Significance
Resistance to current antibiotics is rising worldwide, and replacement molecules are urgently needed. The tryptophanyl–tRNA synthetase inhibitor indolmycin, containing a unique oxazolinone core, was first isolated in 1960, but the molecular basis for its construction is still unknown. Here we report the complete pathway to indolmycin, using a combination of genetic and biochemical approaches. Our work reveals how the oxazolinone core is assembled by the dual action of the ATP-dependent enzyme Ind3, and Ind6, an unusual gatekeeper protein. Our work sets the stage for generating improved microbial production systems for indolmycin and for development of indolmycin-like antibiotics.
Keywords: biosynthesis, antibiotic, indolmycin
Abstract
The bacterial tryptophanyl–tRNA synthetase inhibitor indolmycin features a unique oxazolinone heterocycle whose biogenetic origins have remained obscure for over 50 years. Here we identify and characterize the indolmycin biosynthetic pathway, using systematic in vivo gene inactivation, in vitro biochemical assays, and total enzymatic synthesis. Our work reveals that a phenylacetate–CoA ligase-like enzyme Ind3 catalyzes an unusual ATP-dependent condensation of indolmycenic acid and dehydroarginine, driving oxazolinone ring assembly. We find that Ind6, which also has chaperone-like properties, acts as a gatekeeper to direct the outcome of this reaction. With Ind6 present, the normal pathway ensues. Without Ind6, the pathway derails to an unusual shunt product. Our work reveals the complete pathway for indolmycin formation and sets the stage for using genetic and chemoenzymatic methods to generate indolmycin derivatives as potential therapeutic agents.
Indolmycin (1) is an antibacterial drug produced by bacterial strains from both terrestrial and marine environments, such as Streptomyces griseus ATCC 12648 and Pseudoalteromonas luteoviolacea (1, 2). As a structural analog of l-tryptophan, indolmycin competitively inhibits bacterial tryptophan–tRNA synthetases (TrpRSs) (3). Recently, indolmycin was reported to exhibit potent antibacterial activity against human pathogens, such as Helicobacter pylori (4), and mupirocin-, methicillin-resistant Staphylococcus aureus (5). Although it has been proposed that indolmycin could be tested as a topical treatment for staphylococcal infections, the drug has not yet been tested in humans. Indolmycin congeners, 5-hydroxyindolmycin and 5-methoxyindolmycin, have been obtained by feeding the corresponding tryptophan and indole precursors to the indolmycin producer (6). These derivatives show a moderate increase in antimicrobial activity compared with indolmycin. Besides its biological activity, the unique structure, with a core oxazolinone ring, has attracted interest from synthetic chemists. Several total synthetic routes to indolmycin and its analogs have been reported (7–14); however, commercially available indolmycin is currently generated through fermentation, and yields of ∼20 mg/L are reported in the literature (15).
How indolmycin is synthesized in nature remains largely unknown. In the 1970s, the Floss group established tryptophan, arginine, and methionine as indolmycin precursors and β-methylindolepyruvate and indolmycenic acid as intermediates in the biosynthetic pathway by using isotope-labeled precursors in feeding experiments (15–17). Furthermore, preliminary investigation into the enzymology of the indolmycin biosynthetic pathway resulted in the detection of two enzyme activities from cell-free extracts of S. griseus ATCC 12648. These were attributed to an l-tryptophan aminotransferase and an indolepyruvate C-methyltransferase, which were proposed to catalyze the first two steps of indolmycin biosynthesis (18). However, since then, no work has been done on indolmycin biosynthesis. Thus, its biosynthetic pathway, including the enzymatic machinery driving the construction of the characteristic oxazolinone heterocycle, is still an enigma.
To understand how the oxazolinone ring is assembled and to set the stage for future efforts to improve titers and generate nonnatural derivatives, we undertook the elucidation of the indolmycin biosynthetic pathway. Here we report the isolation of the indolmycin biosynthetic gene cluster from S. griseus ATCC 12648. By combining in vivo gene inactivation and in vitro biochemical assays, the indolmycin biosynthetic pathway is proposed, and enzymes responsible for the remarkable oxazolinone heterocycle formation are deciphered. Moreover, we accomplish the enzymatic total synthesis of indolmycin. Notably, we find that Ind3 catalyzes amide bond formation between indolmycenic acid and dehydroarginine, followed by subsequent cyclization to give two products, both of which contain the characteristic oxazolinone heterocycle. In the presence of a multitasking protein, Ind6, N-demethylindolmycin appears as the dominant product, whereas without Ind6, an off-pathway shunt product is detected as the major product. Ind6 thus serves as a “gatekeeper,” determining the outcome of the whole pathway. In addition, we show that Ind6 also has a chaperone-like activity, solubilizing a dehydroarginine biosynthetic enzyme, Ind5, by forming a protein–protein complex.
Results
Identification of the Indolmycin Biosynthetic Gene Cluster.
