Significance
Inflammatory bowel diseases (IBD), including Crohn’s disease and ulcerative colitis, are intestinal disorders of poorly understood origin and are associated with significant morbidity and mortality. A crucial factor associated with IBD onset is the presence of elevated levels of the proinflammatory cytokine tumor necrosis factor (TNF) in the intestine, signified by the use of anti-TNF therapy to treat patients with Crohn’s disease. Despite its pathogenic relevance, the mechanisms regulating TNF expression and IBD onset remain largely unknown. Here, we show that loss of epigenetic regulation results in the induction of TNF in the intestinal epithelium, leading to a loss of intestinal barrier function and inflammation. Our results suggest that mutations in genes controlling epigenetic regulators can lead to IBD onset.
Keywords: inflammation, Uhrf1, DNA methylation, tumor necrosis factor, zebrafish
Abstract
The intestinal epithelium forms a barrier protecting the organism from microbes and other proinflammatory stimuli. The integrity of this barrier and the proper response to infection requires precise regulation of powerful immune homing signals such as tumor necrosis factor (TNF). Dysregulation of TNF leads to inflammatory bowel diseases (IBD), but the mechanism controlling the expression of this potent cytokine and the events that trigger the onset of chronic inflammation are unknown. Here, we show that loss of function of the epigenetic regulator ubiquitin-like protein containing PHD and RING finger domains 1 (uhrf1) in zebrafish leads to a reduction in tnfa promoter methylation and the induction of tnfa expression in intestinal epithelial cells (IECs). The increase in IEC tnfa levels is microbe-dependent and results in IEC shedding and apoptosis, immune cell recruitment, and barrier dysfunction, consistent with chronic inflammation. Importantly, tnfa knockdown in uhrf1 mutants restores IEC morphology, reduces cell shedding, and improves barrier function. We propose that loss of epigenetic repression and TNF induction in the intestinal epithelium can lead to IBD onset.
Intestinal epithelial cells (IECs) function as a barrier to prevent luminal contents from accessing underlying tissues, and loss of barrier function is a crucial factor leading to the development of inflammatory bowel diseases (IBD) (1). IBD, including Crohn’s disease and ulcerative colitis, are intestinal disorders of poorly understood origin thought to arise from genetic susceptibility, luminal microbiota, immune responses, and environmental factors (2–4). A key element in IBD onset is the up-regulation of the proinflammatory cytokine tumor necrosis factor (TNF) by various cell types including immune cells and IECs. TNF overexpression has been detected in the Paneth cells within the epithelium of human IBD patients (5), and anti-TNF treatments are used successfully to treat patients with Crohn’s disease (6). Previous research in mice has demonstrated that intestinal TNF exposure leads to loss of barrier function (7), and overexpression of TNF in mouse IECs is sufficient to elicit an IBD phenotype (8). Despite its pathogenic relevance, the genetic mechanisms regulating TNF expression and IBD onset remain largely unknown.
Genome-wide association studies have identified numerous susceptibility loci associated with IBD including ubiquitin-like protein containing PHD and RING finger domains 1 (UHRF1) and the DNA methyltransferases DNMT1 and DNMT3a (9, 10), which are genes involved in DNA methylation controlling epigenetic transcriptional repression. Moreover, low concordance rates have been observed in monozygotic twin studies (3), leading to the hypothesis that epigenetic regulation also contributes to IBD pathogenesis. Changes in DNA and histone modifications associated with epigenetic regulation have been detected in IBD patients (3, 4, 9, 11, 12), but direct links to the IBD intestinal pathology have not been established. However, recent work has shown that IEC-specific deletion of the histone deacetylase HDAC3 results in increased susceptibility to intestinal damage and inflammation, although specific molecular targets remain to be identified (13).
In this study, we took a forward genetics approach and found that loss of the maintenance DNA methylation regulator uhrf1 leads to hypomethylation of the tnfa promoter and derepression of tnfa. The expression of tnfa in IECs is exacerbated by microbial stimuli and causes cell shedding, a rapid loss of intestinal barrier function and the recruitment of immune cells. Our data suggest that IBD can be triggered by the loss of epigenetic regulation of tnfa in IECs.
Results
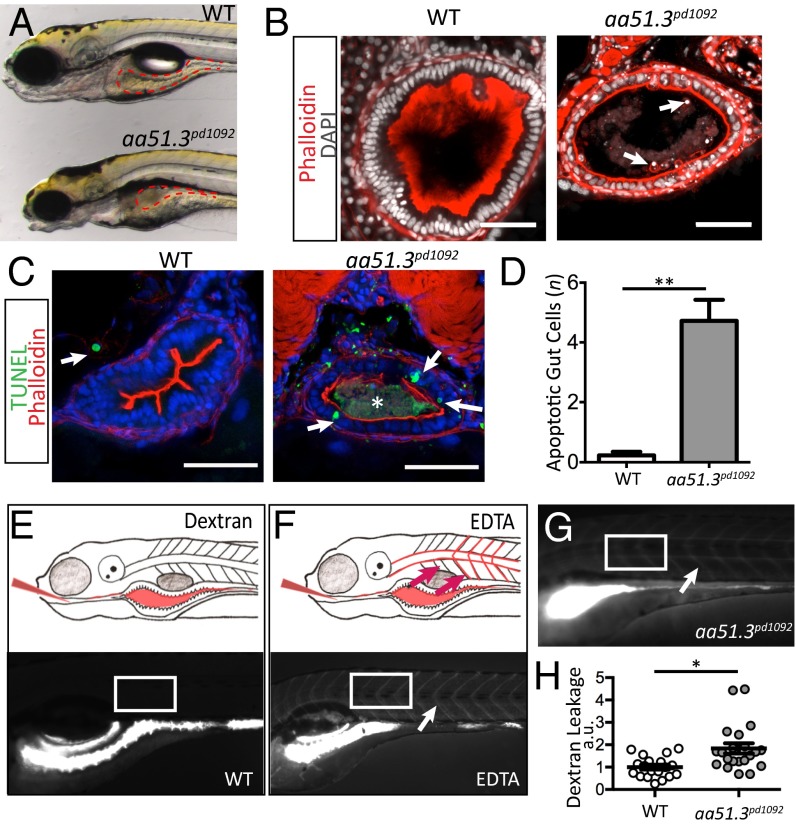
To uncover regulators of intestinal barrier function and inflammation in zebrafish, we performed a forward genetic screen to identify mutants with defects in gut epithelium integrity (14). One of the mutants we identified, aa51.3pd1092, presents a significant disruption of the gut epithelium. At 120 h post-fertilization (hpf), the gut epithelium in control wild-type (WT) siblings has a columnar morphology, thick epithelial folds and a well-developed brush border (Fig. 1 A and B). In contrast, aa51.3pd1092 mutants show a flattened epithelium, reduced brush border, cuboidal epithelial morphology, accelerated shedding of cells from the epithelium and an accumulation of cellular debris within the gut lumen (Fig. 1 A and B). Analysis of cell death revealed a significant increase in apoptotic IECs in mutants in comparison with WT siblings (Fig. 1 C and D). Because accelerated cell shedding can be associated with barrier loss, we assessed whether aa51.3pd1092 mutants display alterations in barrier function. Fluorescent dextran [f-dextran; 4,000 molecular weight (MW)] gavaged orally (15) into the intestinal lumen of WT zebrafish remained exclusively in the lumen 30 min after gavage (Fig. 1 E and H). Disruption of the epithelial barrier through cogavage of EDTA and f-dextran into WT larvae resulted in a rapid, widespread presence of the tracer in the vasculature and intersomitic space (Fig. 1F). Importantly, ∼40% of the aa51.3pd1092 mutants displayed f-dextran in systemic circulation following gavage, indicating that mutants have reduced barrier function (Fig. 1 G and H).
