Yolken et al. (1) claim detection of Acanthocystis turfacea chlorella virus 1 (ATCV-1, gi119953744) in the normal human oropharyngeal viral flora and associate it with altered cognitive function. However, the reported presence of a freshwater algae virus, previously not known to infect other species, was based on a few sequence reads homologous to ATCV-1 identified with BLASTn. These reads span relatively few bases (97–698 bp) per sample, dispersed over a minor fraction (0.03–0.24%) of the 288 kb ATCV-1 genome.
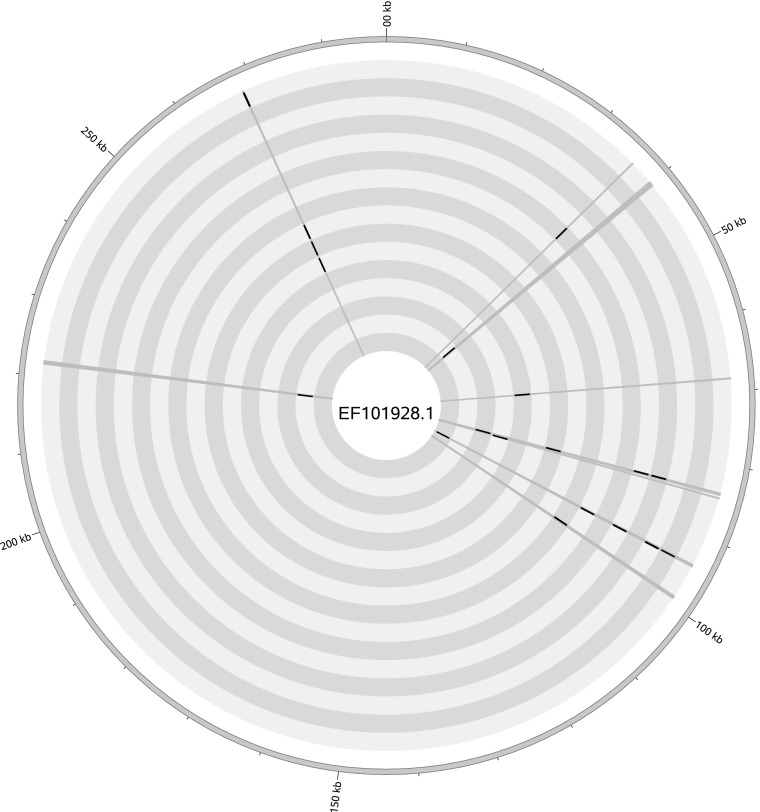
In an unrelated sequencing experiment involving 289 human specimens, we detected traces of ATCV-1 in 16 samples of 7 different types (including skin, colon, bone marrow, and breast) and a nontemplate control. The coverage (0.01–0.05%) and length of homologous regions (50–222 bp) are comparable to those obtained by Yolken et al. (1). In the absence of full coverage of the genome of the ATCV-1 virus, we hypothesize that the scattered presence of homology in so diverse sample types—including a negative control—derived from low-level laboratory contamination. The traces of ATCV-1 identified in our samples appear in close vicinity, suggesting they do not correspond to sporadic artifacts (Fig. 1).
Fig. 1.
Schematic view of ATCV-1-like reads identified in the 16 different samples mapping to the ATCV-1 genome (EF101928.1). The outer circle represents the linear double-stranded DNA genome of ATCV-1. The radial gray lines depict where, in the genome of ATCV-1, there is a matching read from any sample, and the black regions show the sample-specific positions of all BLASTn hits. The inner gray concentric circles serve as a visual guide for each sample.
Nucleic acid contamination from laboratory reagents and kits have previously been documented; for example, NIH-CQV/PHV in silica membranes (2), Legionella DNA in Qiagen DNA extraction kits (3), and murine DNA in extraction columns (4). Consequently, we correlated the presence of ATCV-1 in our samples with metadata from our laboratory procedures. Two laboratory components, Promega RQ1 RNase-Free DNase and Qiagen RNeasy MinElute cleanup, correlated significantly with the positive identification of ATCV-1 in a one-tailed Fisher’s exact test (P values 3.16 × 10−4 and 1.61 × 10−6, respectively) (Table 1).
Table 1.
Association between the detection of ATCV-1 in our sample treatments and metadata describing our laboratory procedures
| Laboratory procedure | P value | V+LP+ | V+LP− | V−LP+ | V−LP− |
| RNEasy MinElute, Qiagen | 1.61 × 10−6 | 13 | 3 | 92 | 324 |
| Promega RQ1 RNase-FREE DNase | 3.16 ×10−4 | 8 | 8 | 49 | 367 |
A one-tailed Fisher’s exact test was used to test significant association between the detection of ATCV-1 in our sample treatments and metadata describing our laboratory procedures. Significant associations are defined by a P value smaller than 0.05 and displayed. The confusion table is also shown. V+/V− designate presence/absence of the virus in the sample and LP+/LP− define whether a particular proceeding was used or not.
In several samples harboring traces of ATCV-1 reads, we concomitantly identified other algae viral species nonhomologous to ATCV-1. These findings support the presence of low-level algae viral sequence contamination.
To confirm their metagenomic findings, Yolken et al. (1) performed quantitative PCR screening. This type of analysis is prone to the same types of artifacts as the metagenomic sequencing, and the results reported by Yolken et al. could similarly originate from low-level contamination.
The discovery of novel viruses or their characterization in new hosts demands thorough investigation and validation beyond the identification of a few sequencing reads (5). Furthermore, the use of well-defined controls and critical awareness of the possible sources of contamination, either common or rare, introduced by the experimental reagents are a prerequisite. Based on our investigation of 289 human samples, we are confident that the presence of ATCV-1 sequences reported by Yolken et al. (1) originates from low-level laboratory contamination and, consequently, are rebutting the reported presence of ATCV-1 in human oropharynx.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yolken RH, et al. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc Natl Acad Sci USA. 2014;111(45):16106–16111. doi: 10.1073/pnas.1418895111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naccache SN, et al. The perils of pathogen discovery: Origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol. 2013;87(22):11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans GE, et al. Contamination of Qiagen DNA extraction kits with Legionella DNA. J Clin Microbiol. 2003;41(7):3452–3453. doi: 10.1128/JCM.41.7.3452-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erlwein O, et al. DNA extraction columns contaminated with murine sequences. PLoS ONE. 2011;6(8):e23484. doi: 10.1371/journal.pone.0023484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalton-Griffin L, Kellam P. Infectious causes of cancer and their detection. J Biol. 2009;8(7):67. doi: 10.1186/jbiol168. [DOI] [PMC free article] [PubMed] [Google Scholar]