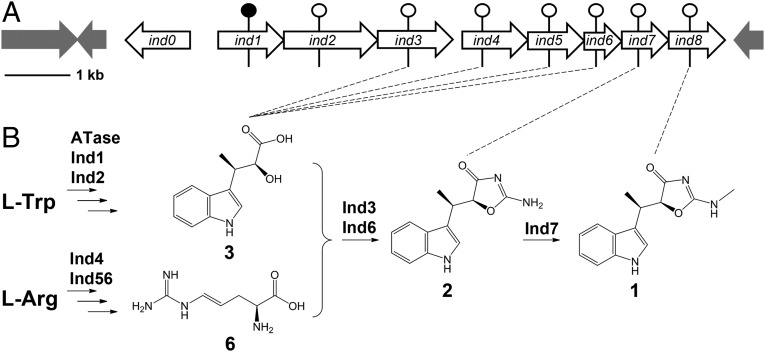
Resistance and biosynthetic genes for a specific secondary metabolite are usually clustered in bacteria. Therefore, to isolate the gene cluster for 1 biosynthesis, we initially screened the genomic library of S. griseus ATCC 12648 by PCR with primers targeting the tryptophanyl–tRNA synthetase gene that confers high-level resistance to 1 (19). However, sequence analysis of the region flanking this gene failed to reveal any genes that might be related to 1 biosynthesis. We thus turned to genome scanning. Through bioinformatics analysis of the resulting genomic data, we identified three putative tryptophanyl–tRNA synthetase (TrpRS) genes throughout the genome. Among them, ind0 drew our attention (Fig. 1A). This gene is located in a putative nine-gene cluster that spans a 10.3-kb DNA region, flanked by transposase genes. The GC content (65.0%) of this putative gene cluster is significantly lower than the average GC content (72.7%) of the genome, suggesting that it might have been acquired from a foreign source by horizontal gene transfer. Sequence analysis of this gene cluster, which we named the ind cluster, predicts that ind4 and ind8 encode pyridoxal 5′-phosphate (PLP)–dependent aminotransferases, ind1 and ind7 encode methyltransferases, ind2 and ind5 encode dehydrogenases, ind3 encodes a phenylacetate–CoA ligase or F390 synthetase, and ind6 encodes a gene of unknown function (Table 1). The presence of aminotransferase and methyltransferase genes is consistent with the previous work showing that a tryptophan aminotransferase and indolepyruvate C-methyltransferase are involved in the indolmycin biosynthetic pathway (18).
Fig. 1.
Indolmycin biosynthetic gene cluster and overall pathway. (A) Indolmycin biosynthetic gene cluster. The black circle indicates the gene was subjected to in-frame deletion, and white circles indicate insertional inactivation. (B) Proposed indolmycin biosynthetic pathway based on this study. Dashed lines indicate the major accumulated metabolite from each gene-inactivation mutant.
Table 1.
Predicted functions of ORFs of the indolmycin biosynthetic gene cluster
| Gene | Size (aa) | Predicted function |
| ind0 | 321 | tryptophanyl–tRNA synthetase |
| ind1 | 329 | SAM-dependent C-methyltransferase |
| ind2 | 570 | NADH-dependent dehydrogenase |
| ind3 | 452 | ATP-dependent amide synthetase |
| ind4 | 383 | PLP-dependent aminotransferase |
| ind5 | 319 | NADH-dependent dehydrogenase |
| ind6 | 229 | unknown |
| ind7 | 237 | SAM-dependent N-methyltransferase |
| ind8 | 361 | PLP-dependent aminotransferase |
To link this gene cluster to 1 biosynthesis, we generated eight mutants (Δind1–ind8) (SI Appendix, Table S1 and Fig. S1 A–F). Metabolic profiling and subsequent structure elucidation revealed that all mutants, with the exception of ∆ind8, had abolished 1 production. Mutants ind3, ind4, ind5, and ind6 had increased indolmycenic acid (3) production, whereas ∆ind7 accumulated N-demethylindolmycin (2) (Fig. 1B; see SI Appendix, Figs. S1 G and H and S2). In vitro analysis of Ind7 with 2 verified that this enzyme is responsible for the last N-methyltransfer reaction to generate 1 (SI Appendix, Figs. S3A and S4A). Neither ∆ind1 nor ∆ind2 produced 3, suggesting that they are involved in 3 biosynthesis. A combination of in vivo and in vitro experiments revealed Ind1 as an indolepyruvate C-methyltransferase and Ind2 as responsible for the NADH-dependent conversion of β-methylindolepyruvate to 3 (SI Appendix, Fig. S4 B and C).
Intriguingly, although earlier studies suggested the involvement of a tryptophan aminotransferase as the first step in the pathway, neither Δind4 nor Δind8 abolished 3 production. To further investigate the possible role of these enzymes in converting tryptophan to indolepyruvate through a PLP-dependent reaction, we carried out in vitro assays. Production of indolepyruvate by Ind4 was barely detectable, and the production by Ind8 was also low (SI Appendix, Fig. S4D). However, another two putative aminotransferases, ORF4725 and ORF2651, were encoded elsewhere in the genome, with end-to-end homology with tryptophan aminotransferase, MarG (20). ORF4725 was able to convert tryptophan to indolepyruvate in in vitro assays, and ORF2651 also showed reasonable turnover. To determine which of these proteins might be expressed during indolmycin biosynthesis, we carried out RT-PCR and observed bands for ind4, ind8, and orf2651, but not for orf4725, suggesting that orf2651 might encode the responsible aminotransferase. Coexpression of orf2651, ind1, and ind2 in Escherichia coli furthermore gave production of 3. Altogether, these results suggest that the responsible tryptophan aminotransferase in S. griseus ATCC 12648 might be ORF2651, but that ORF4725 could be used in its place in engineered systems to generate indolepyruvate (SI Appendix, Fig. S4 E and F).
