEUBAD domains can switch UCH-L5 activity and thus provide either positive or negative regulation. We show how the DEUBAD domain in RPN13 activates UCH-L5 by tuning the conformation of structural elements in UCH-L5, and inhibits in INO80G, where it exploits molecular mimicry and UCH-L5 conformational plasticity to prevent ubiquitin docking and catalysis. We also show how the inhibitory domain in INO80G has retained the ability to activate, by its N-terminal INO80Gshort region, and identify the structural elements in the DEUBAD domains that confer the activating or inhibitory effects on UCH-L5 enzymatic activity. Our data show that this remarkable tuning of activity involves large conformational changes and is mediated by precise positioning of both the UCH-L5 C-terminal ULD and active site CL.
Results
Crystal Structures of Activated and Inhibited UCH-L5
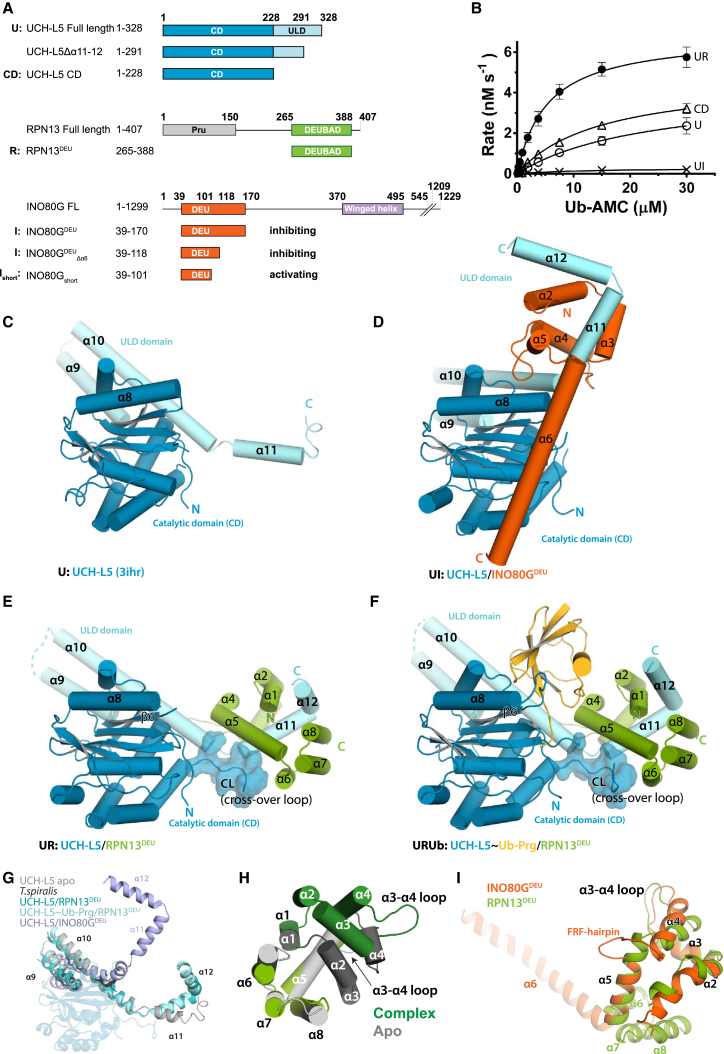
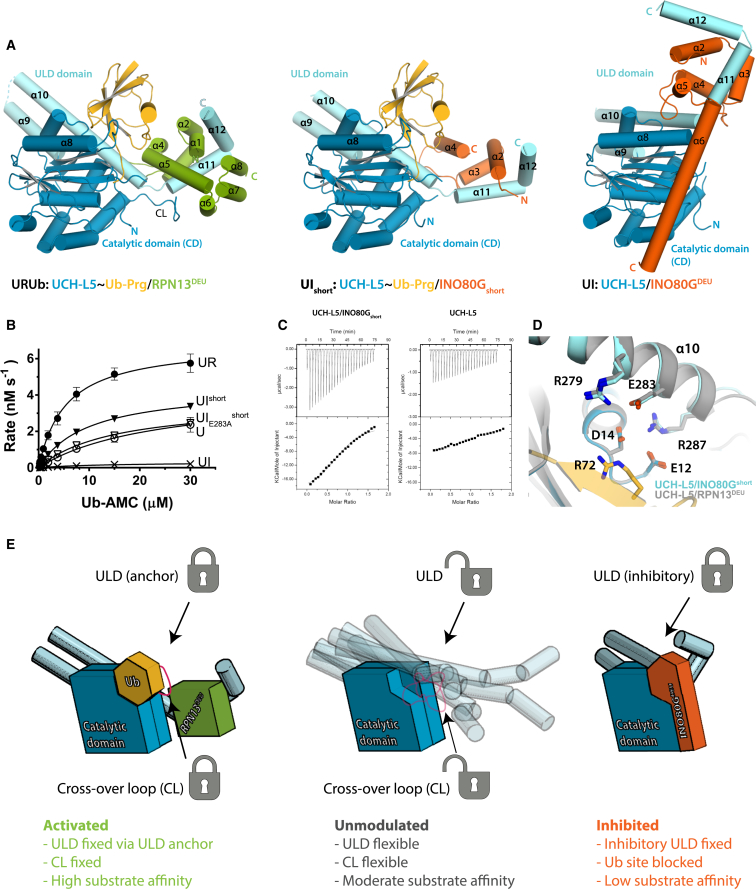
To study the regulation of UCH-L5 by DEUBAD domains, we purified human UCH-L5 in complex with the DEUBAD domains of RPN13 (amino acid [aa] 265–388, referred to as RPN13DEU) and INO80G (aa 39–170, referred to as INO80GDEU) (Figure 1A). We measured the catalytic activity of these complexes towards the minimal substrate ubiquitin-7-amido-4-methylcoumarin (Ub-AMC) (Dang et al., 1998; El Oualid et al., 2010) in comparison to full-length UCH-L5 alone (U) and its isolated CD. In line with previous data, we found that the DEUBAD domain of RPN13 activates UCH-L5 (UR) (Figure 1B; Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006). Since the UCH-L5 CD is more active than the full-length alone, the ULD domain partially inhibits activity (Yao et al., 2006). However, in the presence of RPN13DEU, UCH-L5 is significantly more active than the UCH-L5 CD, and, therefore, RPN13DEU does more than simply remove autoinhibition (Figure 1B). Strikingly, INO80GDEU severely inhibits activity under these conditions (UI) (Figure 1B).
Figure 1.
Crystal Structures UCH-L5/DEUBAD Complexes
(A) Constructs used in this study.
(B) RPN13DEU activates UCH-L5 (UR) while INO80GDEU inhibits UCH-L5 (UI) in Ub-AMC enzyme kinetics. The CD is slightly more active than FL UCH-L5 (U). See Figure 1A for naming codes. Error bars, SD.
(C) Structure of apo UCH-L5 (3ihr). CD, blue; ULD domain, light blue.
(D) Structure of UCH-L5/INO80GDEU (INO80GDEU, orange).
(E) Structure of UCH-L5/RPN13DEU (RPN13DEU, green).
(F) Structure of UCH-L5∼Ub-Prg/RPN13DEU (Ub-Prg, yellow).
