Significance
Pentameric ligand-gated ion channels (pLGICs) mediate neuronal communication in the central nervous system. Upon the neurotransmitter binding, these receptors undergo a rapid conformational change to open an integral ion channel. Mutations impairing the function of pLGICs are known to cause hyperekplexic, myasthenic, and epileptic syndromes. Here, we studied how the local perturbations caused by single mutations result in an alteration of the protein function. Using a chimeric protein assembled by the transmembrane domain of the human glycine receptors fused to the extracellular domain of the bacterial pLGIC GLIC, we performed functional experiments in parallel with X-ray crystallography. On this basis, we propose a molecular mechanism for channel opening that accounts for the phenotypes of several mutants causing hyperekplexia.
Keywords: glycine receptor, hyperekplexia, allostery
Abstract
The glycine receptor (GlyR) is a pentameric ligand-gated ion channel (pLGIC) mediating inhibitory transmission in the nervous system. Its transmembrane domain (TMD) is the target of allosteric modulators such as general anesthetics and ethanol and is a major locus for hyperekplexic congenital mutations altering the allosteric transitions of activation or desensitization. We previously showed that the TMD of the human α1GlyR could be fused to the extracellular domain of GLIC, a bacterial pLGIC, to form a functional chimera called Lily. Here, we overexpress Lily in Schneider 2 insect cells and solve its structure by X-ray crystallography at 3.5 Å resolution. The TMD of the α1GlyR adopts a closed-channel conformation involving a single ring of hydrophobic residues at the center of the pore. Electrophysiological recordings show that the phenotypes of key allosteric mutations of the α1GlyR, scattered all along the pore, are qualitatively preserved in this chimera, including those that confer decreased sensitivity to agonists, constitutive activity, decreased activation kinetics, or increased desensitization kinetics. Combined structural and functional data indicate a pore-opening mechanism for the α1GlyR, suggesting a structural explanation for the effect of some key hyperekplexic allosteric mutations. The first X-ray structure of the TMD of the α1GlyR solved here using GLIC as a scaffold paves the way for mechanistic investigation and design of allosteric modulators of a human receptor.
The glycine receptor (GlyR) belongs to the superfamily of pentameric ligand-gated ion channels (pLGICs), which plays a key role in neuronal communication (1). In humans, this superfamily includes nicotinic acetylcholine (nACh), serotonin (5HT3), and γ-aminobutryic acid (GABAA) receptors. pLGICs are pentamers where each subunit is composed of an extracellular domain (ECD), a transmembrane domain (TMD) made of four helices M1–M4, and an intracellular domain (ICD). Neurotransmitter binding within the ECD triggers receptor activation, resulting in channel opening in the TMD, followed by desensitization (1). These allosteric transitions are at the heart of pLGIC function under normal conditions.
In addition, point mutations altering the conductance of the channel, or more frequently the isomerization constants and/or kinetics between the allosteric states, cause congenital hyperekplexic, myasthenic, and epileptic syndromes (2). Hyperekplexia is a rare inherited neurological disorder characterized by noise- or touch-induced nonepileptic seizures and excessive muscle stiffness. It is mainly caused by mutations in the α1GlyR subunit, which forms functional homomeric receptors as well as heteromeric receptors in association with the βGlyR subunit (3). Although numerous hyperekplexic mutations have been extensively studied in recombinant systems, the molecular basis of their effects has remained elusive.
The past decade has seen decisive progress in the determination of X-ray structures of pLGICs (1). The acetylcholine binding proteins (AChBPs) homologous to the pLGIC-ECDs were first solved (4), followed by two bacterial homologs called ELIC (5) and GLIC (6). GLIC is activated by protons, and its crystallization under conditions favoring the resting (GLIC-pH7) (7) or the active (GLIC-pH4) (6, 8, 9) states revealed its gating mechanism. The ECDs of subunits that are loosely packed at pH 7 come closer according to a “blooming” motion following acidification, in concert with a channel opening that involves a major tilt of the M2 channel-lining helix. Crystallization of several GLIC mutants revealed a locally closed (LC) “GLIC-LC” state resembling the open form but where the M2 helices are arranged like in the GLIC-pH7 form (7, 10, 11). Recently, three structures of eukaryotic pLGICs have been solved: the GluCl receptor from Caenorhabditis elegans (12, 13), the human β3GABAAR (14), and the rat 5HT3R (15), revealing a high conservation of the core structure of pLGICs from bacteria to animals. Interestingly, the GluCl receptor was solved in a closed (13) and an open (12) conformation. Comparison of the two forms shows a reorganization of the ECD resembling that of GLIC, but at the TMD channel opening results from a global tilt of each subunit four-helix bundle that contrasts with the local tilt of M2 found in GLIC.