In Vitro Reconstitution of Oxazolinone Ring Assembly.
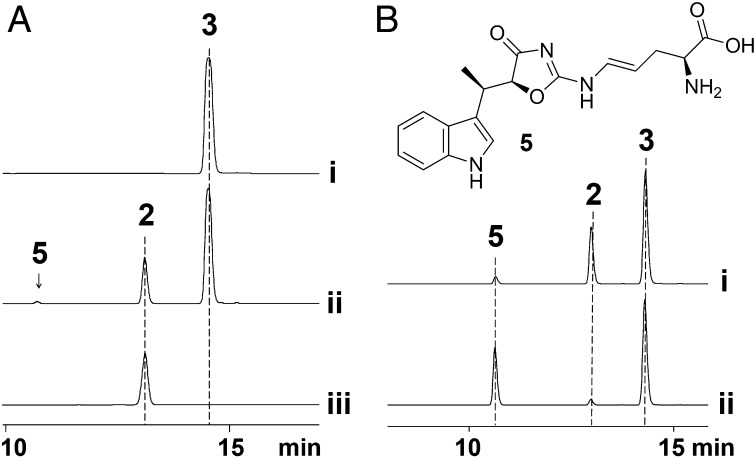
Given that our above in vivo data did not give any insight into the assembly mechanism of the unique oxazolinone ring, we turned to in vitro reconstitution. Mutants ind3–4-5-6 all failed to produce 1 or 2, but generated 3, implicating that some or all of them participate in 2 biosynthesis, using 3 as a precursor. We cloned and expressed these four genes individually, resulting in soluble proteins Ind3, Ind4, and Ind6. After several attempts, we found Ind5 could be expressed in soluble form only when coexpressed with Ind6. Interestingly, His6-tagged Ind5 pulled down untagged Ind6 in approximately equal amounts during the nickel-affinity purification, indicating the formation of an Ind5–Ind6 protein complex, which was verified through mass analysis (SI Appendix, Fig. S3B). Next, we used a one-pot enzymatic synthesis strategy to probe the mechanism for oxazolinone ring assembly. Specifically, 3 and l-arginine (4) were incubated with proteins (Ind3, Ind4, Ind56 complex, Ind6, and Ind8), and all their likely cofactors [α-ketoglutarate (α-KG), PLP, NADH, Mg2+, ATP, and CoA]. HPLC analysis of the reaction mixture revealed two new peaks [retention time (tR) 11.0 and 13.2 min]. The dominant product (tR of 13.2 min) was identified as 2, by comparison with a standard compound (Fig. 2A), demonstrating that all of the components essential for oxazolinone ring formation are included in the reaction mixture. To probe the necessity of each component, they were systemically eliminated from the one-pot assay. The results demonstrated that Ind3–4-5-6, NADH, and ATP are essential for 2 formation, whereas Mg2+ and PLP enhance the production of 2 (SI Appendix, Fig. S4 G and H).
Fig. 2.
Reconstitution of oxazolinone ring assembly in vitro. (A) One-pot enzymatic synthesis of N-demethylindolmycin (2) and indolmycin B (5) from indolmycenic acid (3) and l-Arg (4). (i) Authentic 3, (ii) HPLC analysis of the reaction mixture of the one-pot reaction, (iii) authentic 2. (B) Isolation and characterization of 5. (i) HPLC analysis of products from the one-pot reaction (Ind4 + 4 + PLP + Ind56 + NADH + Ind3 + 3 + ATP + Mg2+), (ii) HPLC analysis of products from the decoupling of Ind4–Ind56- and Ind3-catalyzed reactions (see main text for details). Note that l-Arg (4) (elutes at < 5 min) does not absorb at 225 nm and is therefore not shown. HPLC detection wavelength, 225 nm.
Identification of Indolmycin B as a Shunt Product from the Indolmycin Pathway.
The above one-pot assay established the involvement of Ind3–4-5-6 in oxazolinone ring formation and also gave insight into the timing of the enzymatic reactions. Given that 3 is only consumed when all essential enzymes are present, we reasoned that 3 is used by the enzyme responsible for the last step in the biosynthesis of 2. Among Ind3–4-5-6, Ind3 shows sequence homology to phenylacetate–CoA ligases (PCLs), which usually activate carboxylic acid through ATP-dependent adenylation for subsequent attack by the thiol of CoA (21). Our above one-pot assay had shown that ATP is indispensable, whereas CoA is not. We thus hypothesized that 3 could be activated by Ind3 as an acyl-adenylate intermediate, with subsequent nucleophilic attack by the guanidium–NH2 group of an arginine derivative, for direct amide bond formation. To support this hypothesis, we first performed an Ind4–Ind56-coupled reaction by incubating 4 with Ind4–Ind56, PLP, and NADH. After 4 h, enzymes were heat-inactivated and centrifuged. The resulting supernatant, containing the small molecule pool, was then incubated with Ind3, ATP, Mg2+, and 3. HPLC analysis of the reaction mixture revealed the formation of both 2 and the peak with tR of 11.0 min (Fig. 2B, trace ii). However, surprisingly, 2 was only produced at trace level, whereas the peak with tR of 11.0 min, which was originally generated as a minor product in the one-pot assay (Fig. 2B, trace i), now appeared as the major product. To identify this molecule, we scaled up the above enzymatic reactions and purified the compound by preparative HPLC. Its structure was solved by high-resolution electrospray ionization mass spectrometry (HR-ESI-MS) and extensive NMR spectroscopic analysis, revealing that this molecule, which we name indolmycin B (5), also contains the oxazolinone ring (Fig. 2B and SI Appendix, Fig. S5 A–C).
