Significance
Insulin resistance is a major risk factor for the development of diabetes. Associated increases in hepatic glucose production contribute to this process by promoting compensatory increases in insulin secretion that eventually lead to islet failure. This study characterizes the role of ATF3, a transcriptional repressor of the basic region-leucine zipper (bZIP) family, in mediating the hypoglycemic effects of acute alcohol consumption through the down-regulation of gluconeogenic genes. Based on the ability of ATF3 to displace CREB and its coactivator CRTC2 from CREB-binding sites on gluconeogenic genes, we imagine that small molecules with similar activity may provide therapeutic benefits to individuals with type II diabetes.
Keywords: gluconeogenesis, glucagon, CREB, ATF3, cAMP
Abstract
Increases in circulating glucagon during fasting maintain glucose balance by stimulating hepatic gluconeogenesis. Acute ethanol intoxication promotes fasting hypoglycemia through an increase in hepatic NADH, which inhibits hepatic gluconeogenesis by reducing the conversion of lactate to pyruvate. Here we show that acute ethanol exposure also lowers fasting blood glucose concentrations by inhibiting the CREB-mediated activation of the gluconeogenic program in response to glucagon. Ethanol exposure blocked the recruitment of CREB and its coactivator CRTC2 to gluconeogenic promoters by up-regulating ATF3, a transcriptional repressor that also binds to cAMP-responsive elements and thereby down-regulates gluconeogenic genes. Targeted disruption of ATF3 decreased the effects of ethanol in fasted mice and in cultured hepatocytes. These results illustrate how the induction of transcription factors with overlapping specificity can lead to cross-coupling between stress and hormone-sensitive pathways.
During fasting, pancreatic glucagon maintains circulating glucose concentrations by triggering the cAMP-mediated induction of the gluconeogenic program in liver (1). Increases in cAMP accumulation stimulate the PKA-mediated phosphorylation of CREB, which in turn up-regulates the expression of gluconeogenic genes (2). In parallel, cAMP signaling also promotes the dephosphorylation of CRTC2, a latent cytoplasmic coactivator that translocates to the nucleus and binds to CREB over relevant promoters (3).
Binge drinking can cause life-threatening hypoglycemia owing to the inhibition of hepatic glucose production. As a consequence of ethanol metabolism (4, 5), increases in hepatic NADH production interfere with the conversion of lactate to pyruvate, a major precursor for gluconeogenesis. Whether ethanol directly impacts the fasting-induced metabolic responses is still largely unknown. Recent studies suggest that ethanol also may alter cellular function via increases in stress signaling (6, 7). In the present study, we explored the effects of acute ethanol exposure on expression of the gluconeogenic program in response to glucagon. We found that ethanol inhibits the fasting adaptation in liver by up-regulating a transcriptional repressor that interferes with the CREB/CRTC2 pathway. These results illustrate how the parallel induction of transcription factors with overlapping specificity contributes to cross-coupling between stress and hormone-sensitive pathways.