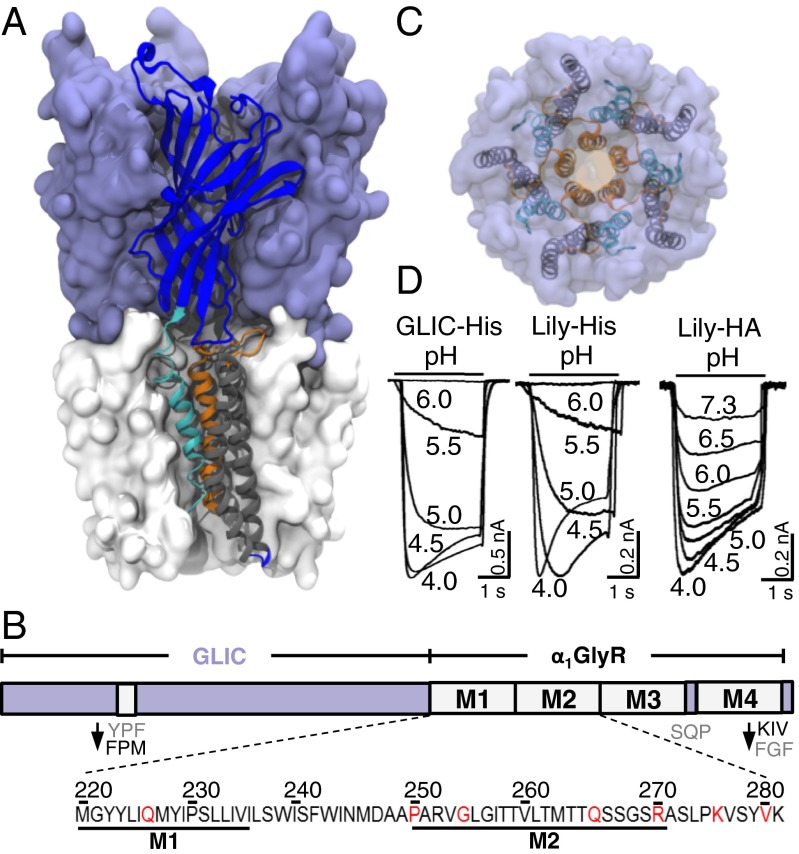
To develop a versatile method for collecting high-resolution structural data of human pLGICs, we used GLIC as a scaffolding protein to host TMDs from other receptors and solve their structure. Indeed, GLIC crystal packings involve mainly interactions between the ECD and the short cytoplasmic M3–M4 loop, but only limited interaction with the TMD, largely shielded from the solvent by the detergent. In previous work, we designed a chimera composed of the ECD of GLIC fused to the TMD of the human α1GlyR, where the ICD of the α1GlyR was replaced by the short linker (SQP motif) found in GLIC (16). In this chimera, mutations Y119F and F121M were introduced (loop 7) and the C-terminal tail of the α1GlyR was substituted for that of GLIC to increase the structural complementarities between the ECD and the TMD. This GLIC-GlyR construct, here called Lily (Fig. 1 A–D), functions as a proton-gated ion channel and displays a transmembrane pharmacology and ion channel properties closely resembling those of the α1GlyR.
Fig. 1.
Overall view of the Lily X-ray structure. (A) Side view of Lily. One chain is represented in a cartoon with the ECD colored in blue, M1 in cyan, M2 and loop M2–M3 in orange, and M3 and M4 in gray. (B) Scheme of the Lily construct with local modifications indicated. The sequence shows the M1 and M2 region, with the mutated residues in red. (C) Upper view of Lily with the same color code. (D) Typical pH-elicited current traces of Lily-His and GLIC-His expressed in S2 cells and Lily-HA expressed in BHK cells. In this and all subsequent figures, horizontal bars indicate the duration of the proton applications with concentrations given as pH units.
Here, we solved the structure of Lily, allowing us to reinvestigate the phenotype of several α1GlyR allosteric mutants and to propose a gating mechanism.
Results
Overexpression of Lily-His in a Functional State.
In contrast to GLIC, we found that Lily does not overexpress in Escherichia coli. We thus expressed Lily in Drosophilia Schneider 2 (S2) cells with a Gly-Gly-(His)10 tag at its C terminus (Fig. 1 and Fig. S1A). We checked that S2 cells expressing both Lily-His and GLIC-His yielded robust proton-elicited currents (Fig. 1D), characterized by EC50s for proton activation of 4.1 ± 0.3 × 10−6 M (pH 5.3) and 4.8 ± 0.6 × 10−6 M (pH 5.3), respectively. At pH 5, application of the channel blocker picrotoxinin (100 μM) yielded a 91 ± 7% inhibition of the Lily-His currents showing the specificity of the response. For the mutational studies shown below, we recorded Lily and GLIC in BHK cells with a C-terminal HA tag (16) that does not alter the electrophysiological response (17), yielding EC50s of 3.3 ± 0.1 × 10−7 M (pH 6.5) and 4.8 ± 0.6 × 10−6 M (pH 5.3), respectively.
X-ray Structure of Lily-His in the Locally Closed Conformation.
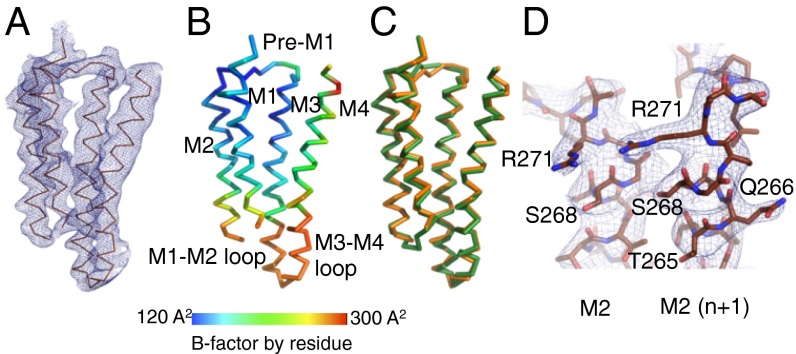
Lily-His was purified in dodecylmaltoside and subjected to extensive crystallization trials. The best crystals were obtained at pH 3 (similar crystals were obtained at pH 4, albeit diffracting to lower resolution). Lily-His was solved at 3.5 Å with I/σ of 1.2 in the highest-resolution shell (Table S1) in a P212121-type crystal packing (Fig. S1 B and C). Noncrystallographic symmetry (NCS) averaging improved the quality of the electron density, allowing the unambiguous reconstruction of the peptide backbone except for the first four N-terminal residues, the short M1–M2 loop (residues 218–222), and the C-terminal histidine tag (Fig. 2 A, B, and D). The quality of the electron density at the bottom of the TMD and at the top of the M3 and M4 helices is lower. At these levels several side chains, representing one-third of the TMD residues, could not be built unambiguously and were omitted in the model (Fig. S2 A and B).
Fig. 2.
Structure of the TMD of a Lily subunit. (A) Cα trace of a TMD monomer of Lily. The blue mesh is the 2mFo-DFc NCS-averaged electron density map contoured at a level of 1.5σ. (B) Same representation as in A colored by residue B factor according to scale shown. (C) Superimposition of the Cα trace of a TMD monomer of Lily (orange) and of GLIC-LC (green). (D) Enlarged representation of the tip of adjacent M2 helices of Lily with protein atoms in sticks. The blue mesh is the 2mFo-DFc NCS-averaged electron density map as defined in A.