Dehydroarginine Production by the Ind4–Ind56-Coupled Reaction.
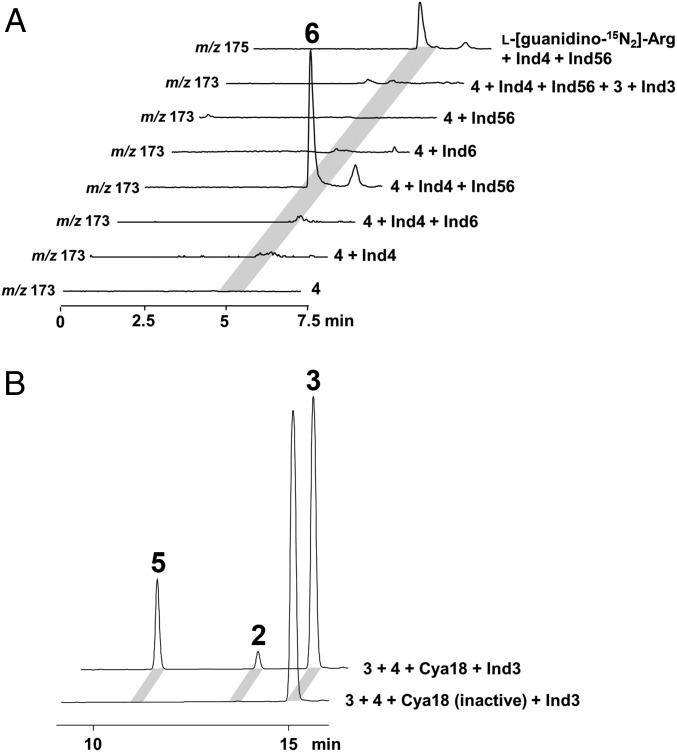
The structure of 5 suggests that its direct precursors might be 3 and 4,5-dehydroarginine. We next investigated the Ind4–Ind56-coupled reaction, which we suspected would generate direct precursor(s) for 2 and 5. In the presence of NADH and PLP, incubation of l-arginine (4) with Ind4 and Ind56 led to the formation of a molecule with mass signal at m/z 173, which is consistent with the [M+H]+ ion for 4,5-dehydroarginine (6). To confirm the product as an arginine derivative and the dependence of its production on both Ind4 and Ind56, we incubated 4 with various combinations of purified enzymes (Ind4, Ind56, Ind6, and Ind3), cofactors, and substrates. Liquid chromatography-mass spectrometry (LC-MS) analysis of these reaction mixtures showed that the compound (m/z 173) is produced only when both Ind4 and Ind56 are present. Also, a mass signal at m/z 175 was observed when l-arginine (4) was replaced with l-[guanidino-15N2]arginine (Fig. 3A). Furthermore, the absence of this ion peak in an assay in which Ind3 was included suggests it could be a substrate of Ind3. To confirm 6 as the product of the Ind4–Ind56-coupled reaction, we set out to purify this compound by scaling up the Ind4–Ind56 reaction and tracking the compound by MS-guided TLC. Although many attempts to separate the product (m/z 173) from the remaining 4 failed, we were able to obtain NMR data for the target molecule (m/z 173) through NMR analysis of a mixture with 4 (SI Appendix, Fig. S5 D–F). All these data are consistent with the structure of 6, supporting 6 as the product of the Ind4–Ind56-coupled reaction.
Fig. 3.
Dehydroarginine (6) production by the Ind4–Ind56-coupled reaction. (A) Selective ion monitoring of 6 production in a reaction containing PLP and NADH along with various combinations of enzymes and substrates. ATP and Mg2+ are added when Ind3 is included. (B) Production of 2 and 5 by incubation of 3 and 4 with Cya18 + Ind3. Both reactions contain α-KG, FeSO4, l-ascorbate, Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), Mg2+, and ATP. Detection wavelength, 225 nm.
Recently, 6 was also reported as a precursor of quinocarcin/SF-1739, and Cya18, an α-ketoglutarate–dependent oxygenase, catalyzes dehydrogenation of 4 to give 6 (22). To further solidify our above result, cya18 was synthesized and expressed in E. coli (SI Appendix, Fig. S3 C and D). Incubation of Cya18 and Ind3 with 4, 3, α-KG, FeSO4, l-ascorbate, Mg2+, and ATP led to the formation of both 2 and 5 (Fig. 3B). The observation that Ind4–Ind56 can be replaced by Cya18 in the one-pot assay confirms 6 as the product of the Ind4–Ind56 reaction.
Ind3 as a Novel Oxazolinone Synthetase and Ind6 as a Gatekeeper Determining the Outcome of Indolmycin Pathway.