Results
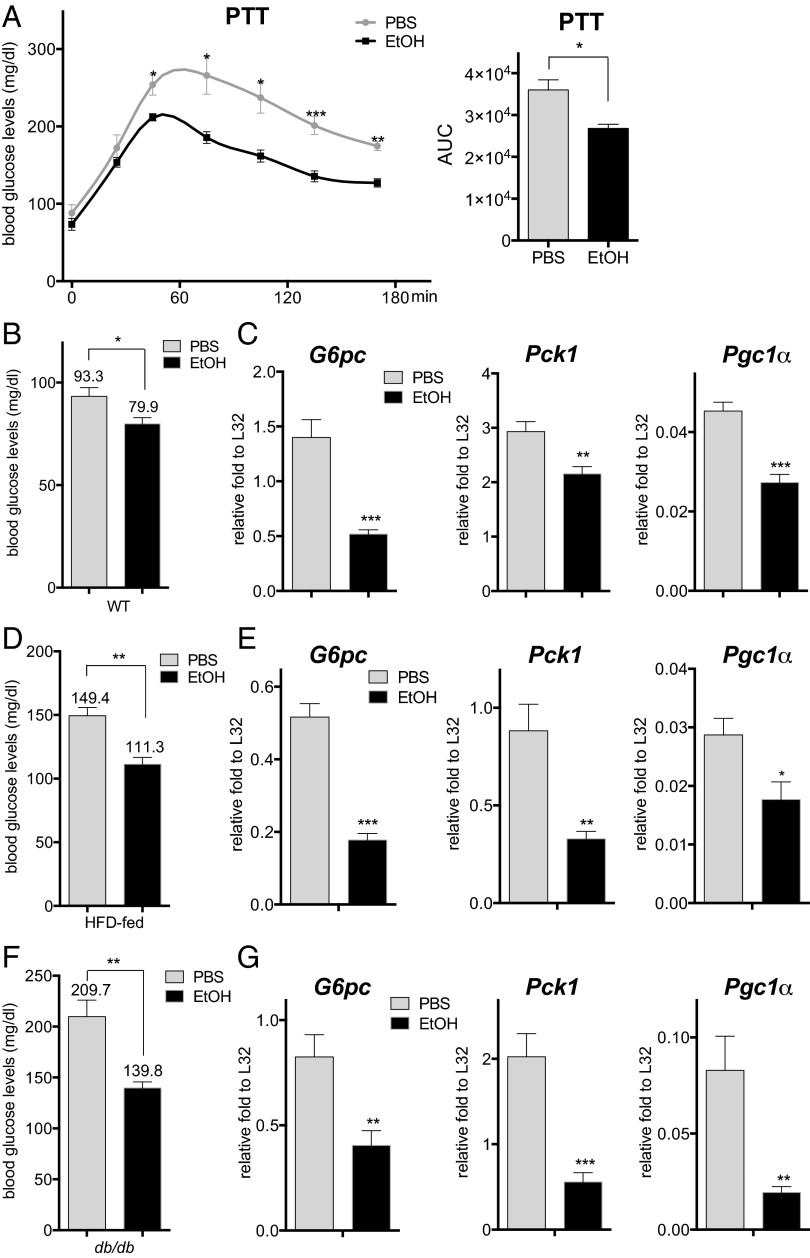
To determine the effects of acute ethanol consumption on the fasting-induced expression of gluconeogenic genes, we treated mice with 4.68 g/kg body weight ethanol and fasted them for 16 h (8, 9). In keeping with its glucose-lowering effects in fasted mice, ethanol administration inhibited hepatic gluconeogenesis as measured by the pyruvate tolerance test (PTT) (Fig. 1 A and B). mRNA amounts for the fasting-inducible gluconeogenic genes glucose-6-phosphatase (G6pc) and phosphoenolpyruvate carboxykinase (Pck1), as well as the gluconeogenic coactivator peroxisome proliferator-activated receptor gamma coactivator 1α (Pgc1α), were down-regulated in ethanol-treated mice (Fig. 1C), suggesting that ethanol may directly block the induction of gluconeogenic genes. Arguing against an effect on glucose sensing per se, circulating glucagon concentrations were comparable in the control and ethanol-treated animals (Fig. S1A).
Fig. 1.