Fig. 1.
aa51.3pd1092 mutants have an IBD-like phenotype. A mutant with excessive intestinal cell shedding and apoptosis and perturbed epithelial morphology was isolated from a forward genetic screen. (A) WT (Upper) and aa51.3pd1092 mutant (Lower) larvae at 120 hpf. (B) Confocal images of WT and aa51.3pd1092 cross-sections. Arrows point to shed cells in the mutant. (C) Confocal images of cross-sections. TUNEL+ cells (green; arrows) in 120 hpf WT and aa51.3pd1092 larvae. Asterisk marks luminal debris. (D) Apoptotic cells in the anterior gut epithelium (n = 13 WT; n = 18 mutants). (E) Schematic of dextran gavage (Upper) and image of 120 hpf WT larva 30 min after gavage (4,000 MW dextran; Lower). (F) Schematic of barrier perturbation after EDTA + dextran cogavage (Upper) and image of 120 hpf WT larva 30 min after gavage (Lower). (G) Dextran gavage in aa51.3pd1092 mutants. Arrow marks extra extraintestinal dextran (F and G). White box indicates region of interest (ROI) measurement (E–G). (H) Quantification of dextran leakage as measured by fluorescence intensity (measured in arbitrary units, a.u.) in WT and mutant larvae after 30 min (n = 19 WT; n = 21 mutants). Mutants show a significant barrier defect. (Scale bars: 50 µm.) Bars represent mean ± SEM. *P < 0.0001, **P < 0.01.
To isolate the mutated locus in aa51.3pd1092, genomic DNA from pools of WT and mutant larvae were subjected to exon capture and sequenced. SNPTrack analysis of the sequencing data (14, 16) revealed a candidate mutation in the splice donor site of exon five in uhrf1, a highly conserved gene that regulates maintenance methylation (Fig. S1 A–A′′). Using reverse transcription PCR (RT-PCR), a band corresponding to exons three through eight for uhrf1 was amplified from WT cDNA but not from aa51.3pd1092 mutants, likely due to nonsense-mediated decay of the transcript (Fig. S1B). Zebrafish mutant for uhrf1 were previously identified in screens for eye and liver defects, although the function of Uhrf1 in the intestine remains unknown (17–20). Crossing the aa51.3pd1092 allele to the previously isolated uhrf1hi3020 allele resulted in noncomplementation, indicating that aa51.3pd1092 is a mutated allele of uhrf1 (Fig. S1 C and D).
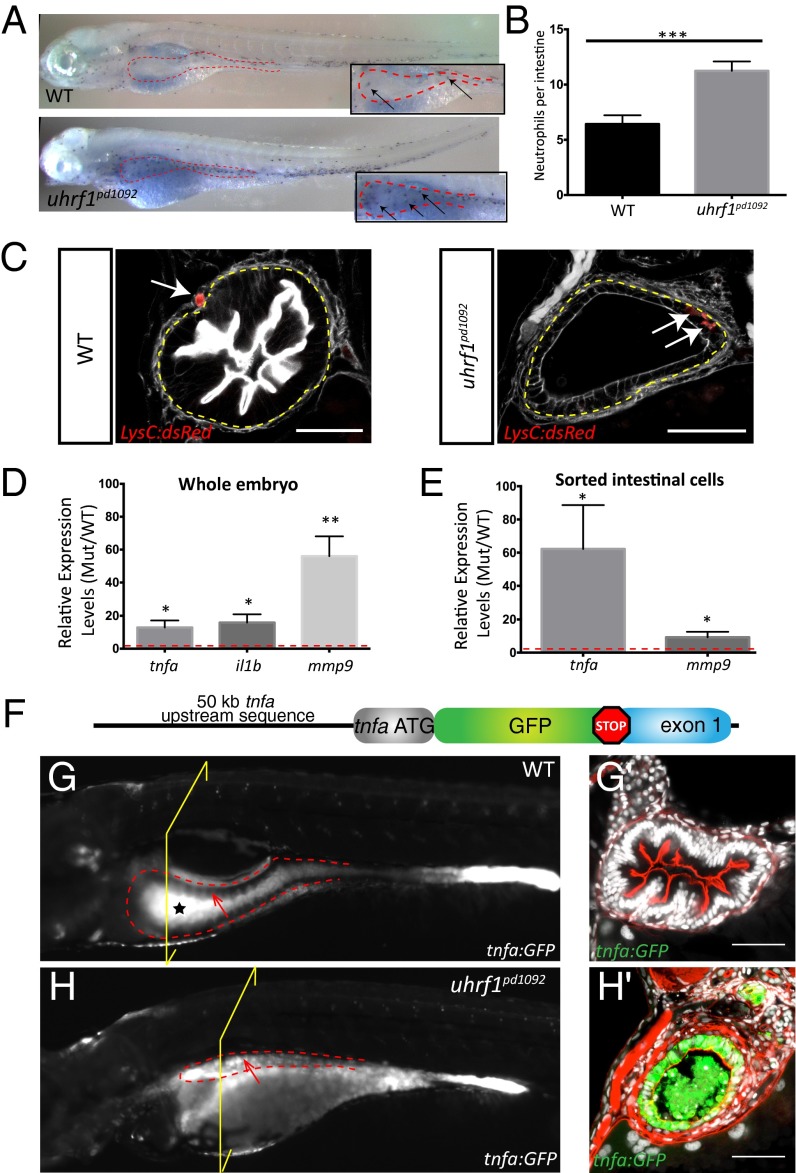
Because accelerated cell shedding and apoptosis are hallmarks of IBD, we wanted to determine whether the immune system is overactive in uhrf1pd1092 mutants. Sudan black staining revealed a significant increase in neutrophil recruitment to the intestine of uhrf1pd1092 mutants in comparison with WT larvae (Fig. 2 A and B). To determine whether the neutrophils were infiltrating the intestinal epithelium, the uhrf1pd1092 line was crossed to Tg(lysC:dsRed) (21), a transgenic line marking neutrophils (22). In 80% of mutants, neutrophils were observed within the gut epithelium whereas they were never observed in the epithelium of WT siblings, but rather in the mesenchyme surrounding the gut (Fig. 2C).