(G) The ULDs are found in different conformations across UCH-L5 structures. The CD is transparent for clarity.
(H) RPN13DEU (green) changes toward an open state upon UCH-L5 complex formation compared to apo RPN13DEU (gray). Helices α1–4 and the α3-4 loop that undergo the largest changes are colored in darker shades.
(I) Superposition of RPN13DEU and INO80GDEU. DEUBAD domains deviate most at FRF hairpin and helix α6. See also Tables 1, S1, S2, and S4 and Figure S1.
We wondered how these related DEUBAD domains achieve such remarkably opposite effects on regulation. To assess this, we performed structural studies on UCH-L5 in complex with DEUBAD domains and compared them with apo UCH-L5 (Burgie et al., 2012; Figure 1C). We determined a crystal structure of UCH-L5 in complex with the inhibitory domain INO80GDEU at 3.7 Å (Figure 1D). Additionally, we determined crystal structures of UCH-L5 in complex with activating RPN13DEU, with and without the suicide inhibitor ubiquitin-propargyl (Ub-Prg) (Ekkebus et al., 2013; Sommer et al., 2013) at 2.3 Å and 2.8 Å, respectively (Figures 1E and 1F). All structures were refined to acceptable statistics (Table 1).
Table 1.
Crystallography Details
| Data Collection | UCH-L5/RPN13DEU | UCH-L5∼Ub-Prg/RPN13DEU | UCH-L5/INO80GDEU | UCH-L5∼Ub-Prg/INO80Gshort | |
|---|---|---|---|---|---|
| Wavelength (Å) | 0.98 | 0.87 | 0.91 | 0.87 | |
| Resolution (Å) | 33.5–2.8 (3.0–2.8) | 45.3–2.3 (2.4–2.3) | 47.7–3.7 (4.1–3.7) | 47.7–3.7 (4.0–3.7) | |
| Space group | P212121 | P212121 | P4122 | C2 | |
| Unit cell | a, b, c (Å) | 56.6, 97.07, 100.6 | 59.34, 98.6, 100.2 | 94.88, 94.88, 132 | 152.1, 137.8, 98.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 102.5, 90 | |
| CC1/2 (%) | 99.8 (72.9) | 99.9 (67.6) | 98.3 (41.8) | 98.4 (72.6) | |
| Rmerge (%) | 7.1 (93.0) | 12.6 (83.2) | 20.9 (73.2) | 19.9 (66.4) | |
| I/σI | 15.3 (2.2) | 9.7 (1.6) | 7.4 (2.4) | 5.8 (1.8) | |
| Completeness (%) | 98.5 (97.3) | 99.8 (98.9) | 99.5 (100) | 97.9 (90.3) | |
| Redundancy | 3.9 (3.7) | 4.1 (4.1) | 6.7 (6.8) | 4.2 (4.0) | |
| Refinement | |||||
| Number of unique reflections | 13,635 | 26,949 | 7,004 | 21,090 | |
| Rwork/Rfree (%) | 23.2/28.4 | 19.5/23.5 | 30.4/35.4 | 23.8/26.6 | |
| Rmsd bond lengths (Å) | 0.003 | 0.005 | 0.004 | 0.008 | |
| Rmsd bond angles (°) | 0.7 | 1.0 | 0.7 | 1.2 | |
| Ramachandran statisticsa (%) (preferred/allowed/not allowed) | 99.2/0.8/0 | 98.5/1.5/0 | 97.7/2.03/0 | 95.02/4.98/0 | |
High-resolution shells in parentheses.
Molprobity.
The resulting structures display striking differences (Figures 1D–1F). Both RPN13DEU and INO80DEU primarily bind the C-terminal ULD domain of UCH-L5, but are positioned radically differently relative to the UCH-L5 CD, which itself hardly changes conformation among all UCH-L5 structures. The differences arise from major changes in orientation of the ULDs relative to the CD (Figure 1G). The ULDs adopt a wide range of positions relative to the CD, even in previously known UCH-L5 structures (Burgie et al., 2012; Maiti et al., 2011; Morrow et al., 2013), suggesting that this element is flexible in solution. Activator and inhibitor may lock this domain in particular conformations.
To allow UCH-L5 binding, RPN13DEU changes conformation compared to the previously determined RPN13DEU apo-state (Chen et al., 2010), by rearranging core helices α1–α4 and the α3-α4 loop (Figure 1H). In this new conformation, the cores of INO80GDEU and RPN13DEU resemble each other, underscoring their common ancestry (Figures 1I and S1A). The C termini of the DEUBAD domains, however, diverge dramatically. Where helices α6–α8 (aa 350–384) form a platform in RPN13DEU, the equivalent region in INO80GDEU forms a single extended helix (α6). Another notable difference between the two DEUBAD domains is a short hairpin (aa 96–103) exclusively present in INO80G, which we named the FRF hairpin. It is inserted between helix α4 and α5 of the DEUBAD domain and is conserved in INO80G orthologs (Figures 1I and S1B).
The structural conservation of the DEUBAD domains is also reflected in their similar binding modes to UCH-L5 (Figures 1D–1F and S1C). In both complexes, the core DEUBAD domains bind primarily to the C-terminal ULD of UCH-L5, where amphipathic helix α11 is clasped by the DEUBAD domains and further stabilized by helix α12 in an extensive hydrophobic interface (Figures S1D and S1E). DEUBAD domain binding requires these helices, since a UCH-L5 variant lacking these (UCH-L5Δα11-12) does not interact with RPN13DEU, whereas the wild-type (WT) UCH-L5 binds tightly (KD = 6 nM) in isothermal titration calorimetry (ITC) (Figure S1F; Table S4).
In short, the conserved DEUBAD domains bind to UCH-L5 but show dramatically different arrangements. In the next sections, we examine how these are achieved and how they can lead to differences in UCH-L5 activity. The structural consequences of ubiquitin binding are discussed in light of the mechanism of activation and inhibition.
DEUBAD Domains Tune UCH-L5 Substrate Affinity
Analysis of UCH-L5 kinetic parameters (Figure 1B; Table S1) reveals that RPN13DEU binding primarily changes the KM of the enzyme, but hardly affects kcat. Therefore, we wondered if changes in substrate affinity of UCH-L5 could explain regulation by DEUBAD domains.
We assessed the ability of UCH-L5 to bind a model substrate in the presence and absence of the DEUBAD domains using ITC. The model substrate Ub-GlySerThr was titrated into active site cysteine (C88A) mutants of the different UCH-L5 complexes. We observed robust binding (KD = 4.5 μM) for UCH-L5/RPN13DEU, whereas binding to UCH-L5 alone, UCH-L5 CD, and UCH-L5/INO80GDEU did not reach saturation, with the UCH-L5/INO80GDEU complex giving the lowest signal (Figures 2A and S2A; Table S3). These results suggest that RPN13DEU enhances UCH-L5 substrate binding, whereas INO80GDEU diminishes it. We validated these results in fluorescence polarization (FP) binding assays, using Ub-LysGlyTAMRA as a model substrate (Geurink et al., 2012), which confirmed that DEUBAD domains tune UCH-L5 activity at the level of substrate binding (Figure 2B).
Figure 2.
Ubiquitin Binding of UCH-L5 Complexes
(A) RPN13DEU increases UCH-L5’s affinity for model substrate Ub-GlySerThr, whereas INO80GDEU decreases the signal in ITC.