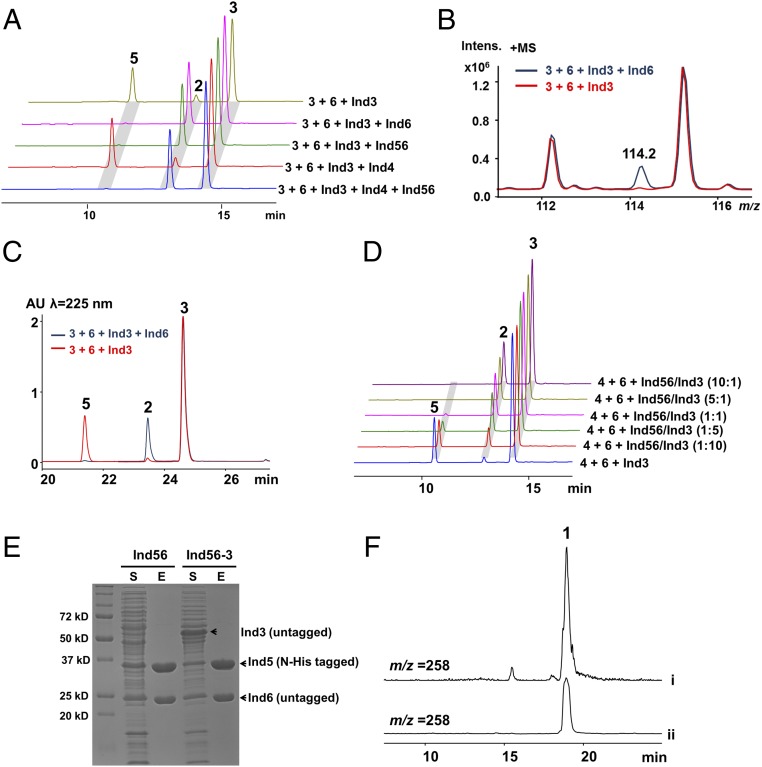
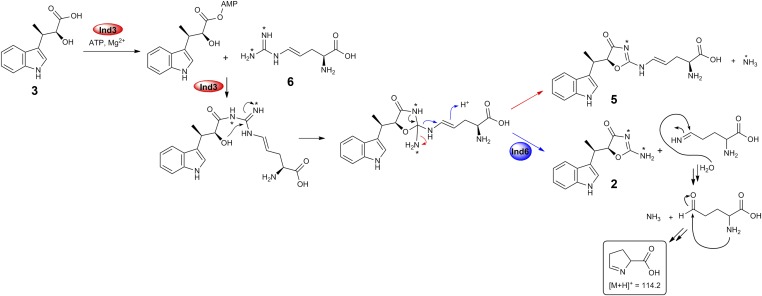
The above results imply that Ind3 is the key enzyme driving the construction of the characteristic oxazolinone ring. Next, we set out to confirm Ind3 as an extending member of PCLs, acting as an unusual ATP-dependent amide synthetase to catalyze the condensation of indolmycenic acid and dehydroarginine. Compounds 3 and 6 (in a mixture with 4) were incubated with Ind3, ATP, and Mg2+. We successfully observed a decrease in 3, 6, and ATP and an increase in 2, 5, and AMP over time (Fig. 4), which supports that 3 is activated by Ind3 as an acyl-adenylate intermediate for subsequent amide bond formation. However, compared with the Ind3–4-5-6-coupled reaction, the ratio of 2 and 5 is altered. The possible involvement of Ind4–5-6 in the Ind3-catalyzed reaction was therefore probed. We separately added Ind4, Ind56, or Ind6 into the Ind3 biochemical assay (Fig. 5A). HPLC analysis revealed that the combination of Ind3 with either Ind6 or Ind56 could alter the ratio of generated 2 and 5, with a dominant production of 2. Collectively, these results demonstrate that Ind3 catalyzes an unusual amide bond formation between 3 and the guanidium–NH2 group of 6, driving oxazolinone ring formation, with the leaving group controlled by Ind6, which prevents the formation of a shunt product 5.
Fig. 4.
Ind3 as an oxazolinone synthetase. (A) LC-MS analysis of the in vitro biochemical reaction of Ind3 at (i) 0 min, (ii) 15 min, (iii) 1 h, and (iv) 2 h. HPLC detection wavelength, 225 nm. (B) Extracted ion chromatogram (EIC) for AMP (m/z 348) from A. (C) EIC for 6 (m/z 173) from A.
Fig. 5.
Ind6 (Ind56) as a gatekeeper determining the ratio of 2 and 5. (A) The involvement of Ind6 in the Ind3-catalyzed oxazolinone ring formation. Incubation of 6 (in a mixture with 4) and 3 with various combinations of enzymes, in the presence of Mg2+and ATP. (B) Identification of the possible leaving group in the Ind3+Ind6 reaction by LC-MS. The ratio of compound (m/z 114.2) production in the Ind3-6 versus Ind3-alone reaction is calculated based on the peak area. (C) Determination of the ratio of 2 in the reaction mixtures of Ind3+Ind6 and Ind3-alone reaction by LC-MS. The ratio of 2 production in the Ind3-6 versus Ind3-alone reaction is calculated based on the peak area at 225 nm. (D) Incubation of 6 (in a mixture with 4) and 3 with various ratios of Ind3/Ind56. (E) Coexpression and purification of Ind5–Ind6 and Ind5–Ind6–Ind3 in Rhodococcus sp. RHA1 (S, soluble fraction; E, elution from nickel–nitrilotriacetic acid column), showing that Ind3 does not form a protein complex with Ind56. (F) In vitro enzymatic total synthesis of indolmycin. EICs for 1 standard compound (i) and ethyl acetate extract from one-pot enzymatic total synthesis of indolmycin (ii).