Fig. 2.
Intestinal inflammation in uhrf1pd1092 mutants. Increased neutrophil recruitment, neutrophil infiltration into the intestinal epithelium, and elevated proinflammatory cytokine expression is observed in the intestine of uhrf1pd1092 mutants. (A) Whole-mount images of WT (Upper) and mutant (Lower) 120 hpf larvae stained with sudan black to mark neutrophils. Insets show a higher magnification view of the intestine (red dotted line), and arrows point to individual neutrophils. (B) Quantification of neutrophils in the intestinal area in WT (black) and mutant (gray) larvae. (n = 21 WT; n = 25). (C) Confocal images of cross-sections showing neutrophil (red) localization in 120 hpf WT and uhrf1pd1092 mutant larvae expressing LysC:dsRed. White arrows indicate neutrophils, which have infiltrated into the intestinal epithelium (yellow dotted line) in mutants but are restricted to the mesenchyme of WT. (Scale bars: 50 µm.) (D) Relative expression (mutant/WT) of proinflammatory genes in whole larvae. All genes are significantly elevated in mutants compared with WT. (E) Relative expression (mutant/WT) of tnfa and mmp9 in sorted intestinal epithelial cells. tnfa is greatly up-regulated in the intestine of mutants. (F) TgBAC(tnfa:GFP) construction schematic. (G and Gʹ) Whole-mount (G) and confocal image of cross-section (Gʹ) of a 120 hpf WT TgBAC(tnfa:GFP) transgenic larvae. The black star in G highlights autofluorescence in the WT gut, whereas the arrow points to the anterior gut epithelium that is devoid of TgBAC(tnfa:GFP) expression. (H and Hʹ) Whole-mount (H) and confocal image of cross-section (Hʹ) of 120 hpf uhrf1pd1092 larva with TgBAC(tnfa:GFP) in intestinal epithelium. The arrow highlights elevated epithelial expression of TgBAC(tnfa:GFP) in the intestine. The yellow bracket indicates the plane of section for G′ and H′. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether the intestinal phenotype in uhrf1pd1092 mutants is associated with increased cytokine-driven inflammation, tnfa, interleukin 1b (il1b), and matrix metallopeptidase 9 (mmp9) expression levels were examined by quantitative real-time PCR (qPCR) in whole WT and mutant larvae. Expression of all three genes was significantly increased in whole larvae (Fig. 2D). Interestingly, when IECs were isolated by fluorescence-activated cell sorting (FACS), tnfa expression was >60-fold increased in mutants compared with WT, whereas mmp9 was moderately elevated (Fig. 2E). These data indicate that tnfa induction is highly specific to the gut, whereas mmp9 is induced mostly in extraintestinal cell types.
To analyze the spatiotemporal dynamics of tnfa up-regulation in vivo, we used BAC recombineering to generate a transgenic reporter of tnfa transcriptional activity, TgBAC(tnfa:GFP) (Fig. 2F). Basal expression in TgBAC(tnfa:GFP) was observed in the eye, brain, dorsal root ganglion neurons, and the posterior gut epithelium (Fig. 2G and Fig. S2). To verify that TgBAC(tnfa:GFP) fish are responsive to inflammatory stimuli in vivo, stable TgBAC(tnfa:GFP) fish were infected with Mycobacterium marinum, a close genetic relative of Mycobacterium tuberculosis and a potent inflammatory stimulus in zebrafish (23, 24). Although sham-infected fish did not up-regulate TgBAC(tnfa:GFP) expression, fish infected with fluorescently labeled M. marinum showed a robust up-regulation of tnfa:GFP in granulomas surrounding the bacteria, with kinetics similar to what has been observed via qPCR and in situ hybridization (23, 25) (Fig. S3).
We next crossed uhrf1pd1092 mutants into the TgBAC(tnfa:GFP) background. At 96 hpf, TgBAC(tnfa:GFP) expression is, as in WT, restricted to posterior IECs in uhrf1pd1092 mutant larvae. The gut epithelium in mutants at this stage is morphologically indistinguishable from WT siblings, has no excessive cell shedding or apoptosis, normal apico-basal polarization, and proper differentiation of intestinal cell fates (Fig. S4). Collectively, these results demonstrate that loss of uhrf1 does not affect the initial development or specification of IECs. Beginning around 103 hpf, uhrf1pd1092 mutants display a significant and specific up-regulation of tnfa:GFP in anterior IECs that grew to a 10-fold induction by 120 hpf (Figs. 2 G, G′, H, and H′ and 3D), well in line with our qPCR data. These data show that the up-regulation of tnfa in the gut epithelium precedes the appearance of defects in epithelial integrity and function in uhrf1pd1092 mutants (Fig. 1 A and B). The late onset of the phenotype is likely due to the presence of maternally loaded uhrf1 at early stages (19). Further supporting the specificity of the intestinal inflammation phenotype observed in uhrf1pd1092 mutants, we also detected a dramatic increase in intestinal expression of the tnfa target NFκB in uhrf1pd1092;Tg(NFκB:EGFP) larvae (Fig. S5).