(B) Validation of binding results using model substrate Ub-LysGlyTAMRA in FP binding assays. Error bars, SD.
(C) Overview of the ubiquitin-binding interface of the UCH-L5∼Ub-Prg/RPN13DEU complex.
(D) Contacts between ubiquitin and RPN13DEU are limited.
(E) Ubiquitin binding in the UCH family is structurally conserved (UCH-L5/RPN13DEU; T.spiralis UCH-L5, 4i6n; UCH-L1, 3ifw; UCH-L3, 1xd3; P.Faliciparum UCH-L3, 2wdt).
(F) The ubiquitin tail is extensively coordinated.
(G) The ubiquitin core is stabilized by three specific exosite contacts. UCH-L5/RPN13DEU (without Ub-Prg) displayed in gray sticks.
(H) Partial melting of UCH-L5 helix α8 relocates Ile216 to contact the ubiquitin Ile36 patch. This interaction is conserved in T.spiralis UCH-L5. See also Table S3 and Figure S2B.
Ubiquitin Binding by the UCH-L5/RPN13DEU Complex
UCH-L5 activation by the RPN13 DEUBAD domain results from enhanced ubiquitin-substrate binding; therefore, we analyzed the details of ubiquitin interaction in the UCH-L5∼UbPrg/RPN13DEU crystal structure (Figure 2C). The presence of Ub-Prg hardly changed the global UCH-L5/RPN13DEU conformation (Figure 1). Direct contact between ubiquitin and RPN13DEU involved a small interface (286 A2) with three hydrogen bonds (Figures 2C and 2D). Moreover, this interface did not affect the position and orientation of ubiquitin on UCH-L5, which resembled the previously solved T.spiralis UCH-L5 ubiquitin complex. In fact, it was identical to the canonical ubiquitin-binding mode found in all UCH family members (Figure 2E).
In practice, ubiquitin binds via its C-terminal tail close to the active site and via its core relatively far from the active site, in a series of so-called exosites. In the UCH-L5 complex, the ubiquitin C-terminal tail adopts an extended conformation. It is buried and positioned by an extensive network of side-chain and backbone interactions with the CD (Figures 2C and 2F).
The binding of the ubiquitin core creates three specific exosite interactions on the UCH-L5 CD. The first involves UCH-L5 Trp36, which rearranges, compared to apo UCH-L5/RPN13DEU, to avoid clashes and to promote a direct contact with ubiquitin Ile44 (Figures 2C and 2G), the primary hydrophobic binding site on ubiquitin.
A second exosite interaction involves UCH-L5 Ile216 at the C terminus of helix α8 (Figure 2G). This helix melts out in UCH-ubiquitin complexes, but is extended in the absence of ubiquitin (Figure S2B). Partial melting of α8 is crucial since it rearranges Ile216 from a buried position to a position in the α8-β6 connecting loop that is compatible with ubiquitin binding, similar to T.spiralis UCH-L5 where Val214 (equivalent to human Ile216) contacts the ubiquitin Ile36 patch (Morrow et al., 2013; Figure 2H). Interestingly, in the UCH-L5/RPN13DEU structure, the C terminus of helix α8 is already disordered in the absence of ubiquitin, indicating that RPN13DEU may affect this region allosterically to facilitate ubiquitin binding.
Finally, the melting of helix α8 and ordering of the α8-β6 connecting loop promotes positioning of Phe218 and formation of a highly conserved pocket that includes UCH-L5 Leu38. This hydrophobic pocket on UCH-L5 allows a snug interaction with the ubiquitin β1-β2 hairpin containing Leu8 and Thr9 (Figure 2G). Formation of this pocket was described for UCH-L1, upon ubiquitin interaction (Boudreaux et al., 2010). In UCH-L1 the Phe214 positioning promotes rearrangement of Phe53 (equivalent to UCH-L5 residues, Phe218 and Phe56), which is necessary to organize the catalytic site conformation. In UCH-L5 this relay is not required, since Phe56 is already positioned such that the catalytic triad is active in the apo-structure. Nevertheless, the conformational change of UCH-L5 Phe218 in this pocket is conserved upon ubiquitin binding, as is the interaction with the ubiquitin β1-β2 hairpin. All three exosite interactions are well conserved in the UCH family, explaining the remarkably similar ubiquitin positioning on the UCH CDs (Figure 2E).
The DEUBAD Domain of RPN13 Activates UCH-L5 by ULD and CL Positioning
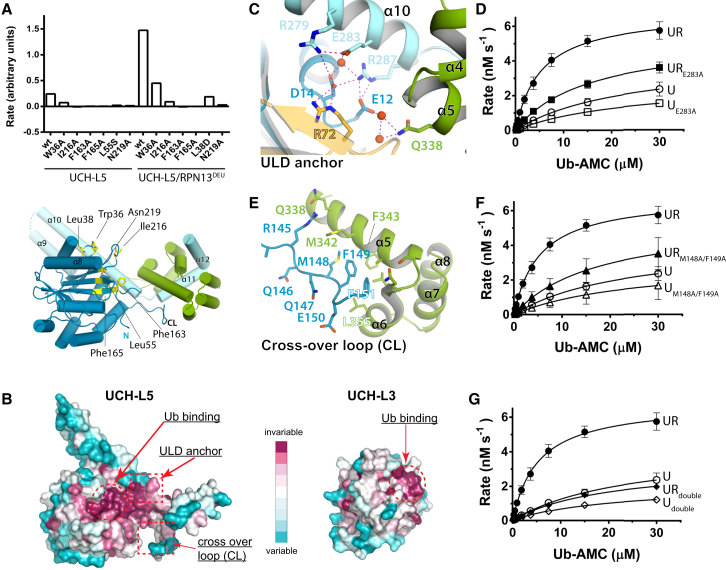
To investigate how RPN13DEU promotes enhanced substrate binding by UCH-L5, we tested the effect of mutations on activity. We first focused on the effect on activation of the ubiquitin-binding residues in UCH-L5. Mutations in these residues lowered the activity substantially, irrespective of the presence of RPN13DEU, indicating that they are primarily important for basic DUB function (Figure 3A). We then tested mutations of RPN13DEU located in the interface with ubiquitin, and found that these provided only a limited contribution to UCH-L5 activation (Figures 2D and S2C).
Figure 3.
Activation Mechanism RPN13DEU
(A) Mutations in the ubiquitin interface severely compromise DUB activity irrespective of RPN13DEU (top). Location mutants (yellow sticks) on UCH-L5 (bottom).
(B) Surface representation of UCH-L3 and UCH-L5 colored by conservation.
(C) UCH-L5 helix α10 is anchored to the CD via an extensive polar network.
(D and F) Compared to WT (UR and U from Figure 1B), UCH-L5 mutants E283A and M148A/F149A cannot be activated by RPN13DEU to the same extent on Ub-AMC. Error bars, SD.
(E) The CL is positioned by RPN13DEU.
(G) Activation of the combined CL and ubiquitin anchor mutants E283A/M148A/F149A is almost completely abrogated compared to WT (UR and U from Figure 1B). Error bars, SD. See also Figure S2 and Tables S1–S4.