We then investigated the possible leaving group in the Ind3–Ind6-coupled reaction. We observed an increased mass signal at m/z 114.2 in the Ind3–Ind6-coupled reaction, which was ∼10-fold higher than that seen in the Ind3-alone reaction (Fig. 5B). The ∼10-fold ratio also corresponds to the ratio of 2 production in the Ind3-6 versus Ind3-alone reaction (Fig. 5C). This mass signal (m/z 114.2) is consistent with 1-pyrroline–5-carboxylic acid, which could easily derive from loss of 2-amino–5-iminopentenoic acid, followed by imine hydrolysis, aldehyde formation, and cyclization (Fig. 6) (23). By contrast, the structure of 5 suggests that after amide bond formation, ammonia derived from the guanidinium side chain of dehydroarginine would instead be used as the leaving group when cyclization occurs. To support this hypothesis, l-[guanidino-15N2]arginine was used to replace l-arginine in the biochemical assay. Using LC-HRMS, an increase of 1 Da in mass for 5 was observed (357.1592–358.1581), whereas the mass for 2 increases by 2 Da (244.1084–246.1029) (SI Appendix, Fig. S6 A and B), confirming that one of the 15N-labeled guanidino nitrogen atoms is lost when 5 is formed, whereas in the case of 2 formation, both 15N-labeled guanidino nitrogen atoms stay (Fig. 6).
Fig. 6.
Proposed mechanism of Ind3/Ind6-catalyzed oxazolinone ring assembly. Red arrows indicate the route to 5, and blue arrows indicate the route to 2.
To further test whether 5 is also produced in vivo, LC-MS was applied to search for 5 (m/z 357, [M+H]+) in the culture broth of S. griseus ATCC 12648. However, no 5 production was detected (SI Appendix, Fig. S6C), suggesting the biosynthesis of 1 is tightly controlled in vivo. The partially overlapped coding regions of ind5 and ind6 suggest they are transcriptional-coupled, and the same amount of Ind5 and Ind6 would be produced in vivo. Based on Ind56 complex formation in the recombinant system, Ind6 most likely exists in the form of an Ind56 complex in S. griseus. To test how much Ind56 (compared with Ind3) is needed to block 5 production and whether the ratio of 2 and 5 could be altered by changing the ratio of Ind3 and Ind56, we used different ratios of Ind3 to Ind56 in the biochemical reactions (Fig. 5D). The production level of 2 rises as the ratio of Ind56:Ind3 is increased, with complete abolishment of 5 at a 5:1 ratio. Overall, our above results unveil a remarkable multitasking protein Ind6. Besides its chaperone-like activity with Ind5, Ind6 also serves as a gatekeeper, directing the biosynthesis of indolmycin, preventing the formation of an off-pathway shunt product 5 (Fig. 6). Interestingly, whereas Ind56 form a stable complex in a recombinant system, no complex formation is observed for Ind3–56 (Fig. 5E).
Enzymatic Total Synthesis of Indolmycin.
Having all of the enzymes that contribute to indolmycin production in hand, we performed an in vitro enzymatic total synthesis of 1. All of the enzymes (Orf4725, Ind1, Ind2, Ind3, Ind4, Ind56 complex, and Ind7) were incubated with l-tryptophan and l-arginine in the presence of cofactors [α-KG, PLP, S-adenosylmethionine (SAM), NADH, ATP, and Mg2+]. After incubation overnight, the production of 1 was successfully detected by LC-MS (Fig. 5F).
Discussion
Indolmycin (1) was first reported in 1960 and various synthetic routes to 1 have been reported since then, but the genetic and molecular basis for its biosynthesis, especially the enzymology associated with assembly of the unusual, core oxazolinone ring, have remained largely unknown. In this study, we report identification of the ind cluster responsible for 1 biosynthesis. Through in vivo gene inactivation, in vitro biochemical assays, and enzymatic total synthesis of 1, the biosynthetic pathway of 1 is proposed.
Our work addresses the underlying biosynthetic mechanism for the unique five-membered oxazolinone ring construction, which we have shown derives directly from an l-tryptophan derivative 3 and an l-arginine derivative 6. Both 3 and 6 are first biosynthesized separately by enzymes encoded by genes within and outside the ind cluster. l-tryptophan is first transaminated to yield indolepyruvate, followed by Ind1-catalyzed methyltransfer from SAM to the β carbon of indolepyruvate to produce β-methylindolepyruvate, which is then reduced by Ind2 to give 3. Separately, l-arginine is converted to 6, catalyzed by Ind4 and the Ind56 complex. An ATP-dependent amide synthetase Ind3 then catalyzes an unusual amide bond formation between 3 and 6, and subsequent cyclization (either catalyzed by Ind3 or occurring spontaneously) gives two products, 2 and 5, both of which contain the characteristic oxazolinone ring. The most intriguing observation is the multifunctional protein, Ind6, which has no characterized homolog. Ind6 apparently serves as a gatekeeper, assisting Ind3 to dominantly produce 2, an intermediate in the biosynthetic pathway of 1. Without Ind6, the Ind3-catalyzed reaction generates an off-pathway side product 5 as the major product. Furthermore, Ind6 also has a chaperone-like activity, earlier in the pathway, as it solubilizes Ind5 by forming an Ind5–Ind6 protein complex. However, as no soluble Ind5 alone could be obtained, a catalytic role played by Ind6 in the Ind4–Ind56-coupled reaction leading to 6 cannot be excluded. The last step of the biosynthetic pathway of 1 involves the Ind7-catalyzed transfer of the methyl group of SAM to the amino group of 2, leading to the final product, 1.