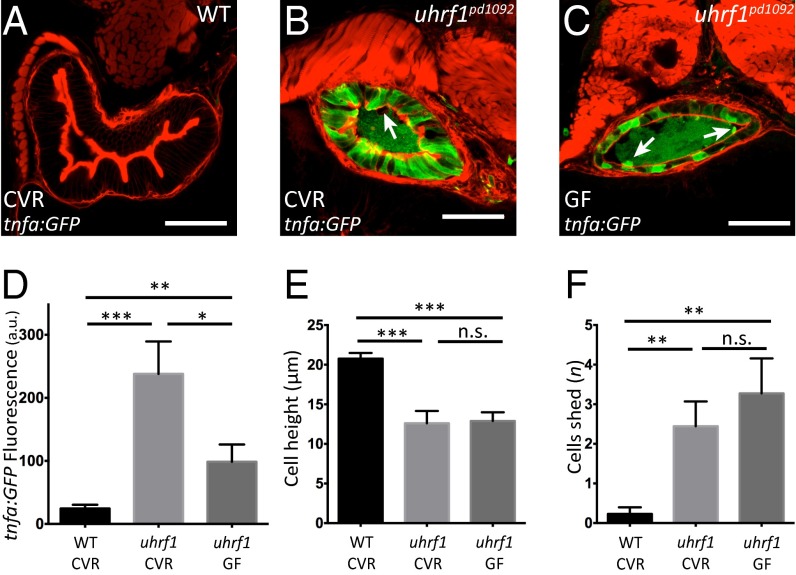
Fig. 3.
Microbiota presence enhances tnfa elevation in the uhrf1pd1092 gut epithelium but is not required for the development of an IBD-like phenotype. (A–C) Confocal images of cross-sections of conventionally raised (CVR) 120 hpf WT (A); CVR 120 hpf uhrf1pd1092 (B), and germ-free (GF) 120 hpf uhrf1pd1092 (C) larvae. The arrows in B and C mark cells being shed from the epithelium. (D) TgBAC(tnfa:GFP) fluorescence (measured in arbitrary units, a.u.) under CVR and GF conditions. (E) Cell height measurement under CVR and GF conditions. Cell height is unchanged between CVR and GF mutants. (F) Quantification of cell shedding. Cell shedding was not decreased in GF mutants. Phalloidin, red. (Scale bars: 50 µm.) (D–F: n = 13 WT CVR; n = 9 mutant CVR; n = 11 mutant GF). Bars represent mean ± SEM. Significant differences between the means were detected after an ANOVA. *P < 0.05, **P < 0.01, ***P < 0.0001, n.s., not significant.
Luminal microbiota have been implicated in IBD pathogenesis (1). To determine whether microbiota contribute to the observed tnfa elevation in IECs, uhrf1pd1092 mutants were derived under germ-free (GF) conditions and compared with conventionally raised (CVR) mutant siblings (26). GF derivation of uhrf1pd1092 mutant larvae reduced IEC TgBAC(tnfa:GFP) expression in comparison with CVR uhrf1pd1092 mutants (Fig. 3 B–D). However, this reduction in TgBAC(tnfa:GFP) expression was still significantly elevated compared with WT (Fig. 3A) and not accompanied by a reduction in cell shedding or an increase in epithelial cell height in the gut (Fig. 3 D–F). These data suggest that although microbial colonization of the intestinal tract contributes to the observed TgBAC(tnfa:GFP) elevation, it is not the driving factor underlying the IBD phenotype in uhrf1pd1092 mutants.
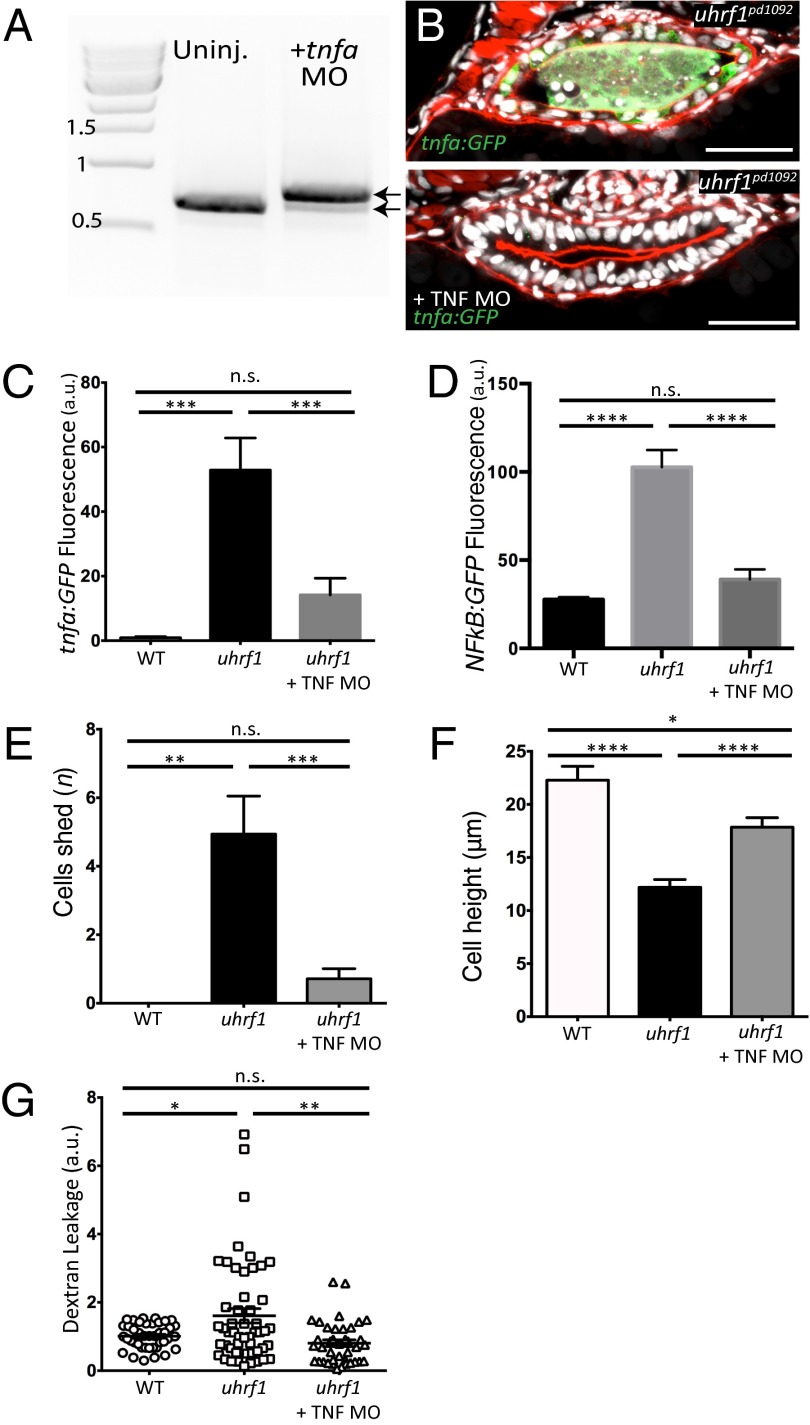
To determine whether elevated tnfa expression underlies the IBD phenotype in uhrf1pd1092 mutants, we obtained and validated a splice-blocking morpholino (27) to knockdown (KD) tnfa expression (Fig. 4A). Efficient KD of tnfa in genotyped uhrf1pd1092 mutants decreased Tg(NFκB:EGFP) fluorescence, cell shedding, increased epithelial cell height, and improved barrier function (Fig. 4 B–G). In agreement with these results, KD of the tnf receptor 1 (tnfr1) also increased epithelial cell height and reduced cell shedding, whereas a control morpholino had no rescue (Fig. S6). Altogether, these data indicate that elevation of tnfa in the intestinal epithelium of uhrf1pd1092 mutants underlies the observed IBD-like phenotype. Interestingly, tnfa KD also resulted in a marked reduction of intestinal tnfa induction in genotyped uhrf1pd1092 mutants, as visualized by the TgBAC(tnfa:GFP) reporter, suggesting that a positive feedback loop enhances tnfa expression in uhrf1pd1092 mutants (Fig. 4 B and C).