To further explore the molecular origins of the activation, we analyzed the evolutionary conservation of surface residues on UCH-L5 with ConSurf (Glaser et al., 2003). A UCH-L5 sequence alignment from species across all major eukaryotic lineages that possess both RPN13 and UCH-L5 was projected onto the UCH-L5 structure. This was compared to an analogous conservation analysis for UCH-L3, a prototype UCH member that lacks the C-terminal ULD domain. We noted several conserved regions. Both UCHs have a conserved surface patch where ubiquitin binds (Figure 3B). Adjacent to this patch, we found a second highly conserved site in UCH-L5 orthologs that is absent in UCH-L3 orthologs. This site centers on Glu283 and anchors the ULD to the CD through a polar interaction network (Figure 3C). The strong conservation of this ULD anchor is intriguing as the area is not directly involved in ubiquitin or RPN13DEU binding.
We assessed the functional importance of the ULD anchor by testing UCH-L5 mutants in Ub-AMC assays. UCH-L5E283A had similar activity to WT but this mutant could not be activated to the same extent as WT by RPN13DEU, mainly due to weaker KM (Figure 3D; Tables S1 and S2). The fact that this E283A mutation does not affect intrinsic UCH-L5 activity, but only the activity of the RPN13DEU complex, strongly suggests that an intact ULD anchor is required for RPN13DEU-dependent activation of UCH-L5. Next we tested the effect of E283A mutation on substrate binding. Using ITC and stopped-flow binding analysis, we found that UCH-L5E283A/RPN13DEU shows decreased affinity for Ub substrates compared to WT complex (Figures S2A and S2D; Table S3). This indicates that RPN13DEU induces a higher affinity for substrates by utilizing the intact ULD anchor.
We then focused our attention on the UCH-L5 CL that is disordered in most UCH-L5 crystal structures. In our complexes, the CL makes contacts with RPN13DEU (Figure 3E) via the highly conserved Met148 and Phe149, partially ordering the loop. Given the importance of the CL for the ability of UCHs to process larger substrates (Popp et al., 2009; Zhou et al., 2012), we made mutants to test whether the interface of the CL with RPN13DEU could affect activity by positioning the loop. In a Ub-AMC assay, UCH-L5M148A/F149A hydrolyzed Ub-AMC comparable to WT, indicating that the mutant was still functional. However, this mutant was only marginally activated by the addition of RPN13DEU (2.7-fold instead of 7-fold in WT), indicating that UCH-L5 activation by RPN13DEU requires an intact CL (Figure 3F; Tables S1 and S2). Interestingly, unlike the ULD anchor mutant, the CL mutant and WT complexes bound model substrates with similar affinities (Figures S2A and S2D; Table S3).
Combining the CL and ULD anchor mutants into UCH-L5double resulted in an almost complete abrogation of RPN13DEU-mediated activation, illustrating that the CL and ULD anchor are the principal regulatory sites used by RPN13DEU (Figure 3G). As none of the UCH-L5 mutants were compromised in RPN13DEU binding, as shown by ITC (Figure S2E; Table S4), we conclude that RPN13DEU exerts its stimulatory effect on UCH-L5 through positioning of the CL and ULD anchor.
Mechanism of UCH-L5 Inhibition by INO80GDEU
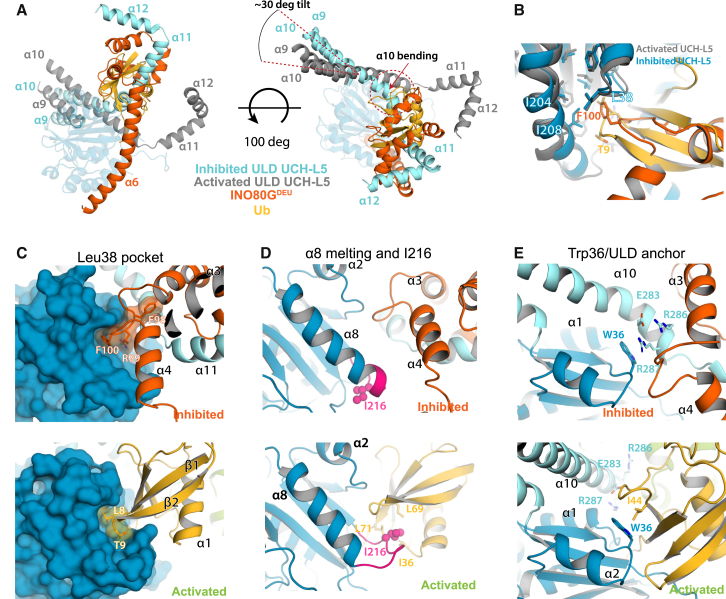
Our binding assays showed that INO80GDEU decreases the affinity of UCH-L5 for substrates (Figures 2A and 2B). To understand this effect, we analyzed how INO80GDEU affects UCH-L5 conformation (Figure 1) in more detail. INO80GDEU alters the ULD domain’s relative position and conformation in two specific ways. First, helix α9 and α10 are tilted by ∼30° compared to the active ULD conformation, and, second, the C-terminal end of helix α10 is bent toward the CD (Figure 4A). As a result, sections of the ULD and INO80GDEU occupy the canonical ubiquitin-binding exosites on UCH-L5 and thus prevent substrate docking (Figure 4A). The blockage of the exosites by the INO80GDEU complex presents a structural rationale for the decreased substrate binding that we observed, and provides a simple yet unexpectedly striking explanation of the INO80GDEU inhibition mechanism.
Figure 4.
Inhibition Mechanism INO80GDEU
(A) The ubiquitin-docking site on UCH-L5 is blocked as a result of INO80GDEU-induced conformational changes of the ULD. RPN13DEU is removed for clarity.
(B and C) The FRF hairpin mimics the ubiquitin β1-β2 hairpin to bind the Leu38 pocket.
(D) In contrast to the activated state, helix α8 in UCH-L5 adopts an extended state in the INO80GDEU complex to bury Ile216.
(E) The ULD anchor interaction is disrupted in the INO80GDEU complex due to ULD tilting and helix α10 bending, establishing intramolecular stacking of Arg287 on Trp36.
Analysis of the UCH-L5/INO80GDEU interface shows how the large conformational changes in UCH-L5 organize novel interfaces where key elements for ubiquitin binding and RPN13DEU-mediated activation are exploited by INO80GDEU to inhibit UCH-L5 activity. First, ULD conformational changes allow INO80GDEU helix α6 to contact the UCH-L5 CD, possibly stabilizing the inhibitory conformation of the ULD (Figures 1D and S1D). Second, in a neat example of molecular mimicry, the INO80G FRF hairpin binds to the UCH-L5 Leu38 pocket in a fashion that resembles the binding of the structurally analogous ubiquitin β1-β2 hairpin to this pocket (Figures 4B and 4C). Next, the C terminus of UCH-L5 helix α8 refolds to make the extra helical turn seen in the apo-structure. As a result, UCH-L5 Ile216 rearranges toward the hydrophobic core, preventing the possibility of the important interaction with the ubiquitin Ile36 patch (Figure 4D). Finally, helix α10 bending in the INO80GDEU complex relocates the ULD anchor residues toward UCH-L5 Trp36, creating a novel intramolecular interface consisting of a cation-π stacking interaction between the indole ring of the Trp36 and Arg287 in UCH-L5 helix α10 (Figure 4E). This relocation simultaneously precludes Trp36 availability for ubiquitin binding and impedes the formation of the intricate polar interaction network in the ULD anchor that is required for UCH-L5 activation by RPN13DEU (Figure 3C). Thus, all elements for ubiquitin binding in UCH-L5 are effectively obscured by INO80GDEU binding.