Lily-His adopts the same locally closed conformation as GLIC-His, with a Cα rmsd between the two structures of 0.68 Å (0.43 Å in the ECD and 0.85 Å in the TMD, Fig. 2C) indicating a quasi-identical backbone conformation of the two proteins, notably of the four transmembrane helices and of the M2–M3 loop. Concerning GLIC-His, solved in the same crystal packing at a similar resolution (3.35 Å) (7), most of the side chains could be modeled from the electron density map. This indicates that, in this crystal, the TMD of the α1GlyR is more flexible than the TMD of GLIC, a feature supported by the high B factor of the TMD of Lily compared with that of its ECD (200 ± 4 Å2 vs. 160 ± 12 Å2). The lower thermostability of Lily-His in detergent solution (42 °C) compared with GLIC-His (52 °C) (Fig. S2C) is consistent with these observations.
Ion channel structure.
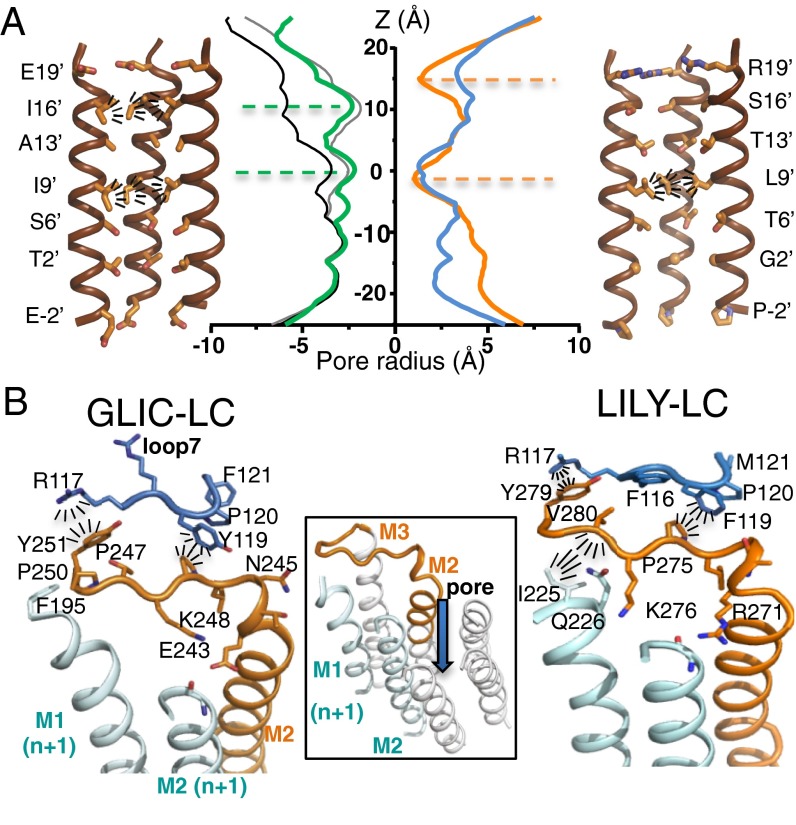
The ion channel is closed in the Lily structure (Fig. 3A): The ring of L261(9′) residues forms a tight hydrophobic constriction of 2.2 Å diameter constituting the gate of the channel (the main barrier to ion translocation). Above, rings of T264(13′) and S268(16′) form a wider hydrophilic pore (6.1 Å and 7.1 Å diameter, respectively). A similar architecture is observed in the closed structure of the GluCl-Apo receptor, in contrast to bacterial cationic pLGICs that display a larger diameter at L261(9′) but carry an extended hydrophobic gate at positions 13′ and 16′ (see Fig. S3A for GluCl-Apo, GLIC-pH7, and ELIC structures). As no second hydrophobic barrier exists, the highly constricted barrier at the L261(9′) level might be necessary to seal the channel and prevent any ion conduction. These data suggest a similar gate in the GlyR and GluCl receptors that are closely related in terms of sequence and ion channel properties. Finally, the upper turn of the M2 helix is partially broken in Lily, carrying the R271(19′) residue that forms a polar/positively charged constriction of 2.8 Å diameter. Below the L261(9′) position, the B factor progressively increases when going down through T258(6′), G254(2′), and P250(−2′), to reach the short disordered M1–M2 loop.
Fig. 3.
Ion channel architecture and interaction networks in Lily and GLIC-LC. (A) Cartoon representation of the pore lined by M2 helices for GLIC-LC (Left) and Lily (Right). The two front helices were removed. Residues lining the pore are shown as sticks. Radiating dashes show side-chain hydrophobic interactions that plug the pore. Center shows the pore radius along the z axis, computed on the full atom model of GLIC-LC (green), GLIC-pH4 (black), GLIC-pH7 (gray), Lily (orange), and GluCl-Apo (blue). (B) Cartoon representation of the interaction network around the M2–M3 loop in GLIC-LC (Left) and Lily (Right) X-ray structures. M2 and the M2–M3 loop of one subunit are colored orange, and loop 7 from the same subunit is colored blue. M1 and M2 helices from the adjacent subunit (n+1) are colored cyan. Radiating dashes represent interactions that are discussed in the text. Center shows the orientation of the view by the addition of the three remaining M2 helices that form the pore.
Interhelix interactions.
The structure reveals a set of side-chain interactions that are specific to the α1GlyR (Fig. 3B). At the tip of M2, the carboxylate of E243(GLIC) and the guanidinium moiety of R271(Lily) point toward the adjacent M2 helix and elicit quaternary hydrogen bonding with the main-chain carbonyl of the 17′ residue. The K248(GLIC)/K276(Lily) (24′) side chain points toward E243(19′) and is well resolved in GLIC, whereas it is resolved in only one of five chains in Lily and points in the opposite direction. This suggests electrostatic attraction/repulsion between K248(GLIC)/K276(Lily)(24′) and E243(GLIC)/R271(Lily)(19′). The tip of M1 interacts with the C terminus of the M2–M3 loop in Lily through hydrophobic interactions between I225 and V280 whereas no such interaction occurs in GLIC.