Fig. 4.
tnfa knockdown rescues the IBD-like phenotype in the uhrf1pd1092 gut. (A) RT-PCR for tnfa from 103 hpf larvae after injection of the tnfa splice-blocking morpholino. The morpholino resulted in the inclusion of the 112-bp intron between exons 1 and 2 (top arrow; bottom arrow marks endogenous product). (B) Confocal images of cross-sections of genotyped uhrf1pd1092 mutant (Upper) and tnfa morpholino-injected uhrf1pd1092 mutant (Lower) larvae at 103 hpf expressing TgBAC(tnfa:GFP). (C) TgBAC(tnfa:GFP) fluorescence quantification (measured in arbitrary units, a.u.) revealed a decrease in intestinal fluorescence of genotyped mutants after morpholino injection. (D) Tg(NFκB:EGFP) fluorescence quantification (measured in arbitrary units, a.u.). Intestinal fluorescence was decreased in genotyped mutants after morpholino injection. (E) Quantification of cell shedding: Cell shedding was decreased in genotyped morpholino-injected mutants. (F) Cell height measurement: Cell height was increased in genotyped morpholino-injected mutants. (G) Dextran leakage quantification. Improved barrier function was observed in morpholino-injected mutants. (C and E: n = 9, WT; n = 16, mutant; n = 21, mutant + morpholino; D: n = 14, WT; n = 7, mutant; n = 16, mutant + morpholino; F: n = 43, WT; n = 53, mutant; n = 40, mutant + morpholino) Phalloidin, red. (Scale bars: 50 µm.) Results are shown as mean ± SEM. Significant differences between the means were detected after an ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., not significant.
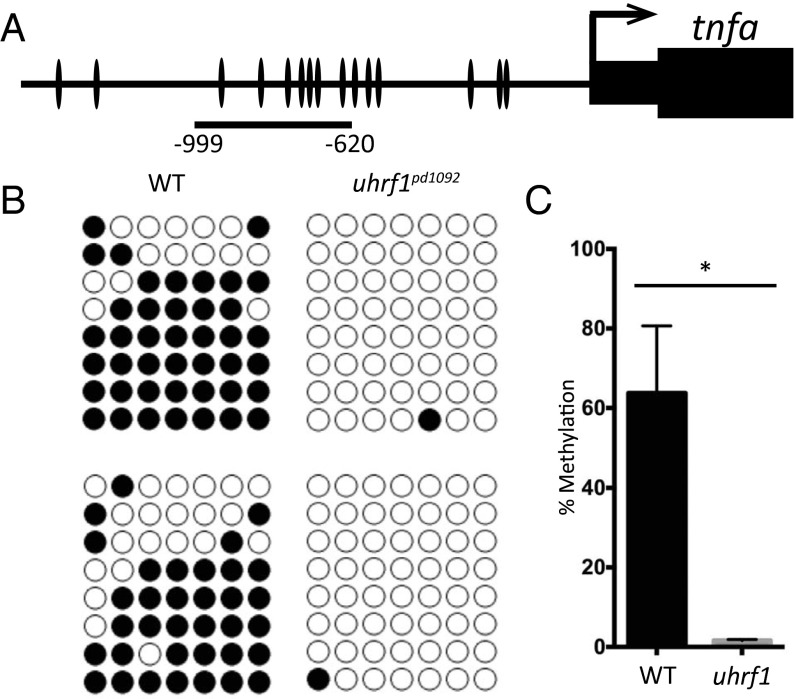
A primary role of Uhrf1 is to recruit DNMT1 to replicating DNA where it acts to maintain methylation patterns at CpG dinucleotides through replication (28–32). In vertebrates, promoters with low CpG density acquire DNA methylation during development (33), which has been correlated with repression of transcriptional activity (34) and demonstrated to cause promoter silencing (35). In zebrafish, uhrf1 expression becomes highly enriched in the gut approximately 96 hpf (19). Given the critical role of uhrf1 in maintaining DNA methylation, we hypothesized that the elevation of tnfa expression observed in uhrf1pd1092 mutants is triggered by hypomethylation of the tnfa promoter. To test this hypothesis, genomic DNA was extracted from IECs isolated via FACS and seven CpG sites proximal to the tnfa promoter were assessed by bisulfite sequencing (Fig. 5A). Whereas these sites were >60% methylated in WT siblings, uhrf1pd1092 mutants had a marked reduction in methylation at all seven sites (Fig. 5 B and C). Similar reductions in methylation signal were observed by using combined bisulfite restriction analysis (COBRA) (Fig. S7). Interestingly, treatment of zebrafish with 5-azacytidine, an inhibitor of methylation at hemimethylated cytidine residues, has been shown to globally reduce CpG methylation in larvae by half without increasing tnfa expression (36). Because tnfa levels do not change, there may be a threshold level of demethylation that must be crossed to allow for activation of tnfa.
Fig. 5.