The FRF Hairpin Creates the Activity Switch in DEUBAD Domains
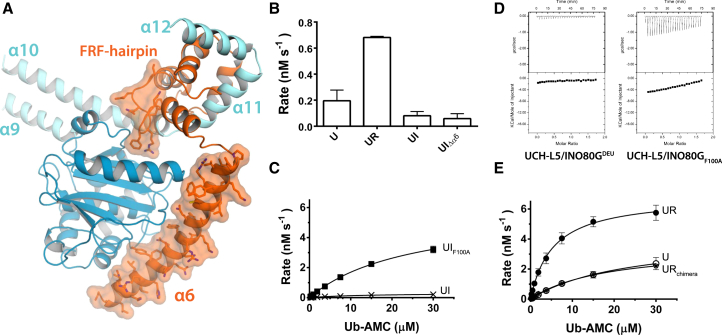
To study what features in INO80GDEU are required to achieve UCH-L5 inhibition, we analyzed its differences with the activating DEUBAD domain of RPN13. The most striking difference between the DEUBAD domains of RPN13 and INO80G is the C-terminal part where three helices α6–α8 in RPN13DEU change to the extended α6 in INO80GDEU that packs against the CD (Figures 5A and S1D). Therefore, we created a shorter version of INO80GDEU, where helix α6 is removed (Figure 1A, residues 39–118, INO80GDEUΔα6). Surprisingly, the truncated protein INO80GDEUΔα6 still inhibited UCH-L5 (Figure 5B), demonstrating that helix α6 is not required for inhibition under these conditions.
Figure 5.
The FRF Hairpin Drives UCH-L5 Inhibition
(A) INO80GDEU differs from RPN13DEU mainly in the FRF hairpin and helix α6 (orange surfaces).
(B) Helix α6 of INO80GDEU is dispensable for inhibition in Ub-AMC assays. Error bars, SD.
(C) Inhibition is completely lost in the INO80GDEU F100A mutant (UIF100A) on Ub-AMC. Error bars, SD.
(D) Mutant complex F100A gains Ub-GlySerThr-binding ability compared to WT INO80GDEU complex (from Figure 3A) in ITC.
(E) Insertion of the FRF hairpin in RPN13DEUchimera abolishes the activation effect of WT RPN13DEU on UCH-L5 (UR and U from Figure 1B). Error bars, SD. See also Figure S3 and Tables S1–S3.
We next assessed the importance of the INO80G FRF hairpin, which is the other major structural difference between the RPN13DEU and INO80GDEU (Figures 1I and 5A). In the hairpin, the side chain of the highly conserved Phe100 (Figure S1B) is accommodated by the UCH-L5 Leu38 pocket (Figure 4C), suggesting that this interaction is important for INO80G function. To address the relevance of this interaction, we made a single-point mutant F100A in INO80GDEU.
This point mutant, INO80GDEUF100A, lost the ability to inhibit UCH-L5 in Ub-AMC assays. In fact, it restored activity to the level of UCH-L5 alone, highlighting the importance of the FRF-hairpin interaction for UCH-L5 inhibition (Figure 5C; Tables S1 and S2). Moreover, the F100A complex gained a significant substrate-binding ability in contrast to the WT INO80GDEU complex (Figure 5D). Loss of the phenylalanine interaction makes the Leu38 pocket available again for binding of the β1-β2 hairpin of ubiquitin (Figure 2G), but most likely also allows helix α10 to revert to its extended state, affecting the ULD position.
The structural changes in the DEUBAD domains may have been a crucial evolutionary event facilitating novel regulatory modes of the DEUBAD domain. To test this we created a chimeric RPN13DEU variant (RPN13DEUchimera) by inserting the INO80G FRF hairpin into the structurally equivalent position in RPN13DEU (Figure S3A). The chimera formed a stable complex with UCH-L5 (Figure S3B), but completely abolished the activation effect. The inserted FRF hairpin was not sufficient to inhibit UCH-L5 to the same extent as INO80GDEU however (Figure 5E; Table S1). In RPN13DEUchimera the presence of the FRF hairpin likely diminishes ubiquitin binding, as it would be overlapping with the ubiquitin-binding site explaining the loss of activation potential. These results demonstrate that the FRF hairpin and its location within the DEUBAD domain have a crucial effect on UCH-L5 activity.
The INO80Gshort Activation Mechanism Also Relies on ULD Positioning
We wondered how lack of the FRF hairpin would affect the stucture of UCH-L5 and INO80G. To this end, we determined the 3.7 Å structure of UCH-L5 in complex with Ub-Prg and INO80Gshort (aa 39–101), a shorter fragment of INO80GDEU (Figure 1A). The artificial INO80Gshort construct has the remarkable capability to activate UCH-L5 in vitro (Yao et al., 2008). INO80Gshort starts at the same residue as INO80GDEU but terminates in the middle of the FRF hairpin and, hence, is predicted not to contain a folded FRF hairpin. Indeed, in the crystal structure, the C-terminal end of INO80Gshort could not be unambiguously modeled, indicating that the FRF hairpin is not formed in this complex.
Strikingly, the UCH-L5∼UbPrg/INO80Gshort crystal structure resembled the activated RPN13DEU complex rather than the inhibited state (Figures 6A, S4A, and S4B). In this complex, the ULD largely reverted to the conformation seen in the activated RPN13DEU complexes with an extended helix α10 (Figure S4C). All conformational changes in UCH-L5 required to create the canonical ubiquitin-binding mode were also in place (Figures S4D and S4E). The structure of INO80Gshort itself and its binding mode to UCH-L5 were moreover identical to INO80GDEU (apart from FRF hairpin and α5-6) and RPN13DEU (Figure S4F).
Figure 6.
Reactivation of the Inhibitor and Model for UCH-L5 Regulation
(A) The crystal structure of UCH-L5∼Ub-Prg/INO80Gshort (middle) largely resembles the UCH-L5/RPN13DEU structures (left from Figure 1F), but not the UCH-L5/INO80GDEU structure (right from Figure 1D).
(B) INO80Gshort (UIshort) can activate UCH-L5, albeit not to RPN13DEU levels (UR, UI, and U from Figure 1B), and depends on the ULD anchor for activation on Ub-AMC. Error bars, SD.
(C) Activation by INO80Gshort correlates with enhanced Ub-GlySerThr binding in ITC compared to WT UCH-L5 (from Figure 3A).
(D) The ULD anchors in the INO80Gshort (light blue) and RPN13DEU (gray) superimpose very well and stabilize the same intramolecular interaction.
(E) Unmodulated UCH-L5 (middle) is characterized by CL and ULD flexibility that limits substrate binding and catalysis. Activation by RPN13DEU (left) limits flexibility by locking the ULD and CL in favorable conformations. INO80GDEU (right) locks the ULD in a conformation incompatible with substrate docking. See also Figure S4 and Tables S1–S3.