Acute ethanol exposure inhibits hepatic gluconeogenesis. (A) PTT of fasted WT mice following treatment with PBS or ethanol (EtOH). AUC values were calculated from PTT assays. The mean BAL was 473 ± 19 mg/dL in the ethanol-treated mice at 30 min after the last ethanol dose. Each bar represents averaged results, n = 4. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (B) Fasting blood glucose concentrations in WT mice following PBS or ethanol treatment. The mean BAL was 498 ± 13 mg/dL. Each bar represents averaged results, n = 12. Error bars indicate SEM. *P < 0.05. (C) Analysis of mRNA amounts for gluconeogenic genes in livers of fasted WT mice following PBS or ethanol treatment. Each bar represents averaged results, n = 5. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Fasting blood glucose concentrations in HFD-fed mice after PBS or ethanol administration. The mean BAL was 452 ± 23 mg/dL. Each bar represents averaged results, n = 5. Error bars indicate SEM. **P < 0.01. (E) mRNA amounts for gluconeogenic genes in livers of fasted HFD-fed mice with PBS or ethanol administration. Each bar represents averaged results, n = 5. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (F) Fasting blood glucose concentrations in db/db mice following PBS or ethanol treatment. The mean BAL was 436 ± 14 mg/dL. Each bar represents averaged results, n = 7. Error bars indicate SEM. **P < 0.01. (G) mRNA amounts for gluconeogenic genes in livers of fasted db/db mice following PBS or ethanol treatment. Each bar represents averaged results, n = 7. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
In addition to effects of glucagon during fasting, a decrease in circulating insulin concentrations also promotes expression of the gluconeogenic program via the dephosphorylation and activation of the forkhead family protein FOXO1 (10). Ethanol administration had no effect on circulating insulin concentrations (Fig. S1B), AKT phosphorylation, or FOXO1 activation, however (Fig. S1 C and D). mRNA amounts for FOXO1 target genes, including insulin-like growth factor binding protein 1 (Igfbp1) and insulin receptor substrate 2 (Irs2), appeared comparable or somewhat elevated in ethanol-treated mice compared to control mice (Fig. S1E). Indeed, ethanol administration actually reduced fasting blood glucose concentrations and gluconeogenic gene expression in the context of dietary and genetic obesity, where increases in insulin resistance otherwise up-regulate hepatic glucose production (Fig. 1 D–G).
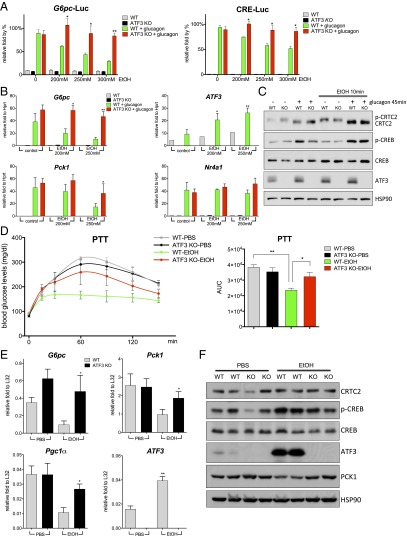
Having seen that ethanol administration has no effect on FOXO1 activity, we evaluated the potential involvement of the CREB/CRTC2 pathway in this process. Similar to its effects in mice, ethanol exposure also inhibited the induction of gluconeogenic genes (G6pc, Pck1, and Pgc1α) by glucagon in primary cultured hepatocytes (Fig. 2A and Fig. S2A). Remarkably, CREB phosphorylation in response to glucagon was unaffected by ethanol (Fig. 2B); CRTC2 dephosphorylation and nuclear translocation also proceeded comparably in control and ethanol-treated cells (Fig. 2 B and C). Despite these effects, however, ethanol treatment disrupted the induction of G6pc-Luc and CRE-Luc reporters in primary hepatocytes exposed to glucagon (Fig. 2D).
Fig. 2.
Ethanol disrupts recruitment of CREB and CRTC2 to gluconeogenic promoters. (A) Effect of a 10-min pretreatment with ethanol (EtOH) on gluconeogenic gene expression in mouse primary hepatocytes exposed to glucagon for 1.5 h. Each bar represents averaged results for three biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (B) Western blot showing the effect of a 10-min pretreatment with ethanol on phosphorylation of CREB and LKB and dephosphorylation of CRTC2 in primary hepatocytes exposed to glucagon for 45 min. CREB and HSP90 served as loading controls. (C) Immunofluorescence staining showing the effect of a 10-min pretreatment with ethanol (250 mM) on the nuclear translocation of CRTC2 in primary hepatocytes exposed to glucagon for 45 min. Red, CRTC2; green, actin; blue, DAPI. (D) Effect of a 10-min pretreatment with ethanol on G6pc-Luc and CRE-Luc reporter activities in primary hepatocytes exposed to glucagon for 5 h. Each bar represents averaged results for three biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (E) ChIP assays showing the effects of a 10-min pretreatment with ethanol (250 mM) on the recruitment of CREB, phospho-CREB, and CRTC2 to CREB-binding sites over gluconeogenic (G6pc, Pck1) promoters in primary hepatocytes exposed to glucagon for 1 h. The 36b4 ribosomal protein gene served as a negative control. Each bar represents averaged results for two biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Based on its ability to down-regulate cAMP-inducible promoters, we considered the possibility that ethanol may inhibit recruitment of CREB or CRTC2 to relevant binding sites. In chromatin immunoprecipitation (ChIP) assays of cultured primary hepatocytes, exposure to glucagon increased the amounts of phospho (Ser133)-CREB as well as CRTC2 over CREB-binding sites on gluconeogenic promoters (G6pc, Pck1); however, costimulation with ethanol reduced phospho-CREB and CRTC2 occupancy (Fig. 2E).