ECD/TMD coupling interface.
The coupling region essentially consists of intrasubunit interactions between the lower part of the ECD (loop 2, loop 7, and pre-M1) and the upper part of the TMD (M2–M3 loop). Ten of 15 interfacial residues are conserved between GLIC-LC and Lily (Fig. S3B), resulting in the conservation of key interactions, notably hydrophobic stacking of P247(GLIC)/P275(Lily) with F116 and Y(GLIC)/F(Lily)119 (Fig. 3B). In both structures, the side chain of L246(GLIC)/L274(Lily) points toward hydrophobic residues from the TMD. In the M2–M3 loop, the phenol moiety of Y251(GLIC)/Y279(Lily) is stacked with the guanidinium moiety of R117. This overlapping set of interactions at the ECD/TMD interface provides a structural explanation for the compatibility of the GLIC-ECD and the α1GlyR-TMD to form a functional channel.
Nonconserved GLIC/Lily residues are T253/K281, Y254/A282, Y119/F119, F121/M121, and Y194/M220 (Fig. S3B). They do not impair the allosteric coupling, but certainly modulate the functional properties. For instance, we showed that performing the F119Y and M121F mutations on Lily results in an acceleration of the desensitization kinetics (16). Extensive mutagenesis work has shown that the entire interface is involved in fine tuning of the allosteric response to agonist (18, 19).
Electrophysiological Analysis of Allosteric Mutants in Lily.
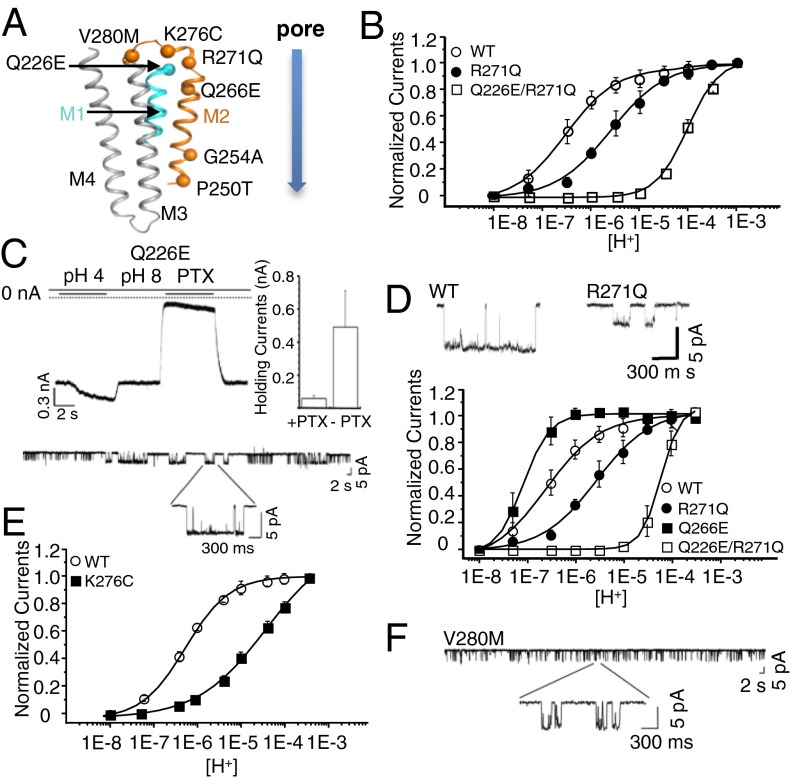
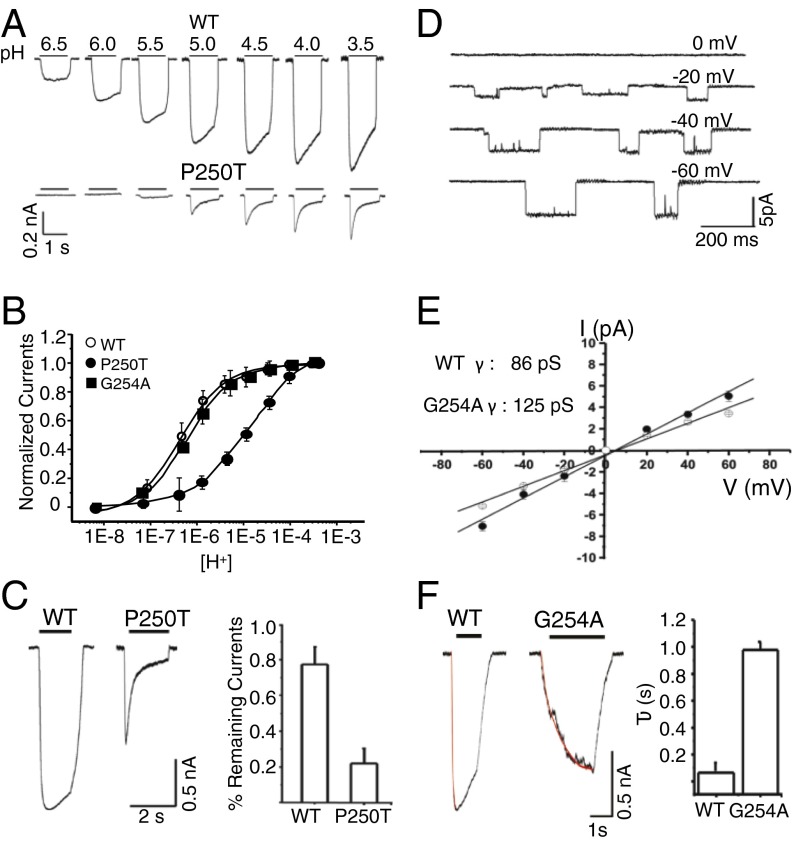
We reinvestigated the effect of well-characterized α1GlyR mutations by patch-clamp electrophysiology on Lily-HA and α1GlyR in BHK cells. We selected seven mutants, localized in various regions of the TMD: upper part of M1 behind the gate (Q226E), lower part of the channel (P250T and G254A), upper part of the channel (Q266E and R271Q), and M2–M3 loop (K276C and V280M) (Fig. 4A). Except for Q266E and G254A, all mutations are associated with autosomal dominant hyperekplexia.