Enzyme kinetics analysis confirmed that INO80Gshort activates UCH-L5 on Ub-AMC (Figure 6B). The activation effect correlates with increased affinity for substrates, since UCH-L5 binds substrates better in the presence of INO80Gshort in ITC-binding assays (Figure 6C). These results stress that DEUBAD domains mainly modulate activity by tuning substrate affinity.
The similarity in structure to the RPN13DEU complex suggests that INO80Gshort makes use of the same activation mechanism. To test this hypothesis, we used the UCH-L5E283A ULD anchor mutant, asking whether loss of the ULD anchor would also affect the activation in this case. We found that the UCH-L5E283A/INO80Gshort mutant complex was compromised in Ub-AMC hydrolysis (Figure 6B; Tables S1 and S2), reverting to the activity observed for UCH-L5 alone. This finding indicates the importance of the ULD anchor (Figure 6D) for INO80Gshort activation, and it suggests that ULD positioning in general is a major feature of the activation. INO80Gshort, containing only the helices α2–α4 of the DEUBAD domain, can activate UCH-L5, demonstrating that the core DEUBAD fold is already sufficient to bind and provide modest activation. The INO80Gshort complex does not attain UCH-L5/RPN13DEU activity levels however. Most likely this is because it lacks helix α5, which RPN13DEU uses to position the CL (Figure S4G).
Collectively, the UCH-L5/INO80Gshort structure analysis reconciled all our previous findings. First, it confirmed that the FRF hairpin is the crucial factor for inhibition, since absence of this element resulted in loss of inhibition. Second, loss of the FRF hairpin destabilized the inhibitory ULD conformation, causing it to snap back to a substrate-binding-competent conformation. Third, the core DEUBAD fold was sufficient to provide the basic UCH-L5 activation function. Like RPN13 it executed activation by stabilizing the substrate-binding-competent conformation of the ULD through the ULD anchor.
Discussion
Our data show how UCH-L5 activity can be modulated by DEUBAD domains present in RPN13 and INO80G through remarkably large conformational changes. Functionally, the activity of UCH-L5 is tuned at the level of substrate affinities, where RPN13DEU increases the affinity for substrates and INO80GDEU dramatically decreases substrate affinity. Our structural and biochemical analyses indicate that RPN13DEU achieves this effect by precise positioning of the ULD anchor and CL. The direct contact between RPN13DEU and ubiquitin furthermore confers a mild activation effect on UCH-L5 by stabilizing ubiquitin. Conversely, INO80GDEU exploits molecular mimicry and ULD conformational plasticity to prevent ubiquitin docking and catalysis. We showed which structural changes in DEUBAD domains may have been instrumental for the evolution of different modes of UCH-L5 regulation.
The proposed evolutionary conservation of the DEUBAD domains (Sanchez-Pulido et al., 2012) was confirmed by structural analyses in this study. The DEUBAD domains appear to be modular. The core DEUBAD fold (α1–α4) is shared among INO80GDEU, RPN13DEU, and INO80Gshort, and is responsible for binding to UCH-L5 and modest activation. Accessory structural modules in RPN13DEU (helix α5 that positions the CL) and INO80GDEU (FRF hairpin) lead to full activation or inhibition of UCH-L5. The modular nature of the DEUBAD domains explains their versatility as regulatory domains.
A key feature of UCH-L5 activity modulation is the conformational plasticity of the ULD that is found in a variety of conformations in the available UCH-L5 crystal structures (Figure 1G). This plasticity and the current data are consistent with a model where the UCH-L5 ULD can adopt a multitude of possible conformations in a dynamic fashion when free in solution (Figure 6E). As a consequence of the ULD’s proximity to the ubiquitin docking site, some of these conformations are sterically incompatible with ubiquitin binding while others allow efficient ubiquitin binding. The DEUBAD domain in RPN13DEU and INO80GDEU restricts ULD conformational plasticity by preferentially stabilizing specific conformations. RPN13DEU activates and increases the affinity for substrates by fixing the ULD into a substrate-binding-competent conformation, using the ULD anchor. Additionally, full activation is achieved by the stabilization of the ubiquitin orientation by RPN13DEU and correct positioning of the CL. On the other hand, INO80GDEU binds to UCH-L5 and uses ULD conformational flexibility to dock its unique inhibitory FRF hairpin into the Leu38 pocket. This interaction fixes the ULD in such a way that the ubiquitin-binding site is blocked by INO80GDEU and the ULD. In the process, key molecular elements for ubiquitin binding and RPN13-dependent activation are masked or disrupted, effectively inhibiting activity.
This regulatory model may co-exist with possible roles of the UCH-L5 oligomeric state for regulation of its activity (Burgie et al., 2012; Jiao et al., 2014; Yao et al., 2006). As the enzyme concentrations under our assay conditions were low, it is unlikely that we captured these phenomena. However, we cannot exclude that additional layers of regulatory complexity may exist in cells that involve UCH-L5 oligomeric states.
Our structures of UCH-L5/RPN13DEU explain the basic mechanisms of activation by the RPN13 DEUBAD module. It will be interesting to investigate the additional activation that is required to hydrolyze K48-polyubiquitin in the proteasomal 19S regulatory particle. It is conceivable that efficient K48-polyubiquitin hydrolysis will only take place after steps that are possibly related to proper positioning and unfolding of the compact K48-polyubiquitin chains. A key step here will be the identification of the minimal proteasomal complex required to perform chain hydrolysis.
The UCH-L5/INO80GDEU structure may provide novel approaches in unraveling the enigmatic role of UCH-L5 and INO80G in INO80 chromatin-remodeling complexes. INO80G is a key factor in embryonic stem cells and knock out leads to loss of pluripotency (Wang et al., 2014). Of specific interest is why UCH-L5 is kept in an inhibited state in the INO80 complex (Yao et al., 2008). A possibility is that INO80 controls UCH-L5 in a temporal manner, where in some circumstances UCH-L5 is inhibited while under other circumstances post-translational modifications (PTMs) and/or conformational changes release the inhibition and activate UCH-L5, allowing for additional layers of regulation. We have already seen in INO80Gshort that the core DEUBAD fold has the intrinsic ability to activate UCH-L5 and all that is required for INO80GDEU to relieve inhibition is disruption of the FRF hairpin. Such relief of inhibition would be important for the recently reported UCH-L5/INO80G role in DNA double-strand-break response, since UCH-L5 catalytic activity is required for proper DNA end resection (Nishi et al., 2014).
A unique element of the INO80GDEU domain is the extended helix α6. This helix packs against the CD close to the active site and, therefore, initially was thought by us to confer INO80GDEU inhibitory function. In our in vitro assays, this element was dispensable for inhibition, but this may be different in a cellular context where the additional contacts between this helix and the CD may further stabilize the inactivated state. An interesting feature of helix α6 is the presence of a large solvent-exposed positively charged patch. We speculate that this patch may be important in cells as a binding platform for INO80 chromatin-remodeling factors. As the equivalent region in RPN13DEU folds into a helical platform, it also may be possible that, under some conditions, driven by PTMs for example, α6 could refold into a conformation seen in RPN13 to meet functional requirements. DUB activity regulation by PTMs such as phosphorylation have been shown previously to be important for DUBA (Huang et al., 2012).