We performed gene profiling studies of primary hepatocytes to identify nuclear factors that disrupt CREB signaling in this setting. In keeping with its effects on the gluconeogenic program, exposure to ethanol inhibited 18% (31/169) of glucagon-inducible genes, several of which (Sik1, Pde4d, and Klf4) have functional CREB-binding sites (Fig. 3 A and B). Within the list of ethanol-inducible genes, we identified activating transcription factor 3 (ATF3) as a top hit (Fig. 3C). Indeed, cotreatment with ethanol increased ATF3 mRNA and protein amounts in primary hepatocytes exposed to glucagon, as well as in livers of fasted mice (Fig. 3 D and E). In keeping with the ability for p38 and JNK pathways to stimulate ATF3 expression (11, 12), exposure of primary hepatocytes to ethanol increased the phosphorylation and activation of both kinases (Fig. S2B). A member of the basic region-leucine zipper (bZIP) family of transcription factors, ATF3 has been shown to function as a transcriptional repressor (13). Based on its ability to recognize CREB-binding sites, we considered that ATF3 might compete with CREB for occupancy over gluconeogenic promoters. Supporting this idea, exposure to ethanol increased ATF3 recruitment to CREB-binding sites on the G6pc and Pck1 promoters in primary hepatocytes cotreated with glucagon (Fig. 3F).
Fig. 3.
Ethanol stimulates expression of ATF3. (A) Gene profile analysis of primary hepatocytes following exposure to ethanol (EtOH). Pie charts show the proportion of cellular genes up- or down-regulated by ≥1.5-fold by a 1-h pretreatment with ethanol under basal conditions and following exposure to glucagon for 1.5 h. (B) Heat map showing the effects of ethanol on the top 40 scoring glucagon-inducible genes in primary hepatocytes. (C) Heat map showing the top 40 scoring ethanol-inducible genes in primary hepatocytes. (D) (Left) Effects of a 10-min pretreatment with ethanol on ATF3 RNA expression in primary hepatocytes exposed to glucagon for 1.5 h. Each bar represents averaged results for three biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (Right) Western blot showing the effects of a 10-min pretreatment with ethanol on ATF3 protein amounts in primary hepatocytes exposed to glucagon for 1 h. CREB served as a loading control. (E) Western blot of hepatic ATF3 protein amounts in fasted WT mice treated with PBS or ethanol. HSP90 served as a loading control. (F) ChIP assays showing the effects of a 10-min pretreatment with ethanol (250 mM) on the recruitment of ATF3 to CREB-binding sites at G6pc and Pck1 promoters in primary hepatocytes exposed to glucagon for 1 h. Each bar represents averaged results for two biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
If ATF3 mediates inhibitory effects of ethanol, then targeted disruption of this gene should rescue CREB activity in this setting. In contrast to WT hepatocytes, ethanol had no inhibitory effect on G6pc-Luc and CRE-Luc reporter activities in ATF3 knockout (ATF3−/−) cells (Fig. 4A). Indeed, ATF3−/− hepatocytes also were resistant to the inhibitory effects of ethanol on glucagon-dependent increases in mRNA amounts for G6pc and Pck1 (Fig. 4 B and C).
Fig. 4.