Fig. 4.
Phenotypes of the Q226E, Q266E, R271Q, K276C, and V280M mutants on Lily. (A) Cartoon representation of a Lily subunit with the Cα of mutated residues shown as spheres. (B) Normalized dose–response curves in the whole-cell configuration. Lower traces show single-channel recording (−60 mV and pH 6.0) of outside-out patches for WT and R271Q. (C) Sample trace for Q226E mutant showing the leak current observed at pH 8.0, the proton-elicited current, and the inhibition of the leak current by 100 μM picrotoxin (PTX) (quantified in the bar graph where error bars represent ±SD). Lower panel shows single-channel recording (−60 mV and pH 8.0) of outside-out patches of Q226E. (D) Dose–response curves recorded in the whole-cell configuration. (E) Dose–response curves recorded in the whole-cell configuration. (F) Single-channel recording (−60 mV and pH 8.0) of outside-out patches for the V280M mutant.
On Lily, R271Q produces a 10-fold increase in EC50 for protons with a 1.5-fold decrease in maximal currents and a reduction in unitary conductance (from 86 ± 2 pS in WT to 56 ± 3 pS) (Fig. 4B and Fig. S4E). On the α1GlyR, we observe a more marked 450-fold increase in EC50 for glycine (Fig. S4 A–C), in agreement with previous works that also report a reduced single-channel conductance (20, 21). The phenotype of R271Q is thus similar in Lily and α1GlyR, although a stronger effect on the agonist EC50 is found for α1GlyR. Interestingly, the R271 residue in Lily appears close enough to elicit long-range electrostatic interaction with the K33 residue from the ECD, whereas a Thr is found at this position in the α1GlyR. The different side-chain environment may contribute to the difference in phenotypes between both receptor types.
Recently, Q226E was reported to produce spontaneously open channels (22, 23). On Lily, this mutation equally produces constitutive opening: (i) Immediately after the whole-cell clamping at pH 8.0, cells expressing Q226E generate high-leak currents (∼600 pA) that are blocked by 100 μM of picrotoxin; application of a more acidic solution further increases the currents, but the bad shape of the cells precluded repetitive stimulations and measurement of dose–response curves; and (ii) single-channel recording at pH 8.0 shows repetitive spontaneous openings (Fig. 4C), whereas no spontaneous activity is observed for Lily-WT.
On the α1GlyR, mutant cycle analysis suggests that Q226E stabilizes the active state through enhanced electrostatic attraction to R271 (22). We investigated this possibility on Lily. We observe that Q226E/R271Q no longer displays spontaneous currents, but a 570-fold increase in the EC50 for protons compared with WT (Fig. 4B and Fig. S4F). It is not possible to measure the EC50 of Q226E, but assuming a value around pH 8 as judged from single-channel traces, mutant cycle analysis yields strong energetic coupling between Q226E and R271Q (15 kJ⋅mol−1). This suggests that both positions interact through electrostatic forces to stabilize the open conformation. On the α1GlyR, R271Q and Q226E/R271Q both produce an increase in EC50, but the extent of their effects is significantly different from that found in Lily, pointing this time to a near-additive effect of R271Q and Q226E (Figs. S4G and S5). We also introduced a glutamate at a different position near R271. On Lily, Q266E produces a 3.3-fold increase in sensitivity to protons (Fig. 4D). Combining Q266E with R271Q results in a 230-fold decrease in proton sensitivity, again indicating energetic coupling between the two positions (10 kJ⋅mol−1). However, performing the same set of mutations on the α1GlyR also points to an additive effect due to the different R271Q phenotype (Fig. S5 D–F). The quantitative differences between the phenotypes of the R271Q and Q226E/R271Q in Lily and the α1GlyR may tentatively be accounted by the fact that, for Lily, proton activation may involve the titration of transmembrane residues interacting with the E226/R271 cluster. Indeed, recording of the α1GlyR R271Q and Q226E/R271Q mutants at pH 5 indicates a significant alteration of the dose–response curves (Fig. S5 A–C). In conclusion, the analysis of Lily shows coupling energy between Q226E/R271Q and to a lesser extent Q266E/R271Q, but these features are not unraveled on the α1GlyR possibly due to a different mode of activation.
In the M2–M3 loop, we found that K276C on Lily produces a 120-fold increase in EC50 and a large reduction of the maximal currents (Fig. 4E and Fig. S4 D and E). This phenotype is similar to what is found in the α1GlyR (24, 25). Likewise, V280M (26) was reported to produce a strong gain of function; we observed a phenotype similar to Q226E bearing spontaneous activity at pH 8 (Fig. 4F).
At the bottom of the channel, P250T confers reduced glycine sensitivity and increases the rate of desensitization on the α1GlyR (Table S2) (27). First, on Lily, P250T produces a very similar phenotype characterized by (i) a marked increase in desensitization kinetics, with current traces displaying a 20% vs. an 80% decay upon 1-s application of proton at pH 4 for WT and P250T, respectively (Fig. 5 A and C); (ii) a 46-fold increase in EC50 for protons (Fig. 5B); and (iii) a 5-fold reduction of the maximal currents. Second, the α2GlyR displays slow kinetics of activation and a large conductance in the 100- to 120-pS range (28). The M2 helices of α1 and α2 GlyRs are identical except at position 254 (Lily numbering) (α1Gly and α2Ala). We thus performed the G254A mutation on Lily (Fig. 5 D–F), showing a marked reduction of the rate of activation evaluated by following the onset of the whole-cell current traces (at pH 5.0 τact = 950 ± 62 ms vs. 64 ± 6.6 ms for G254A and WT, respectively). Moreover, the G254A mutant shows an increase in single-channel conductance (125 pS vs. 86 pS).
