Significance
Familial dysautonomia (FD) is caused by missplicing of the IκB kinase complex-associated protein (IKAP) gene, which results in the skipping of exon 20, especially in neurons. FD would be treatable if exon 20 inclusion were increased correctly to reestablish correct splicing. Here, we have established a dual-color splicing reporter that recapitulates FD-type splicing. By using this reporter, we have identified a small chemical compound, named rectifier of aberrant splicing (RECTAS), that rectifies the aberrant splicing of FD. RECTAS promotes both exon 20 inclusion and the product IKAP expression in cells of patients with FD. Furthermore, we have demonstrated that modification levels of wobble uridine residues of several tRNAs are reduced in FD cells and that RECTAS can recover not only tRNA modifications but also cell viability of FD cells.
Keywords: RNA disease, splicing, small molecules, tRNA modification, neurodegenerative disease
Abstract
Familial dysautonomia (FD), a hereditary sensory and autonomic neuropathy, is caused by missplicing of exon 20, resulting from an intronic mutation in the inhibitor of kappa light polypeptide gene enhancer in B cells, kinase complex-associated protein (IKBKAP) gene encoding IKK complex-associated protein (IKAP)/elongator protein 1 (ELP1). A newly established splicing reporter assay allowed us to visualize pathogenic splicing in cells and to screen small chemicals for the ability to correct the aberrant splicing of IKBKAP. Using this splicing reporter, we screened our chemical libraries and identified a compound, rectifier of aberrant splicing (RECTAS), that rectifies the aberrant IKBKAP splicing in cells from patients with FD. Here, we found that the levels of modified uridine at the wobble position in cytoplasmic tRNAs are reduced in cells from patients with FD and that treatment with RECTAS increases the expression of IKAP and recovers the tRNA modifications. These findings suggest that the missplicing of IKBKAP results in reduced tRNA modifications in patients with FD and that RECTAS is a promising therapeutic drug candidate for FD.
The inhibitor of kappa light polypeptide gene enhancer in B cells, kinase complex-associated protein (IKBKAP) gene encodes the 150-kDa IKK complex-associated protein (IKAP). IKAP is currently known as elongator protein 1 (ELP1), an integral component of the human Elongator complex, which was originally identified in Saccharomyces cerevisiae and shown to be well conserved among species (1). Although multiple functions of IKAP/ELP1 in JNK signaling, neuronal development during embryogenesis, exocytosis, and actin cytoskeleton regulation have been reported (reviewed in refs. 2, 3), yeast genetic analyses have shown that the Elongator complex is also required for the formation of the C5-substituent of 5-carbamoylmethyl (ncm5), 5-methoxycarbonylmethyl (mcm5), and its derivatives at the wobble uridine in tRNAs recognizing purine-ending codons (4, 5). Most recently, it was demonstrated that conditional IKAP/Elp1 KO in mouse testes results in male infertility by disrupting meiotic progression, along with the reduction of modified nucleosides [5-methoxycarbonylmethyl uridine (mcm5U), 5-carbamoylmethyl uridine (ncm5U), and 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U)] of total tRNAs in the testes (6). These modifications are highly likely to play critical roles in the maintenance of translational fidelity, suggesting that the defects in these modifications lead to the mistranslation of various proteins.
Familial dysautonomia (FD; Riley–Day syndrome), an autosomal recessive neurodegenerative disease, is characterized by impaired development and progressive degeneration of the sensory and autonomic nerves. Patients who have FD exhibit various symptoms, including cardiovascular instability, recurrent pneumonia, vomiting/dysautonomic crisis, gastrointestinal dysfunction, decreased sensitivity to pain and temperature, and defective lacrimation. FD is a very common disorder in the Ashkenazi Jewish population, with a carrier frequency of 1 in 27. More than 99% of patients who have FD harbor a homozygous mutation in intron 20 (IVS20 + 6T > C: FD mutation) of IKBKAP (7). This mutation reduces base pairing with U1 small nuclear ribonucleic protein (snRNP), resulting in the skipping of exon 20 (8, 9), which, in turn, causes a frameshift and the generation of a premature termination codon (PTC) in exon 21 of IKBKAP mRNA (10). Interestingly, this mutation does not completely abolish the inclusion of exon 20 in pre-mRNA splicing; indeed, WT mRNA is expressed in patients who have FD. The skipping ratio of exon 20 varies among different tissues in patients with FD, with the lowest production of exon 20-containing IKBKAP mRNA observed in neuronal tissues, which is a likely cause of FD (10). These findings suggest that splicing of IKBKAP and IKAP expression could potentially be manipulated, offering the promise of therapeutic approaches. Several attempts have been made to search for therapeutic chemical compounds that promote exon 20 inclusion in patients with FD by quantifying RT-PCR analysis in patient cells or reporter-transfected cells (11–16). Even with the most potent splicing modifier, the previously identified plant cytokinin kinetin, however, the effect is not sufficiently strong or specific to promote exon 20 inclusion in IKBKAP (15, 16).