As the key enzyme responsible for the oxazolinone heterocycle assembly, Ind3 is predicted to belong to the adenylate-forming domain (AFD) –class-I superfamily (cd04433) (24). Ind3 also shows sequence homology to various phenylacetate–CoA ligases (PCLs). An AMP-binding motif (119TGGSSGTTVRV129) and the sequences 258DEIGSE263 and 306YRQGD310, which are conserved in PCLs and match motifs II and III in acyl-adenylate–forming enzymes (25), can be identified in Ind3. However, PCLs catalyze an ATP-dependent two-step reaction to first activate the carboxylate substrate as adenylates and then transfer the carboxylate to the pantetheine group of CoA. Thus, Ind3 is functionally distinct from PCLs and more similar to a group of amide ligase or synthetase enzymes, such as NovL, McbA, and XimA, involved in the biosynthesis of novobiocin, marinacarbolines, and xiamenmycin, respectively. All of these enzymes activate carboxylates for nucleophilic attack by donor amino groups to bring together monomers (26–28). However, phylogenetic analysis reveals that Ind3 is distinct from those enzymes (SI Appendix, Fig. S7). A more closely related enzyme to Ind3 is DdaG (29), which adenylates fumarate and then uses 2,3-diaminopropionate as a nucleophile, using either the α-NH2 or β–NH2 group, to generate a precursor to the dapdiamides. Ind3 catalyzes instead an unusual amide bond between indolmycenic acid and the NH2 group of the guanidinium side chain of dehydroarginine and also may be responsible for the subsequent oxazolinone ring closure. Although information on the catalytic function of Ind3 can be surmised from our experimental work and bioinformatics analysis, little information is available regarding the mechanism of Ind6 at this stage. The ability of Ind6 to chaperone the soluble expression of Ind5 and, separately, to act as a gatekeeper controlling the outcome of Ind3 catalysis, suggests a highly unusual mechanism of action. To provide insight into the detailed mechanism, structural investigation of Ind6 is currently underway.
In conclusion, we have now established the long-awaited biosynthetic pathway to indolmycin. Our work reveals that oxazolinone synthetase Ind3 catalyzes an unusual amide bond formation between 3 and the guanidium–NH2 group of 6, followed by either catalyzed or spontaneous C–O bond formation to complete the oxazolinone heterocycle. A multitasking protein, Ind6, controls the leaving group during oxazolinone ring formation and thus directs indolmycin biosynthesis. This work sets the stage to use genetic and chemoenzymatic methods to generate indolmycin derivatives and higher-production strains for this unique antibiotic.
Methods
Detailed descriptions of bacterial strains, plasmids, and culture conditions; genomic library construction and genome scanning; generation of Streptomyces mutant strains; metabolic analysis; purification and structure elucidation; cloning, expression, and purification of recombinant proteins; RNA isolation and RT-PCR; in vitro biochemical assays; heterologous production of methylindolpyruvate and indolmycenic acid in E. coli; enzymatic synthesis and isolation of indolmycin B and 4,5-dehydroarginine; and enzymatic total synthesis of indolmycin are provided in the SI Appendix, Methods, and the SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This work was funded by Grand Challenges Canada and the National Sciences and Engineering Council of Canada. Y.-L.D. is supported by a Michael Smith Foundation for Heath Research Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data for the indolmycin gene cluster, orf2651, and orf4725 have been deposited with GenBank (accession nos. KM596502, KP262402, and KM596503).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419964112/-/DCSupplemental.
References
- 1.Marsh WS, Garretson AL, Wesel EM. PA 155 A, B, and X antibiotics produced by a strain of Streptomyces albus. Antibiot Chemother. 1960;10:316–320. [PubMed] [Google Scholar]
- 2.Vynne NG, Månsson M, Nielsen KF, Gram L. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar Biotechnol (NY) 2011;13(6):1062–1073. doi: 10.1007/s10126-011-9369-4. [DOI] [PubMed] [Google Scholar]
- 3.Werner RG, Thorpe LF, Reuter W, Nierhaus KH. Indolmycin inhibits prokaryotic tryptophanyl-tRNA ligase. Eur J Biochem. 1976;68(1):1–3. doi: 10.1111/j.1432-1033.1976.tb10758.x. [DOI] [PubMed] [Google Scholar]
- 4.Kanamaru T, et al. In vitro and in vivo antibacterial activities of TAK-083, an agent for treatment of Helicobacter pylori infection. Antimicrob Agents Chemother. 2001;45(9):2455–2459. doi: 10.1128/AAC.45.9.2455-2459.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurdle JG, O’Neill AJ, Chopra I. Anti-staphylococcal activity of indolmycin, a potential topical agent for control of staphylococcal infections. J Antimicrob Chemother. 2004;54(2):549–552. doi: 10.1093/jac/dkh352. [DOI] [PubMed] [Google Scholar]