The ULD is conserved in UCH family member BAP1 that is activated by the ASXL1 DEUBAD domain to deubiquitinate H2A (Scheuermann et al., 2010). Because of the strong conservation of key elements between UCH-L5/RPN13DEU and BAP1/ASXL1, we anticipate that the ASXL1 DEUBAD domain employs similar strategies to activate BAP1. Both BAP1 and ASXL1 are important cancer drivers. BAP1 is a key tumor suppressor that is mutated in a number of cancers where loss of BAP1 is associated with poor prognosis and tumor aggressiveness (Carbone et al., 2013; White and Harper, 2012). Our crystal structures have valuable implications for BAP1 function in its cellular roles and pathogenesis.
The mechanisms of DUB regulation that we have described are different from those in previously studied DUB regulators. UAF1 increases the basicity of the USP1 catalytic histidine, increasing its potency as a general base (Villamil et al., 2012). Likewise, incorporation of Ubp8 in the SAGA complex stabilizes the catalytic center, also facilitating catalysis (Köhler et al., 2010; Samara et al., 2010). Activation of USP7 by GMPS against a minimal substrate changes only kcat (Faesen et al., 2011). All of these differ from UCH-L5 where a major part of the activity modulation involves tuning substrate affinities, rather than actual catalytic steps.
Whereas inhibition of DUBs by proteins is still a rare phenomenon, inhibition of general proteases by proteins has been well described (Dubin, 2005; Rzychon et al., 2004). Serpins inhibit serine proteases by irreversibly trapping the acyl-enzyme intermediate. Additionally, general cysteine proteases can be inhibited by cystatins and related proteins by occupying active site clefts. Instead, INO80G functions as an exosite inhibitor where not the active site cleft but an exosite, in this case the ubiquitin-core-docking site, is blocked. Enzyme exosite targeting by a naturally evolved inhibitor could provide powerful clues about the most efficient way for DUB inhibition, and may thus be promising from a pharmaceutical perspective.
Experimental Procedures
Plasmids and Cloning
Human UCH-L5, RPN13, and INO80G cDNA were subcloned from the HAP1 cell line. RPN13DEUchimera was purchased as a synthetic construct. All constructs were cloned into the pGEX or pET bacterial expression vectors of the NKI LIC suite (Luna-Vargas et al., 2011).
Protein Expression and Purification
All protein variants and protein complexes were (co-) expressed in E.coli. UCH-L5 and variants were purified using glutathione S-transferase (GST) affinity purification (GSH 4B sepharose, GE Healthcare) followed by a desalting (HiPrep 26/10, GE Healthcare) and final size-exlusion chromatography step (Superdex S200, GE Healthcare). UCH-L5 complexes were purified similarly except for an additional first nickel purification step. RPN13DEU alone was expressed in E.coli and purified using nickel affinity chromatography, desalting, and size-exclusion chromatography (Superdex S75, GE Healthcare).
Ub-AMC Enzymatic Assays
Enzyme activity was followed as release of fluorescent AMC from the quenched Ub-AMC substrate, providing a direct readout of DUB activity. Michealis-Menten parameters were determined using 1 nM enzyme while varying the substrate concentration. Initial rates were plotted against substrate concentration and fitted to the Michealis-Menten model using non-linear regression in Prism 6. In single-concentration experiments, 1 nM enzyme was allowed to react with 1 μM substrate. Activity was quantified by calculating the initial rates.
FP Binding Assays
Binding assays between UCH-L5 complexes and model substrate Ub-LysGlyTAMRA were performed by measuring FP. Model substrate (5 nM) was incubated at 25°C with varying amounts of different UCH-L5 complexes to obtain binding curves. To prevent substrate hydrolysis, inactive C88A mutants of UCH-L5 were used.
Stopped-Flow Fluorescent Polarization Binding Assay
Pre-steady-state binding events between UCH-L5 (C88A) complexes and Ub-LysGlyTAMRA were monitored in stopped-flow fluorescent polarization experiments. Varying concentrations of UCH-L5 variants were injected together with 20 nM (final concentration) Ub-KGTAMRA, after which fluorescent polarization was followed during 10 s. Association binding traces were fitted to a one-phase exponential model in Prism 6 to obtain kobs. The kobs values were plotted against protein concentration to estimate kon, koff, and KD.
ITC
ITC experiments were performed in a VP-ITC Microcal calorimeter at 25°C. In 10 μl injections, 450 μM UbGlySerThr was titrated into 45 μM UCH-L5 (C88A) or 110 μM UCH-L5 into 12.5 μM RPN13DEU. Data were fitted to a one-site-binding model with the manufacturer’s Origin software.
Structure Determination
Data collection was done at the European Synchrotron Radiation Facility and Swiss Light Source at 100K. Images were integrated with XDS (Kabsch, 2010) and merged/scaled with Aimless (Evans and Murshudov, 2013), followed by molecular replacement with Phaser (McCoy et al., 2007). Model refinement was carried out by Phenix (Adams et al., 2010), autoBUSTER (Smart et al., 2012), and Refmac (Murshudov et al., 1997), and models were built using COOT (Emsley et al., 2010). All structure figures were generated using PyMOL.
Author Contributions
D.D.S. designed, performed, and analyzed all experiments and wrote the manuscript. W.J.v.D. and D.D.S. expressed and purified proteins. F.E.O. and R.E. performed ubiquitin-conjugate synthesis. H.O. supervised ubiquitin-conjugate research. T.K.S. supervised and designed research and wrote manuscript. All authors critically read the manuscript.
Acknowledgments
We thank B8 members and Haico van Attikum for discussion and critical reading of the manuscript. We thank Marcello Clerici and Flora Groothuizen for advice in structure determination, Tatjana Heidebrecht for assistence in crystal harvesting, Vincent Blomen and Thijn Brummelkamp for the HAP1 cDNA library, Paul Geurink for Ub-LysGlyTAMRA, and beamline scientists at Swiss Light Source and European Synchrotron Radiation Facility for data collection assistance. This work was supported by European Research Council advanced grant 249997 and Netherlands Organization for Scientific Research ECHO 700.59.009. H.O. and F.E.O. are cofounders and shareholders of UbiQ.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Accession Numbers
The Protein Data Bank accession number for UCH-L5/RPN13DEU is 4UEM, for UCH-L5∼Ub/RPN13DEU is 4UEL, for UCH-L5/INO80GDEU is 4UF5, and for UCH-L5∼Ub/INO80Gshort is 4UF6.