The tnfa promoter is hypomethylated in uhrf1pd1092 gut epithelial cells. (A) Schematic of zebrafish tnfa promoter, with individual CpG dinucleotides indicated. The region used for bisulfite sequencing spans −620 bp to −999 bp. (B) Biological replicates of bisulfite sequencing data from WT (n = 8 clones each) and mutant (n = 8 clones each) samples. (C) Quantification of DNA methylation levels from biological replicates. Bars represent mean ± SEM; *P < 0.05.
To further substantiate that loss of the methylation function of Uhrf1 underlies the elevated intestinal tnfa, we obtained a previously published dnmt1 mutant allele (dnmt1s872) (20). Upon examination of this mutant, we noticed a close resemblance to both another mutant we had recovered in our genetic screen (cc39.3pd1093) and to uhrf1pd1092 (Fig. S8 A and B). Complementation analyses revealed cc39.3pd1093 is a mutated allele of dnmt1 (Fig. S8A). Importantly, when the TgBAC(tnfa:GFP) line was crossed into dnmt1pd1093 mutants, we observed a specific and robust increase in intestinal tnfa at 103 hpf, similar to what we found in uhrf1pd1092 mutants (Fig. S8 C–E).
While other genes are also hypomethylated in uhrf1pd1092 mutants (20), we found that knockdown of tnfa was sufficient to rescue the IBD-associated phenotypes in IECs. Thus, hypomethylation of the tnfa promoter and subsequent overexpression of tnfa is a predominant contributor to the IBD phenotype in uhrf1pd1092 mutants. Here, we propose that loss of DNA methylation and epigenetic repression at the tnfa promoter leads to intestinal barrier loss, inflammation, and IBD (Fig. S9).
Discussion
In this study, we found that loss of function of the epigenetic regulator uhrf1 results in the induction of tnfa in IECs. This elevation of tnfa is quickly followed by excessive IEC shedding, loss of barrier function, and immune infiltration, which are hallmarks of IBD. This inflammatory scenario is further exacerbated by the presence of microbes and positive feedback of Tnfa stimulating further activation of tnfa in IECs. In IBD patients, TNF expression has been detected in Paneth cells within the intestine by using in situ hybridization (5), and in various immune cell types by qPCR (2). However, these techniques have limited sensitivity or spatial resolution and do not allow direct live detection of TNF activation. Using a transgenic reporter—TgBAC(tnfa:GFP) —we were able to visualize for the first time to our knowledge the cellular source of Tnfa during intestinal inflammation. Our data clearly demonstrate that in uhrf1pd1092 mutants, IECs express tnfa before the onset of an IBD-like phenotype, thus placing IECs at the forefront of IBD onset. IEC involvement is consistent with previous work demonstrating that TNF expression in mouse enterocytes is sufficient to trigger an intestinal inflammation phenotype similar to that of Crohn’s disease (8).
The prevailing view on IBD onset is that the immune response to luminal microbiota is a key event in the establishment of inflammation in the gut (2). However, our results clearly demonstrate that although the microbiota contribute to the exacerbation of the inflammatory condition and also likely play a significant role in chronic disease establishment, IBD onset can occur in the absence of microbial input upon derepression of tnfa. Therefore, understanding the mechanisms regulating TNF expression under basal conditions (i.e., absence of microbe-driven inflammation) is key for uncovering the genetic basis of host susceptibility.
Given the potency of TNF, it was intriguing to find tnfa expressed in the distal zebrafish intestine under basal—uninflamed—conditions (Fig. 2G). We detected a similar pattern of expression in the mouse intestine (Fig. S10). However, when we compared tnfa expression levels detected by our transgenic reporter and qPCR, we found some differences that suggest posttranscriptional regulation also controls tnfa levels. Whereas in the anterior gut of uhrf1pd1092 mutants both methods yield similar levels of transcript induction, in the distal gut we detected significantly lower levels of the native transcript compared with tnfa:GFP under basal conditions. These differences likely result from posttranscriptional regulation of the native transcript, mediated by the AU-rich regulatory elements (ARE) of the 3′UTR (8), which are conserved across species. Additionally, it is also possible that the distal gut in both zebrafish and mice is somewhat refractory to TNF. Thus, the fine regulation of TNF levels in the intestine, and likely also across other tissues, depends on a balance of transcriptional inputs, stimulated by proinflammatory signals from luminal microbiota, repressed by DNA methylation, and subject to posttranscriptional regulation of the transcript. Identifying the molecular machinery controlling TNF stability through its ARE and how this machinery is regulated by genetic and environmental factors will be key for understanding the onset and outcome of inflammatory conditions such as IBD.
Numerous genetic studies have tried to identify genes implicated in IBD onset, and more than 160 susceptibility loci have been implicated in IBD, yet these loci only account for a small fraction of disease heritability (10). Accumulating evidence has also pointed to epigenetics as an important factor (3). Importantly, genome-wide association studies of Crohn’s disease and ulcerative colitis patients have identified SNPs near UHRF1, DNMT1, and DNMT3a (37, 38), suggesting that alterations in these genes may lead to IBD. Although alternative roles for UHRF1 including cell cycle regulation and histone ubiquitylation have been identified (39–41), up-regulation of tnfa in the intestine of both uhrf1pd1092 and dnmt1pd1093 mutants strongly suggests that it is the loss of DNA methylation at CpG dinucleotides at the tnfa promoter that leads to elevated tnfa expression.
Our work illustrates a simple and direct mechanism in which epigenetics and environmental factors such as microbiota converge on TNF to trigger IBD onset. The ability to combine forward genetics and live imaging makes the zebrafish model a powerful tool to dissect the mechanisms controlling TNF expression and IBD onset.
Materials and Methods
Fish Stocks.
Zebrafish were maintained at 28 °C and mated as described (42). The following zebrafish lines were used for this work: AB/TL, EK, Tg(lysC:dsRed)nz50 (21), uhrf1hi3020 (17, 18) Tg(NFkB:EGFP)nc1 (43), Tg(−4.5ifabp:DsRed)pd1000 (43), TgBAC(anxa2b-RFP)pd1113, TgBAC(cldn15la-GFP)pd1034 (qPCR) (44), TgBAC(tnfa:GFP)pd1028, and aa51.3pd1092 (this study) and cc39.3pd1093 (this study).
Mutagenesis, Genetic Screen, and Mapping.