Fig. 5.
Phenotypes of the P250T and G254A mutants on Lily. (A) Sample trace for WT and P250T. (B) Normalized glycine dose–response relationships. (C) Typical traces (Left) and corresponding bar graph (Right) (error bars as ±SD) showing the percentage of current remaining after 1-s application of a pH 4 solution. (D and E) Single-channel amplitudes of G254A at different potentials (pH 6.0). (F) Sample traces of G254A. Kinetics of current onset were fitted with a monoexponential curve (bar graph, error bars are ±SD).
Overall, except for mutants R271Q and Q226E/R271Q, these data point to a similar phenotype when equivalent mutations are performed on Lily and on α1GlyR.
Discussion
A Modular Architecture for pLGICs.
The early observation that the α7nAChR(ECD)-5HT3R(TMD+ICD) chimera was functional was the first evidence that the ECD and TMD may constitute independent tertiary modules (26). The α7nAChR(ECD)-α1GlyR(TMD+ICD) (18), the AChBP-5HT3(TMD+ICD) (29), and recently the ELIC(ECD)-α7nAChR(TMD) chimera (30) were subsequently reported to be functional, the ECD retaining “native-like” pharmacology for agonists and the TMD native-like channel selectivity. It is noteworthy that extensive mutations at the ECD-TMD interface were required to produce properly folded and/or functional AChBP-5HT3(TMD+ICD) and ELIC(ECD)-α7nAChR(TMD) chimeras.
A conserved modular architecture of pLGICs from bacteria to humans is now established by all available X-ray structures. Interestingly, the two domains of Lily, when expressed alone, were previously found to self-assemble. First, we showed that the isolated ECD of GLIC folds as a soluble monomer and its X-ray structure confirmed it retains the usual β-sandwich fold (31). Second, an ensemble of 15 models of the α1GlyR TMD expressed alone and refolded in lipidic micelles has been solved by a combination of electron microscopy (EM) and NMR (32). The former shows in majority a pentameric assembly and the latter a bundle of four helices for each chain. However, these models show a marked difference from the Lily TMD structure, a feature possibly due to lack of structural constraints applied by the ECD (discussion in Fig. S6 A and B).
These data suggest a maturation pathway of Lily that would explain the striking compatibility between ECDs and TMDs from different pLGICs, even when separated by billions of years of evolution. The ECD and TMD of Lily would fold autonomously, followed by the assembly of the TMD into a pentamer, triggering subsequent pentamerization of the ECD. This scheme may be extended to other pLGIC subtypes because other domains were found to self-assemble, including the ECD of the α1nAChR (33) and the isolated TMDs of the α4β2 and α7 (34) nAChRs.
A Plausible Gating Mechanism of the TMD of the α1GlyR.
Our electrophysiological analysis showed that the TMD of Lily displays an ion channel indistinguishable from that of the α1GlyR in terms of conductance and selectivity and a pharmacology for general anesthetics, alcohols, and ivermectin similar to that of the α1GlyR (16). We show here that the phenotypes for key allosteric mutations, scattered at different locations within the TMD, are similar on the α1GlyR and Lily. In addition, those phenotypes involve all aspects of the activation transition, including (i) increase or decrease of the isomerization constant between resting and active states [causing decreased agonist sensitivity (R271Q and K276C) and constitutive opening (Q226E and V280M), respectively], (ii) alteration of activation kinetics (in G254A), and (iii) single-channel conductance (R271Q and G254A). This indicates that those residues experience similar changes in microenvironment during activation in both Lily and the α1GlyR. Concerning desensitization, our data also suggest a preserved mechanism due to the conservation of the phenotype of P250T. These data suggest a similar gating reorganization of the TMD in Lily and in the α1GlyR.
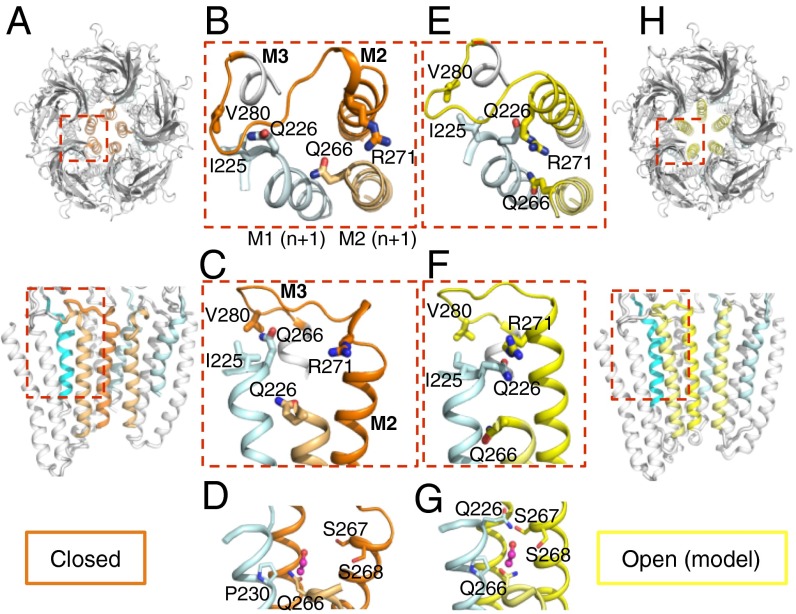
Here, Lily is expressed as a functional channel and crystallized in the LC conformation. For GLIC, the LC form corresponds globally to the open form, but a concerted bending of the upper part of the all M2 helices obstructs the pore by forming a tightly packed bundle, along with a revolving motion of the M2–M3 loop that is similar to what is observed in the pH 7 conformation. Therefore, the conformation of the TMD in the LC form is similar to that in the pH 7 resting form, with the exception of the M1 helix showing a 10° hinge motion in its upper half. We built a “completed X-ray structure” model of the TMD of Lily by adding the missing side chains and the backbone atoms of the five missing residues to the X-ray structure. We verified that a homology model of Lily based on GLIC-pH7 shows a TMD conformation (Fig. S6B) similar to that of the completed X-ray structure (Fig. 6 A–C).