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shami A., Jhaver K.G., Vogel P., Wilkins C., Humphries J., Davis J.J., Xu N., Potter D.G., Gerhardt B., Mullinax R. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS ONE. 2010;5:e13654. doi: 10.1371/journal.pone.0013654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreaux D.A., Maiti T.K., Davies C.W., Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc. Natl. Acad. Sci. USA. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie S.E., Bingman C.A., Soni A.B., Phillips G.N., Jr. Structural characterization of human Uch37. Proteins. 2012;80:649–654. doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M., Yang H., Pass H.I., Krausz T., Testa J.R., Gaudino G. BAP1 and cancer. Nat. Rev. Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lee B.-H., Finley D., Walters K.J. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Fu D., Xi J., Ji Z., Liu T., Ma Y., Zhao Y., Dong L., Wang Q., Shen X. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 2012;57:2310–2317. doi: 10.1007/s10620-012-2181-9. [DOI] [PubMed] [Google Scholar]
- Chia N.-Y., Chan Y.-S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.-S. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Clague M.J., Barsukov I., Coulson J.M., Liu H., Rigden D.J., Urbé S. Deubiquitylases from genes to organism. Physiol. Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D’Andrea A.D. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Dang L.C., Melandri F.D., Stein R.L. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- Dubin G. Proteinaceous cysteine protease inhibitors. Cell. Mol. Life Sci. 2005;62:653–669. doi: 10.1007/s00018-004-4445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkebus R., van Kasteren S.I., Kulathu Y., Scholten A., Berlin I., Geurink P.P., de Jong A., Goerdayal S., Neefjes J., Heck A.J.R. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 2013;135:2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oualid F., Merkx R., Ekkebus R., Hameed D.S., Smit J.J., de Jong A., Hilkmann H., Sixma T.K., Ovaa H. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew. Chem. Int. Ed. Engl. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen A.C., Dirac A.M.G., Shanmugham A., Ovaa H., Perrakis A., Sixma T.K. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Fang Y., Mu J., Ma Y., Ma D., Fu D., Shen X. The interaction between ubiquitin C-terminal hydrolase 37 and glucose-regulated protein 78 in hepatocellular carcinoma. Mol. Cell. Biochem. 2012;359:59–66. doi: 10.1007/s11010-011-0999-7. [DOI] [PubMed] [Google Scholar]
- Fang Y., Fu D., Tang W., Cai Y., Ma D., Wang H., Xue R., Liu T., Huang X., Dong L. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim. Biophys. Acta. 2013;1833:559–572. doi: 10.1016/j.bbamcr.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Geurink P.P., El Oualid F., Jonker A., Hameed D.S., Ovaa H. A general chemical ligation approach towards isopeptide-linked ubiquitin and ubiquitin-like assay reagents. ChemBioChem. 2012;13:293–297. doi: 10.1002/cbic.201100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Goldstein A.M. Germline BAP1 mutations and tumor susceptibility. Nat. Genet. 2011;43:925–926. doi: 10.1038/ng.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang O.W., Ma X., Yin J., Flinders J., Maurer T., Kayagaki N., Phung Q., Bosanac I., Arnott D., Dixit V.M. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat. Struct. Mol. Biol. 2012;19:171–175. doi: 10.1038/nsmb.2206. [DOI] [PubMed] [Google Scholar]
- Jiao L., Ouyang S., Shaw N., Song G., Feng Y., Niu F., Qiu W., Zhu H., Hung L.W., Zuo X. Mechanism of the Rpn13-induced activation of Uch37. Protein Cell. 2014;5:616–630. doi: 10.1007/s13238-014-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M., Ogishima S., Miyamoto T., Miyashita A., Kuwano R., Nakaya J., Tanaka H. Identification of unstable network modules reveals disease modules associated with the progression of Alzheimer’s disease. PLoS ONE. 2013;8:e76162. doi: 10.1371/journal.pone.0076162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Schneider M., Cabal G.G., Nehrbass U., Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Köhler A., Zimmerman E., Schneider M., Hurt E., Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Lee K.K., Florens L., Swanson S.K., Washburn M.P., Workman J.L. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Vargas M.P.A., Christodoulou E., Alfieri A., van Dijk W.J., Stadnik M., Hibbert R.G., Sahtoe D.D., Clerici M., Marco V.D., Littler D. Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J. Struct. Biol. 2011;175:113–119. doi: 10.1016/j.jsb.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Maiti T.K., Permaul M., Boudreaux D.A., Mahanic C., Mauney S., Das C. Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme. FEBS J. 2011;278:4917–4926. doi: 10.1111/j.1742-4658.2011.08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen O., Arpalahti L., Rantanen V., Hautaniemi S., Holmberg C.I. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013;3:1980–1995. doi: 10.1016/j.celrep.2013.05.012. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S., Ottosen S., Izrael-Tomasevic A., Arnott D., Lamkanfi M., Lee J., Liu J., O’Rourke K., Dixit V.M., Wilson A.C. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow M.E., Kim M.I., Ronau J.A., Sheedlo M.J., White R.R., Chaney J., Paul L.N., Lill M.A., Artavanis-Tsakonas K., Das C. Stabilization of an unusual salt bridge in ubiquitin by the extra C-terminal domain of the proteasome-associated deubiquitinase UCH37 as a mechanism of its exo specificity. Biochemistry. 2013;52:3564–3578. doi: 10.1021/bi4003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nishi R., Wijnhoven P., le Sage C., Tjeertes J., Galanty Y., Forment J.V., Clague M.J., Urbé S., Jackson S.P. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 2014;16:1016–1026. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M.W., Artavanis-Tsakonas K., Ploegh H.L. Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J. Biol. Chem. 2009;284:3593–3602. doi: 10.1074/jbc.M807172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.-B., Ouyang S.-Y., Li C.-J., Miao S., Wang L., Goldberg A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzychon M., Chmiel D., Stec-Niemczyk J. Modes of inhibition of cysteine proteases. Acta Biochim. Pol. 2004;51:861–873. [PubMed] [Google Scholar]
- Samara N.L., Datta A.B., Berndsen C.E., Zhang X., Yao T., Cohen R.E., Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Kong L., Ponting C.P. A common ancestry for BAP1 and Uch37 regulators. Bioinformatics. 2012;28:1953–1956. doi: 10.1093/bioinformatics/bts319. [DOI] [PubMed] [Google Scholar]
- Scheuermann J.C., de Ayala Alonso A.G., Oktaba K., Ly-Hartig N., McGinty R.K., Fraterman S., Wilm M., Muir T.W., Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Womack T.O., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G., van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- Sommer S., Weikart N.D., Linne U., Mootz H.D. Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorg. Med. Chem. 2013;21:2511–2517. doi: 10.1016/j.bmc.2013.02.039. [DOI] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamil M.A., Chen J., Liang Q., Zhuang Z. A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry. 2012;51:2829–2839. doi: 10.1021/bi3000512. [DOI] [PubMed] [Google Scholar]
- Wang L., Du Y., Ward J.M., Shimbo T., Lackford B., Zheng X., Miao Y.-L., Zhou B., Han L., Fargo D.C. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Harper J.W. Cancer. Emerging anatomy of the BAP1 tumor suppressor system. Science. 2012;337:1463–1464. doi: 10.1126/science.1228463. [DOI] [PubMed] [Google Scholar]
- Wicks S.J., Haros K., Maillard M., Song L., Cohen R.E., Dijke P.T., Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Wicks S.J., Grocott T., Haros K., Maillard M., ten Dijke P., Chantry A. Reversible ubiquitination regulates the Smad/TGF-beta signalling pathway. Biochem. Soc. Trans. 2006;34:761–763. doi: 10.1042/BST0340761. [DOI] [PubMed] [Google Scholar]
- Yao T., Song L., Xu W., DeMartino G.N., Florens L., Swanson S.K., Washburn M.P., Conaway R.C., Conaway J.W., Cohen R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S.K., Washburn M.P., Florens L., Conaway R.C., Cohen R.E., Conaway J.W. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.R., Zhang Y.H., Liu S., Song A.X., Hu H.Y. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.