The aa51.3pd1092 mutant was identified from a published three-generation forward genetic N-ethyl-N-nitrosurea screen (14). Isolation of the affected locus in the aa51.3pd1092 mutant was performed by using the Agilent SureSelect solution-based zebrafish exon-capture kit (14) and SNPTrack for linkage analysis (16). A splice site mutation in uhrf1 was confirmed via amplification of uhrf1 exons three through eight by using RT-PCR (Invitrogen) with the following primers: uhrf1 exon 3, 5′-ACGACATCGTTCAGCTGTTG-3′; uhrf1 exon 8, 5′-AGTCTTCGTCGTCGGGTATG-3′. Mutant allele genotyping was performed by AvaI digestion using the following primers: uhrf1-5′F: 5ʹ-CACCCCAATTTCCTCTTGAA-3′ʹ; uhrf1-3′R: 5′-CAATCATCAGCACTTTTAATAGCTT-3′.
BAC Recombineering and Transgenesis.
A BAC containing the upstream sequence and first exon of tnfa (CH211-176D22) was modified as described (45). Recombination was performed to replace the tnfa start site with GFP by using the following homology primers: tnfa 5ʹ homology, 5ʹ-CCTTATCAAAAGCATTTACACTTGTAGAATCTTTAAGACATACAGCAATTATGGTGAGCAAGGGCGAGGAG-3ʹ; tnfa 3ʹ homology, 5ʹ-TAAAGGCAACTCTCCTTCTTCAACATCCAAAAACGCCCGACTCTCAAGCTTATTGGAGCTCCACCGCGGTG-3ʹ. The tnfa:GFP BAC was linearized with XhoI and injected into one-cell stage embryos to generate TgBAC(tnfa:GFP)pd1028.
Barrier Assay.
Oral microgavage of 103 and 120 hpf WT and aa51.3pd1092 mutant larvae was performed as described with a few modifications (refs. 15 and 46 and SI Materials and Methods).
Immunofluorescence and TUNEL.
Transverse agarose cross-sections were cut on a Leica VT1000S vibratome and stained as described (ref. 47 and SI Materials and Methods). TUNEL was then performed by using the ApopTag Red in situ apoptosis detection kit (Millipore).
Embryo Dissociation and FACS.
Embryo dissociation and FACS was performed as described (44) with slight modifications (SI Materials and Methods and ref. 48).
RNA Isolation and qPCR.
RNA isolation and cDNA synthesis for qPCR was performed as described (44). qPCR was performed by using a Roche LightCycler carousel-based system and the Roche LightCycler FastStart DNA MasterPLUS SYBR Green I. For each zebrafish qPCR experiment, samples were run as technical duplicates from six independent experiments for tnfa on sorted cells, five independent experiments for mmp9 on sorted cells, and four independent experiments for whole embryos. Expression levels were normalized to elfa.
For mouse qPCR experiments, RNA was isolated from epithelial sheets from the mouse proximal, medial and distal small intestine, and the colon, and cDNA was synthesized as previously described (44).
M. marinum Infection.
TgBAC(tnfa:GFP) incrosses were infected at 48 hpf through caudal vein injections by using single-cell suspensions of M. marinum (strain M, carrying msp12:tdTomato) or mock infected with an arterial injection of 7H9 media alone as described (23, 24). Larvae were imaged at 3 d after infection by using a Zeiss Axio Observer.
Gnotobiotic Husbandry.
Methods for derivation and maintenance of gnotobiotic zebrafish have been described (26). Fish were derived germ free and compared with conventionally raised clutchmates; animals were not fed during this experiment.
Morpholino Knockdown.
The antisense splice-blocking morpholino against tnfa (5′-GCAGGATTTTCACCTTATGGAGCGT-3′; GeneTools) was previously validated and its efficacy was confirmed for this study (Fig. 4A) (27). The tnfa morpholino was diluted to 0.11 mM, and embryos were injected at the one-cell stage with 3 nL (total: 3 ng per embryo) in the aa51.3pd1092;TgBAC(tnfa:GFP) background. Larvae were then fixed at 103 hpf to analyze gut morphology or gavaged at 103 hpf to assay barrier function. The previously published and validated tnfr morpholino (5′-CTGCATTGTGACTTACTTATCGCAC-3′; GeneTools) (49, 50) was diluted to a concentration of 0.33 mM and 2 nL (total: 5.5 ng per embryo) injected into each embryo. A standard control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′; GeneTools) injected at the same concentration as the tnfa morpholino.
Bisulfite Sequencing.
Bisulfite conversion of genomic DNA was performed by using the EZ DNA Methylation Direct kit (Zymo Research). The region of the tnfa promoter that we selected meets the criteria for a region of low to intermediate CpG density, while at the same time encompassing the majority of CpG dinucleotides within 1 kb upstream of the tnfa transcriptional start site. The arrangement of CpGs in this region is highly similar to the architecture observed in the human TNF promoter (51). The zebrafish tnfa promoter (−999 to −620 bp) was amplified from bisulfite converted DNA by using the following primers: GGGAATTGGTATTGGTTTTATT and CAATTTAATACRATTTACTCCTCAC followed by the primers GGTTGTTTTAATTTAAATTTGGG and ATTTACTCCTCACCTAATAA. The amplified DNA was purified and cloned into the pGEM-T easy vector (Promega), and 10 clones were sequenced for WT and mutant in biological duplicate. QUMA (52) was used to analyze biological replicates of the bisulfite sequencing data.
COBRA was performed by digesting the amplified DNA with TaqαI (−721 bp). The TaqαI cut site is maintained following bisulfite conversion only if the site is methylated. Therefore, digestion of the amplified DNA indicates methylation at this site.
Statistical Analysis.
Data are presented as the mean ± SEM. Zebrafish studies were not performed in a blinded fashion. Statistical analyses were performed by using Prism, version 6.0e (GraphPad Software). Statistical differences in barrier function, epithelial cell height, cell shedding, cell death, and TgBAC(tnfa:GFP) fluorescence were analyzed by using a Student’s t test after an ANOVA was performed to confirm statistically significant differences among the means for each experiment. For qPCR, graphs represent fold change in mutant versus WT expression levels. A Student’s t test was performed to determine statistical significance. A P value <0.05 was set as the threshold for statistical significance.