ATF3 mediates the inhibitory effects of ethanol on hepatic gluconeogenesis. (A) Effects of a 10-min pretreatment with ethanol (EtOH) on G6pc-Luc and CRE-Luc reporter activities in primary hepatocytes isolated from WT or ATF3−/− mice in response to a 5-h exposure to glucagon. Each bar represents averaged results for three biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (B) Effects of a 10-min pretreatment with ethanol on gluconeogenic gene expression in primary hepatocytes, WT or ATF3−/−, in response to a 1.5-h exposure to glucagon. Each bar represents averaged results for three biological replicates, assayed three times each. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Western blot showing the effects of a 10-min pretreatment with ethanol on the phosphorylation of CREB, dephosphorylation of CRTC2, and ATF3 expression in primary hepatocytes, WT and ATF3−/−, exposed to glucagon for 45 min. CREB and HSP90 served as loading controls. (D) PTT assays performed on fasted WT or ATF3−/− mice treated with PBS or ethanol. AUC values were calculated from PTT assays. The mean BAL was 450 ± 15 mg/dL in WT mice and 447 ± 6 mg/dL in ATF3−/− mice. Each bar represents averaged results for three biological replicates. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (E) Analysis of mRNA amounts of gluconeogenic genes in livers of fasted WT and ATF3−/− mice treated with PBS or ethanol. The mean BAL was 406 ± 14 mg/dL in WT mice and 371 ± 18 mg/dL in ATF3−/− mice. Each bar represents averaged results for five biological replicates. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (F) Western blot showing the effects of ethanol on ATF3 and PCK1 protein amounts in livers of fasted WT and ATF3−/− mice treated with PBS or ethanol. HSP90 served as a loading control.
Having seen that targeted disruption of the ATF3 gene restores CREB activity in cultured hepatocytes exposed to ethanol, we tested the extent to which a loss of ATF3 rescues hepatic gluconeogenesis in fasted mice. In contrast to control animals, exposure to ethanol had only a modest effect on fasting blood glucose concentrations in ATF3−/− mice (Fig. S3). Moreover, hepatic gluconeogenesis, as measured by the PTT, remained elevated in fasted ATF3−/− mice exposed to ethanol (Fig. 4D). Indeed, ethanol had only modest effects on mRNA and protein amounts for gluconeogenic genes in ATF3−/− mice, demonstrating the importance of this factor in mediating the effects of ethanol on hepatic glucose production (Fig. 4 E and F).
Discussion
During fasting, increases in circulating glucagon up-regulate the gluconeogenic program by stimulating the CREB/CRTC2 pathway. Fasting-associated decreases in insulin signaling also trigger activation of FOXO1, which functions in parallel with CREB to modulate gluconeogenic gene expression (3, 10). Hepatic glucose production is elevated in diabetes, reflecting increases in insulin resistance and in circulating glucagon concentrations that constitutively activate both CREB and FOXO1 (1). Remarkably, acute ethanol administration lowers hepatic gluconeogenesis even in the context of insulin resistance.
We found that ethanol administration attenuates the gluconeogenic program by triggering the production of ATF3, a member of the bZIP family of transcription factors. Originally identified based on its ability to recognize a CREB-binding site, ATF3 has been shown to function as a transcriptional repressor in a variety of cell contexts (13, 14). Indeed, exposure to cAMP up-regulates ATF3 expression in a number of tissues, suggesting that this factor may function as a part of a negative feedback loop that inhibits target gene expression by displacing CREB from its binding sites. Although ethanol appears to up-regulate ATF3 through the induction of stress signaling (Fig. S2B) (7), exposure to cAMP also appears to be important for full induction of this repressor (Figs. 3D and 4B), pointing to cross-talk between stress and cAMP-signaling pathways.