Fig. 6.
Proposed gating mechanism of the TMD of the α1GlyR. (A) Cartoon representation of the X-ray closed structure of Lily in upper and side views. In the side view, the two front subunits were removed. The M2 and M2–M3 loop of a single subunit are colored orange, the two adjacent M2s are colored pale orange, and the neighboring M1 from the adjacent subunit is colored cyan. (B–D) Enlarged view of the structure of Lily with the same color code and selected side chains shown in sticks. (E–H) Same representation as in A–D of the open-pore homology model of Lily based on GLIC-pH4, with M2s and the M2–M3 loop colored yellow. (D and G) Closed and open models of Lily with an EtOH molecule in a pink sphere positioned as in the GLIC(F238A)-EtOH cocrystal structure, which overlaps with the EtOH potentiating binding site of the α1GlyR. Note that in the open model EtOH would elicit multiple stabilizing interactions with neighboring residues as shown by dashes, whereas no such interactions are observed in the closed form.
Concerning the open state, the available structural data of pLGICs show a similar TMD structure of GLIC and of the GluCl receptor. GLIC and the GluCl receptor are phylogenetically close to the α1GlyR (sharing respectively 34% and 26% amino acid identity at the TMD), supporting the idea that they are good models for the open form of the α1GlyR at the TMD. We therefore built a homology model of the open conformation of Lily based on GLIC-pH4. We chose GLIC-pH4 because its open channel structure was not constrained by ivermectin.
Assuming a common gating pathway between GLIC and the α1GlyR, we can use the completed X-ray structure and the homology open model as templates to investigate the activation transition of the α1GlyR TMD (Fig. 6). According to this “Lily” gating model, channel opening is caused by a tilt of the upper half of each M2 helix. In the process, M2 comes closer to M3 from the same subunit and to M1 from the adjacent subunit. This mechanism is consistent with a wealth of biochemical and mutational data collected on various pLGICs (7). In particular, it accounts for the phenotype of cross-linked double-cysteine mutants, which show that M2 comes closer to M3 during activation in both GLIC and the α1GlyR (10, 35).
Structural Interpretation of Allosteric Mutant Phenotypes.
For Lily, electrophysiological data point to an electrostatic interaction between R271 and E226 to stabilize the open state. In the open model of Lily, both residues are close enough to interact through a salt bridge, whereas they are far away in the closed form, suggesting that E226 stabilizes the open channel although “pulling” R271, at the tip of M2, away from the fivefold symmetry axis to interact with M1. On the α1GlyR, the Q226E and R271Q mutations do not reveal coupling energy between the two positions, but a series of other mutations support the above conclusion (22). Data were previously interpreted on the basis of a speculative gating model between ELIC (closed) and GLIC (open) (22, 23), where M2 and M3 tilt altogether as a rigid block. Here we provide a different gating model supported by a combination of electrophysiological, X-ray, and modeling data that mainly involve motion of M2 alone (Fig. 6 C and F and Fig. S7).
In addition, the V280 residue is an interesting reporter as it is located in the M2–M3 loop that undergoes a key revolving motion during gating. V280 interacts with I225 from the upper part M1, a helix portion directly linked to the ECD and involved in signal transduction. In the α1GlyR, the spontaneous currents of V280M were correlated with an increased volume of the side chain (22). In the gating model, the upper part of M1 moves away from M3 in the course of channel opening, resulting in an increased distance between V280 and I225 (a Cβ–Cβ distance increase from 5.7 Å to 6.6 Å). Bulkier residues at position 280 are thus expected to stabilize the open form, providing a structural explanation for the observed phenotypes (Fig. 6 B and E).
We further show a loss-of-function phenotype for the K276C mutation on Lily, whereas the homologous K248C mutation on GLIC has an opposite effect, yielding a 20-fold increase in proton sensitivity (10). This conserved Lys side chain points in different orientations in both structures (Fig. 3B). In GLIC, it projects toward the upper part of M2, possibly making a salt bridge with E243, whereas in Lily, it projects between helices M1 and M2 from the adjacent subunit, possibly because of electrostatic repulsion with R271. The markedly different orientations of this otherwise conserved residue provide a rationale for the opposite phenotypes observed by electrophysiology.
Conclusion
The receptor engineering approach based on functional chimeras presented here allowed us to solve the structure of the TMD of the α1GlyR and should be easily extended to other pLGICs. The Lily structure may help the drug design of allosteric effectors acting at the α1GlyR TMD. For instance, comparison of the open and closed models of Lily shows a reorganization of the intersubunit pocket that is lined by the S268 and A288 residues and mediates allosteric potentiation of the α1GlyR by general anesthetics and ethanol (36). The pocket markedly narrows down in the open-channel form, providing a plausible mechanism whereby it contracts around the effector to facilitate activation (Fig. 6 D and G). We also demonstrate that Lily is a pertinent model for studying the molecular mechanisms underlying allosteric mutations that may be ultimately beneficial for patients suffering hyperekplexia.
Materials and Methods
Lily was expressed and purified as in ref. 7. Crystals were obtained using vapor diffusion. The (6–8 mg/mL) protein was mixed in a 1:1 ratio with reservoir solution containing 16–20% (vol/vol) PEG 2000MME, 50 mM NiCl2, 4% (vol/vol) DMSO, 11% (vol/vol) ethylene glycol, and 0.1 M NaAcetate, pH 3.0. Coordinates and structure factors were deposited in the Protein Data Bank (PDB) with PDB ID 4X5T. Whole-cell and single-channel patch-clamp electrophysiology was performed as previously described (16). Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge financial support from the Fondation de la Recherche Médicale (G.M.-C.), the Direction des Applications de la Recherche et des Relations Industrielles (Institut Pasteur), the Agence Nationale de la Recherche (“Nicochimera”), the “Initiative d’Excellence” (Grant “Dynamics of Energy Transducing Membranes: Biogenesis and SupraMolecular Organization,” ANR-11-LABX-0011 to S.M., A.T., and M.B.), and the Fondation Pierre-Gilles de Gennes pour la Recherche (S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4X5T).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417864112/-/DCSupplemental.