- 6.Werner RG, Demain AL. Directed biosynthesis of new indolmycins. J Antibiot (Tokyo) 1981;34(5):551–554. doi: 10.7164/antibiotics.34.551. [DOI] [PubMed] [Google Scholar]
- 7.Sutou N, Kato K, Akita H. A concise synthesis of (−)-indolmycin and (−)-5-methoxyindolmycin. Tetrahedron Asymmetry. 2008;19(15):1833–1838. [Google Scholar]
- 8.Takeda T, Mukaiyama T. Asymmetric total synthesis of indolmycin. Chem Lett. 1980;9(2):163–166. [Google Scholar]
- 9.Hasuoka A, Nakayama Y, Adachi M, Kamiguchi H, Kamiyama K. Development of a stereoselective practical synthetic route to indolmycin, a candidate anti-H. pylori agent. Chem Pharm Bull (Tokyo) 2001;49(12):1604–1608. doi: 10.1248/cpb.49.1604. [DOI] [PubMed] [Google Scholar]
- 10.Akita H, Kawaguchi T, Enoki Y, Oishi T. Formal total synthesis of (-)-indolmycin. Chem Pharm Bull (Tokyo) 1990;38(2):323–328. [Google Scholar]
- 11.Dirlam JP, Clark DA, Hecker SJ. New total synthesis of (±)-indolmycin. J Org Chem. 1986;51(25):4920–4924. [Google Scholar]
- 12.Witty DR, et al. Synthesis of conformationally restricted analogues of the tryptophanyl tRNA synthetase inhibitor indolmycin. Tetrahedron Lett. 1996;37(17):3067–3070. [Google Scholar]
- 13.Shue Y-K. Total synthesis of (±) indolmycin. Tetrahedron Lett. 1996;37(36):6447–6448. [Google Scholar]
- 14.Preobrazhenskaya MN, et al. Total synthesis of antibiotic indolmycin and its stereoisomers. Tetrahedron. 1968;24(19):6131–6143. doi: 10.1016/s0040-4020(01)96345-8. [DOI] [PubMed] [Google Scholar]
- 15.Hornemann U, Hurley LH, Speedie MK, Floss HG. The biosynthesis of indolmycin. J Am Chem Soc. 1971;93(12):3028–3035. doi: 10.1021/ja00741a035. [DOI] [PubMed] [Google Scholar]
- 16.Hornemann U, Hurley LH, Speedie MK, Guenther HF, Floss HG. Biosynthesis of the antibiotic indolmycin by Streptomyces griseus. C-Methylation at the β-carbon atom of the tryptophan side-chain. J Chem Soc Chem Commun. 1969;6:245–246. [Google Scholar]
- 17.Zee L, Hornemann U, Gloss HG. Further studies on the biosynthesis of the antibiotic indolmycin in Streptomyces griseus. Biochem Physiol Pflanzen. 1975;168:19–25. [Google Scholar]
- 18.Speedie MK, Hornemann U, Floss HG. Isolation and characterization of tryptophan transaminase and indolepyruvate C-methyltransferase. Enzymes involved in indolmycin biosynthesis in Streptomyces griseus. J Biol Chem. 1975;250(19):7819–7825. [PubMed] [Google Scholar]
- 19.Vecchione JJ, Sello JK. A novel tryptophanyl-tRNA synthetase gene confers high-level resistance to indolmycin. Antimicrob Agents Chemother. 2009;53(9):3972–3980. doi: 10.1128/AAC.00723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou Y, et al. Stereospecific biosynthesis of β-methyltryptophan from (L)-tryptophan features a stereochemical switch. Angew Chem Int Ed Engl. 2013;52(49):12951–12955. doi: 10.1002/anie.201306255. [DOI] [PubMed] [Google Scholar]
- 21.Koetsier MJ, Jekel PA, van den Berg MA, Bovenberg RAL, Janssen DB. Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem J. 2009;417(2):467–476. doi: 10.1042/BJ20081257. [DOI] [PubMed] [Google Scholar]
- 22.Hiratsuka T, et al. Core assembly mechanism of quinocarcin/SF-1739: Bimodular complex nonribosomal peptide synthetases for sequential mannich-type reactions. Chem Biol. 2013;20(12):1523–1535. doi: 10.1016/j.chembiol.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Vogel HJ, Davis BD. Glutamic γ-semialdehyde and Δ1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline. J Am Chem Soc. 1952;74(1):109–112. [Google Scholar]
- 24.Schmelz S, Naismith JH. Adenylate-forming enzymes. Curr Opin Struct Biol. 2009;19(6):666–671. doi: 10.1016/j.sbi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Said Mohamed M. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J Bacteriol. 2000;182(2):286–294. doi: 10.1128/jb.182.2.286-294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, et al. Characterization of the xiamenmycin biosynthesis gene cluster in Streptomyces xiamenensis 318. PLoS ONE. 2014;9(6):e99537. doi: 10.1371/journal.pone.0099537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steffensky M, Li S-M, Heide L. Cloning, overexpression, and purification of novobiocic acid synthetase from Streptomyces spheroides NCIMB 11891. J Biol Chem. 2000;275(28):21754–21760. doi: 10.1074/jbc.M003066200. [DOI] [PubMed] [Google Scholar]
- 28.Ji C, et al. Chemoenzymatic synthesis of β-carboline derivatives using McbA, a new ATP-dependent amide synthetase. Tetrahedron Lett. 2014;55(35):4901–4904. [Google Scholar]
- 29.Hollenhorst MA, Clardy J, Walsh CT. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry. 2009;48(43):10467–10472. doi: 10.1021/bi9013165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.