ATF3 is thought to inhibit gene expression by binding as a homodimer; it also can modulate transcription as a heterodimer with members of the bZIP family, including c-Jun and JunD (15). Although our studies indicate that ATF3 is necessary for the inhibitory effects of ethanol on the gluconeogenic program, other bZIP proteins also may participate in this process by associating with ATF3. Following its induction, ATF3 was found to block expression of gluconeogenic genes, but other CREB target genes, such as NR4A1, remained unaffected (Fig. S2A and Fig. 4B). The apparent selectivity of ATF3 may reflect subtle differences in either CREB-binding sites or local chromatin structure that favor ATF3 recruitment to these promoters. Future studies of ATF3 complexes that bind to promoters for CREB target genes should provide greater insight into this process.
In addition to its effects in liver, ATF3 also has been shown to modulate the activity of pancreatic islet and white adipose tissues (16, 17). Islet morphology is disrupted in transgenic mice expressing ATF3 in β cells, owing in part to increases in apoptosis. ATF3 expression in white adipose tissues also may modulate circulating glucose concentrations by inhibiting the production and release of adiponectin. Future studies should reveal the extent to which these tissues also contribute to effects of ethanol on glucose homeostasis.
Materials and Methods
Mouse Strains, Ethanol Administration, and in Vivo Analyses.
The WT C57BL/6J, high-fat diet (HFD)-fed, and db/db mice were purchased from The Jackson Laboratory. The ATF3−/− FVB/N mice have been described previously (18). All mice were adapted to their environment for at least 1 wk before the study.
The mice were fasted for 3 h, after which ethanol was administered in four doses, with a 30-min interval between doses. The first ethanol dose was administered as an i.p. injection at 0.93 g/kg body weight, and the subsequent doses were administered by oral gavage at 1.25 g/kg body weight (8). Blood alcohol level (BAL) was measured at 30 min after the last ethanol dose (19).
The mice were then fasted for 16 h for blood glucose tests, and liver tissue was collected for RNA and protein analysis. The doses of ethanol given to the HFD-fed and db/db mice were reduced to 60% of those given to the WT mice. Ethanol was administered to the FVB/N mice through one i.p. injection at 0.93 g/kg body weight and three oral gavages at 0.75 g/kg body weight.
All animal procedures were performed following protocols approved by from The Salk Institute’s Animal Care and Use Committee.
PTT.
The PTT was performed as described previously (20). In brief, control and ethanol-treated mice were fasted for 16 h and then injected i.p. with sodium pyruvate (2 g/kg body weight). Glucose concentrations were measured at the indicated time points.
Cell Culture.
Mouse primary hepatocytes were isolated as described previously (21) and cultured in Medium 199. Hepatocytes were pretreated with ethanol at the indicated concentrations for 10 min and exposed to glucagon (100 nM).
RNA Analyses, Immunoblotting, and Immunofluorescent Staining.
Cellular RNA was isolated from mouse liver tissue or primary hepatocytes using the Qiagen RNeasy Kit. mRNA levels were measured and immunoblot and immunofluorescent staining assays performed as described previously (22).
ChIP Assay.
Cultured mouse primary hepatocytes were plated in 150-mm plates and pretreated with 250 mM ethanol for 10 min, then exposed to glucagon for 1 h. ChIP assays were performed as described previously, with minimal modification (23). In brief, the treated hepatocytes were cross-linked with 1% formaldehyde for 15 min. Cross-linking reactions were stopped with 0.125 M glycine. Cross-linked cells were washed in PBS three times and stored at −80 °C before use. Fragmented, precleared chromatin lysate was incubated overnight with indicated antibodies.
Luciferase Reporter Assay.
Cultured mouse primary hepatocytes were infected with Ad-G6pc-Luc or Ad-CRE-Luc for 48 h, pretreated with ethanol at indicated concentrations for 10 min, and then exposed to glucagon for 5 h. Luciferase assays were performed as described previously (24).
Gene Profiling Analyses.
Mouse primary hepatocytes were plated in 60-mm plates and pretreated with PBS or 250 mM ethanol for 1 h, and then exposed to glucagon for 1.5 h. RNA was isolated and analyzed using the Affymetrix GeneChip Mouse Gene 1.0 ST Array following the manufacturer’s instructions. The top 40 glucagon-induced and ethanol-induced genes were chosen for heatmap analysis using matrix2png software as described previously (25).
Statistical Analyses.