Barrier assay, antibodies and imaging approaches, embryo dissociation, and FACS procedures and qPCR primers are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Burris, E. Becker, and N. Blake for zebrafish care; J. Gross and P. Crosier for zebrafish lines; A. Navis and K. Ellis for experimental support; B. Hogan and members of the M.B. laboratory for helpful discussions throughout this project; and K. Poss, B. Sartor, Antonio Giraldez, and C. Eroglu for critical reading of the manuscript. This work was supported by NIH New Innovator Award DP2 3034656 and Grand Challenges Explorations Grant OPP1108132 from the Bill & Melinda Gates Foundation (to M.B.); L.M. was supported by Duke Multidisciplinary Fellowship in Pediatric Lung Disease Grant 5T32HL098099-02 and NIH National Research Service Award F32-DK098885-01A1; J.L.C. was supported by NIH NRSA F32-DK094592; J.F.R. was supported by NIH Grants R01-DK081426 and P01-DK094779; R.W.B. and D.M.T. were supported by an NIH Director's New Innovator Award 1DP2-OD008614; and M.G.G. was supported by March of Dimes Foundation Grant 5-FY12-93.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424089112/-/DCSupplemental.
References
- 1.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 2.Sartor RB. Mechanisms of disease: Pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3(7):390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 3.Jenke AC, Zilbauer M. Epigenetics in inflammatory bowel disease. Curr Opin Gastroenterol. 2012;28(6):577–584. doi: 10.1097/MOG.0b013e328357336b. [DOI] [PubMed] [Google Scholar]
- 4.Scarpa M, Stylianou E. Epigenetics: Concepts and relevance to IBD pathogenesis. Inflamm Bowel Dis. 2012;18(10):1982–1996. doi: 10.1002/ibd.22934. [DOI] [PubMed] [Google Scholar]
- 5.Lala S, et al. Crohn’s disease and the NOD2 gene: A role for paneth cells. Gastroenterology. 2003;125(1):47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 6.Targan SR, et al. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn’s disease. Crohn’s Disease cA2 Study Group. N Engl J Med. 1997;337(15):1029–1035. doi: 10.1056/NEJM199710093371502. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, et al. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am J Pathol. 2005;166(2):409–419. doi: 10.1016/s0002-9440(10)62264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roulis M, Armaka M, Manoloukos M, Apostolaki M, Kollias G. Intestinal epithelial cells as producers but not targets of chronic TNF suffice to cause murine Crohn-like pathology. Proc Natl Acad Sci USA. 2011;108(13):5396–5401. doi: 10.1073/pnas.1007811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: The emerging role of epigenetics. Gastroenterology. 2013;145(2):293–308. doi: 10.1053/j.gastro.2013.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham DB, Xavier RJ. From genetics of inflammatory bowel disease towards mechanistic insights. Trends Immunol. 2013;34(8):371–378. doi: 10.1016/j.it.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nimmo ER, et al. Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis. 2012;18(5):889–899. doi: 10.1002/ibd.21912. [DOI] [PubMed] [Google Scholar]
- 12.Cooke J, et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18(11):2128–2137. doi: 10.1002/ibd.22942. [DOI] [PubMed] [Google Scholar]
- 13.Alenghat T, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature. 2013;504(7478):153–157. doi: 10.1038/nature12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan S, et al. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development. 2013;140(21):4445–4451. doi: 10.1242/dev.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchiaro JL, Rawls JF. Microgavage of zebrafish larvae. J Vis Exp. 2013;(72):e4434. doi: 10.3791/4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leshchiner I, et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22(8):1541–1548. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amsterdam A, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101(35):12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross JM, et al. Identification of zebrafish insertional mutants with defects in visual system development and function. Genetics. 2005;170(1):245–261. doi: 10.1534/genetics.104.039727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci USA. 2007;104(5):1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tittle RK, et al. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev Biol. 2011;350(1):50–63. doi: 10.1016/j.ydbio.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang CT, et al. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe. 2012;12(3):301–312. doi: 10.1016/j.chom.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobin DM, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140(5):717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takaki K, Davis JM, Winglee K, Ramakrishnan L. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat Protoc. 2013;8(6):1114–1124. doi: 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clay H, et al. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. 2007;2(1):29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3(12):1862–1875. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Muñoz A, et al. Evolutionary conserved pro-inflammatory and antigen presentation functions of zebrafish IFNγ revealed by transcriptomic and functional analysis. Mol Immunol. 2011;48(9-10):1073–1083. doi: 10.1016/j.molimm.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 29.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 30.Sheaffer KL, et al. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28(6):652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mudbhary R, et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25(2):196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgia S, Kanji M, Bhushan A. DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes Dev. 2013;27(4):372–377. doi: 10.1101/gad.207001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 34.Smith ZD, Meissner A. DNA methylation: Roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 35.Han H, et al. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum Mol Genet. 2011;20(22):4299–4310. doi: 10.1093/hmg/ddr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews RP, et al. TNFalpha-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish S-adenosylhomocysteine hydrolase. Development. 2009;136(5):865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jostins L, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obata Y, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15(6):571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 40.Nishiyama A, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502(7470):249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 41.Chu J, et al. UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is required for zebrafish embryogenesis. Mol Biol Cell. 2012;23(1):59–70. doi: 10.1091/mbc.E11-06-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Univ Oregon Press; Eugene, OR: 2000. [Google Scholar]
- 43.Kanther M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141(1):197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvers AL, Ryan S, Scherz PJ, Huisken J, Bagnat M. Single continuous lumen formation in the zebrafish gut is mediated by smoothened-dependent tissue remodeling. Development. 2014;141(5):1110–1119. doi: 10.1242/dev.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navis A, Marjoram L, Bagnat M. Cftr controls lumen expansion and function of Kupffer’s vesicle in zebrafish. Development. 2013;140(8):1703–1712. doi: 10.1242/dev.091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldsmith JR, Cocchiaro JL, Rawls JF, Jobin C. Glafenine-induced intestinal injury in zebrafish is ameliorated by μ-opioid signaling via enhancement of Atf6-dependent cellular stress responses. Dis Model Mech. 2013;6(1):146–159. doi: 10.1242/dmm.009852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9(8):954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 48.Bjerknes M, Cheng H. Methods for the isolation of intact epithelium from the mouse intestine. Anat Rec. 1981;199(4):565–574. doi: 10.1002/ar.1091990412. [DOI] [PubMed] [Google Scholar]
- 49.Espín-Palazón R, et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 2014;159(5):1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espín R, et al. TNF receptors regulate vascular homeostasis in zebrafish through a caspase-8, caspase-2 and P53 apoptotic program that bypasses caspase-3. Dis Model Mech. 2013;6(2):383–396. doi: 10.1242/dmm.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gowers IR, et al. Age-related loss of CpG methylation in the tumour necrosis factor promoter. Cytokine. 2011;56(3):792–797. doi: 10.1016/j.cyto.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Kumaki Y, Oda M, Okano M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008;36(Web Server issue):W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.