References
- 1.Corringer PJ, et al. Structure and pharmacology of pentameric receptor channels: From bacteria to brain. Structure. 2012;20(6):941–956. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Galzi JL, Edelstein SJ, Changeux J. The multiple phenotypes of allosteric receptor mutants. Proc Natl Acad Sci USA. 1996;93(5):1853–1858. doi: 10.1073/pnas.93.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies JS, et al. The glycinergic system in human startle disease: A genetic screening approach. Front Mol Neurosci. 2010;3(3):8. doi: 10.3389/fnmol.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rucktooa P, Smit AB, Sixma TK. Insight in nAChR subtype selectivity from AChBP crystal structures. Biochem Pharmacol. 2009;78(7):777–787. doi: 10.1016/j.bcp.2009.06.098. [DOI] [PubMed] [Google Scholar]
- 5.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452(7185):375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 6.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457(7225):111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 7.Sauguet L, et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci USA. 2014;111(3):966–971. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauguet L, et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 2013;32(5):728–741. doi: 10.1038/emboj.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457(7225):115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 10.Prevost MS, et al. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol. 2012;19(6):642–649. doi: 10.1038/nsmb.2307. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2013;110(46):18716–18721. doi: 10.1073/pnas.1313156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512(7514):333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474(7349):54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512(7514):270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassaine G, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512(7514):276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- 16.Duret G, et al. Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc Natl Acad Sci USA. 2011;108(29):12143–12148. doi: 10.1073/pnas.1104494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bocquet N, et al. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007;445(7123):116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- 18.Grutter T, et al. Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc Natl Acad Sci USA. 2005;102(50):18207–18212. doi: 10.1073/pnas.0509024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smart TG, Paoletti P. Synaptic neurotransmitter-gated receptors. Cold Spring Harb Perspect Biol. 2012;4(3):a009662. doi: 10.1101/cshperspect.a009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajendra S, et al. Startle disease mutations reduce the agonist sensitivity of the human inhibitory glycine receptor. J Biol Chem. 1994;269(29):18739–18742. [PubMed] [Google Scholar]
- 21.Rajendra S, et al. Mutation of an arginine residue in the human glycine receptor transforms beta-alanine and taurine from agonists into competitive antagonists. Neuron. 1995;14(1):169–175. doi: 10.1016/0896-6273(95)90251-1. [DOI] [PubMed] [Google Scholar]
- 22.Bode A, Lynch JW. Analysis of hyperekplexia mutations identifies transmembrane domain rearrangements that mediate glycine receptor activation. J Biol Chem. 2013;288(47):33760–33771. doi: 10.1074/jbc.M113.513804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bode A, et al. New hyperekplexia mutations provide insight into glycine receptor assembly, trafficking, and activation mechanisms. J Biol Chem. 2013;288(47):33745–33759. doi: 10.1074/jbc.M113.509240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch JW, et al. Identification of intracellular and extracellular domains mediating signal transduction in the inhibitory glycine receptor chloride channel. EMBO J. 1997;16(1):110–120. doi: 10.1093/emboj/16.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lape R, Plested AJR, Moroni M, Colquhoun D, Sivilotti LG. The α1K276E startle disease mutation reveals multiple intermediate states in the gating of glycine receptors. J Neurosci. 2012;32(4):1336–1352. doi: 10.1523/JNEUROSCI.4346-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eiselé JL, et al. Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 1993;366(6454):479–483. doi: 10.1038/366479a0. [DOI] [PubMed] [Google Scholar]
- 27.Saul B, et al. Novel GLRA1 missense mutation (P250T) in dominant hyperekplexia defines an intracellular determinant of glycine receptor channel gating. J Neurosci. 1999;19(3):869–877. doi: 10.1523/JNEUROSCI.19-03-00869.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bormann J, Rundström N, Betz H, Langosch D. Residues within transmembrane segment M2 determine chloride conductance of glycine receptor homo- and hetero-oligomers. EMBO J. 1993;12(10):3729–3737. doi: 10.1002/j.1460-2075.1993.tb06050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouzat C, et al. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 2004;430(7002):896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- 30.Tillman TS, Seyoum E, Mowrey DD, Xu Y, Tang P. ELIC-α7 Nicotinic acetylcholine receptor (α7nAChR) chimeras reveal a prominent role of the extracellular-transmembrane domain interface in allosteric modulation. J Biol Chem. 2014;289(20):13851–13857. doi: 10.1074/jbc.M113.524611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nury H, et al. Crystal structure of the extracellular domain of a bacterial ligand-gated ion channel. J Mol Biol. 2010;395(5):1114–1127. doi: 10.1016/j.jmb.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Mowrey DD, et al. Open-channel structures of the human glycine receptor α1 full-length transmembrane domain. Structure. 2013;21(10):1897–1904. doi: 10.1016/j.str.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat Neurosci. 2007;10(8):953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 34.Bondarenko V, et al. NMR structures of the human α7 nAChR transmembrane domain and associated anesthetic binding sites. Biochim Biophys Acta. 2014;1838(5):1389–1395. doi: 10.1016/j.bbamem.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobo IA, Trudell JR, Harris RA. Cross-linking of glycine receptor transmembrane segments two and three alters coupling of ligand binding with channel opening. J Neurochem. 2004;90(4):962–969. doi: 10.1111/j.1471-4159.2004.02561.x. [DOI] [PubMed] [Google Scholar]
- 36.Mihic SJ, et al. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389(6649):385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.