GraphPad Prism 6 software was used to analyze P values based on at least two independent experiments in three independent assays. The two-tailed unpaired t test was used to compare the differences between two groups. A P value < 0.05 was considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgments
We thank S. Bae for technical support. Funding for this work was provided by the NIH (Grants R01 DK049777, R01 DK083834, and R01 DK091618), the Clayton Foundation for Medical Research, the Leona M. and Harry B. Helmsley Charitable Trust, and the Kieckefer Foundation. W.-W.T. was supported in part by NIH Grant F32 DK096778.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424641112/-/DCSupplemental.
References
- 1.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 3.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber CS. Metabolism of alcohol. Clin Liver Dis. 2005;9(1):1–35. doi: 10.1016/j.cld.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Cederbaum AI. Alcohol metabolism. Clin Liver Dis. 2012;16(4):667–685. doi: 10.1016/j.cld.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy LE. Molecular aspects of alcohol metabolism: Transcription factors involved in early ethanol-induced liver injury. Annu Rev Nutr. 2004;24:55–78. doi: 10.1146/annurev.nutr.24.012003.132258. [DOI] [PubMed] [Google Scholar]
- 7.Shukla SD, Pruett SB, Szabo G, Arteel GE. Binge ethanol and liver: New molecular developments. Alcohol Clin Exp Res. 2013;37(4):550–557. doi: 10.1111/acer.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Wu D, Wang X, Cederbaum AI. Cytochrome P4502E1, oxidative stress, JNK, and autophagy in acute alcohol-induced fatty liver. Free Radic Biol Med. 2012;53(5):1170–1180. doi: 10.1016/j.freeradbiomed.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathews S, Xu M, Wang H, Bertola A, Gao B. Animals models of gastrointestinal and liver diseases. Animal models of alcohol-induced liver disease: Pathophysiology, translational relevance, and challenges. Am J Physiol Gastrointest Liver Physiol. 2014;306(10):G819–G823. doi: 10.1152/ajpgi.00041.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin HV, Accili D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 2011;14(1):9–19. doi: 10.1016/j.cmet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandnes D, et al. Induction of LRF-1/ATF3 by vasopressin in hepatocytes: Role of MAP kinases. Cell Physiol Biochem. 2010;25(4-5):523–532. doi: 10.1159/000303056. [DOI] [PubMed] [Google Scholar]
- 12.Lu D, Chen J, Hai T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem J. 2007;401(2):559–567. doi: 10.1042/BJ20061081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: Is modulation of inflammation a unifying component? Gene Expr. 2010;15(1):1–11. doi: 10.3727/105221610x12819686555015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7(4-6):321–335. [PMC free article] [PubMed] [Google Scholar]
- 15.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759(1-2):4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Qi L, et al. Adipocyte CREB promotes insulin resistance in obesity. Cell Metab. 2009;9(3):277–286. doi: 10.1016/j.cmet.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman MG, et al. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol Cell Biol. 2004;24(13):5721–5732. doi: 10.1128/MCB.24.13.5721-5732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolford CC, et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J Clin Invest. 2013;123(7):2893–2906. doi: 10.1172/JCI64410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, et al. Prolonged exposure to intermittent alcohol vapors blunts hypothalamic responsiveness to immune and non-immune signals. Alcohol Clin Exp Res. 2000;24(1):110–122. [PubMed] [Google Scholar]
- 20.Wang Y, et al. Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature. 2012;485(7396):128–132. doi: 10.1038/nature10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo SH, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437(7062):1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 22.Ravnskjaer K, et al. Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. J Clin Invest. 2013;123(10):4318–4328. doi: 10.1172/JCI69035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai WW, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van de Velde S, Hogan MF, Montminy M. mTOR links incretin signaling to HIF induction in pancreatic beta cells. Proc Natl Acad Sci USA. 2011;108(41):16876–16882. doi: 10.1073/pnas.1114228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai WW, et al. PRMT5 modulates the metabolic response to fasting signals. Proc Natl Acad Sci USA. 2013;110(22):8870–8875. doi: 10.1073/pnas.1304602110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.