We have developed a dual-color splicing reporter system combining two different fluorescent proteins. By using this system, we have succeeded in identifying both cis-elements and trans-acting factors of alternative splicing events in worms, mice, and cultured cells (17, 18). Other groups have also prepared single construction-based, dual-color splicing reporters with specific genes independently (19, 20). These reports highlight the advantage of a dual-color splicing reporter for studying splicing. In the present study, we applied our dual-color splicing reporter system to screen small chemicals that could affect the aberrant splicing of IKBKAP. Our system, which we named the splicing reporter assay for disease genes with dual color (SPREADD), recapitulated and visualized both normal and aberrant splicing patterns in cultured cells. Using our newly developed SPREADD, we screened chemical libraries and found a small molecule that corrects the abnormal splicing of the IKBKAP gene. This molecule was also able to increase IKAP expression in fibroblasts derived from patients with FD at much lower concentrations than the concentration at which kinetin is active. Furthermore, this molecule, named rectifier of aberrant splicing (RECTAS), recovered the level of ncm5U, mcm5s2U, and 5-(carboxyhydroxymethyl) uridine methyl ester (mchm5U) in wobble positions of tRNAs that are typically reduced in cells of patients with FD.
Results
Visualization of FD-Type Abnormal Splicing of IKBKAP Pre-mRNA in Cultured Cells.
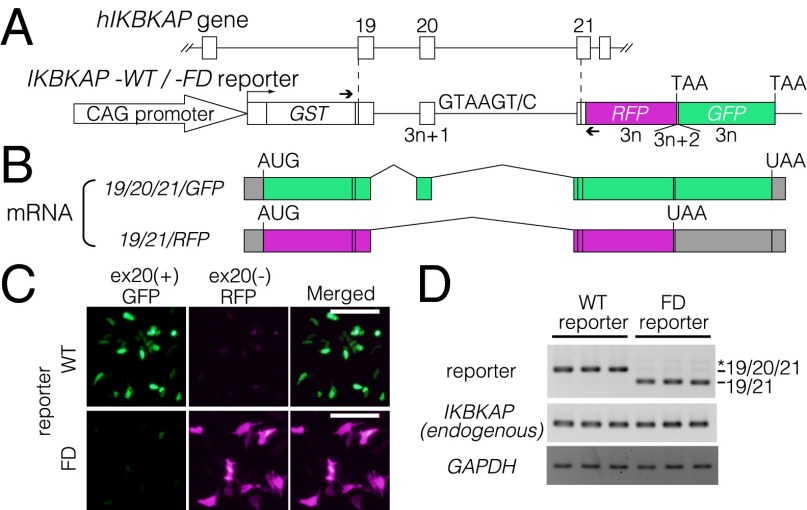
To identify small molecules that efficiently correct aberrant splicing in the IKBKAP gene of patients who have FD, we constructed a dual-color fluorescence splicing reporter that reflects the splicing of exon 20. In the WT reporter construct, the human IKBKAP gene fragment spanning exon 19 to exon 21 was fused upstream of the cDNAs of the fluorescent proteins monomeric red fluorescent protein (mRFP) and enhanced green fluorescent protein (EGFP). These cDNAs encoding the fluorescent proteins were fused to each other in tandem with different ORFs (Fig. 1A). We also prepared another IKBKAP reporter plasmid that carries a point mutation equivalent to the FD mutation (Fig. 1A). With this reporter system, the normal-type splicing (exon 20 inclusion) leads to expression of a GFP-fusion protein. In this case, the RFP cDNA portion is translated with a frame different from the original RFP protein without any stop codons. On the other hand, FD-type abnormal splicing (exon 20 exclusion) results in the production of an RFP fusion protein and the ORF is discontinuous to GFP cDNA (Fig. 1B). To evaluate whether our human IKBKAP (hIKBKAP) splicing reporter recapitulates abnormal splicing in patients with FD, we introduced these reporters into human neuroblastoma SH-SY5Y cells. When the WT reporter was transfected, the GFP signal was predominantly observed, whereas the RFP signal was clearly detected with the FD-type splicing reporter as expected (Fig. 1C). We confirmed the splicing patterns of the reporters by RT-PCR, and observed that exon 20 was predominantly included with the WT but almost entirely excluded with the FD type (Fig. 1D), whereas the splicing pattern of endogenous IKBKAP pre-mRNA was the exon 20-inclusion type in this cell line. Thus, our WT- and FD-type reporters reflect normal- and abnormal-type splicing, respectively, and the splicing reporter system was named the SPREADD.
Fig. 1.
Visualization of disease-specific splicing using the SPREADD system. (A) Schematic representation of the IKBKAP splicing reporter. Primers used to amplify transcripts of the IKBKAP reporter are indicated by arrows. (B) mRNAs derived from the reporters are shown schematically. The predicted ORFs are indicated in green and magenta for exon 20 inclusion and exclusion, respectively. (C) Microscopic analysis of cells of SH-SY5Y cells expressing either the IKBKAP-WT reporter (Top) or IKBKAP-FD reporter (Bottom). Projection images of 19/20/21-GFP, 19/21-RFP, and merged images of the same fields are shown. (Scale bars: 100 μm.) (D) RT-PCR analysis of mRNAs derived from SH-SY5Y cells expressing the IKBKAP-WT reporter or IKBKAP-FD reporter. An asterisk indicates the PCR product corresponding to a hybrid of 19/20/21 and 19/21.
Identification of a Small Molecule That Improves Exon 20 Inclusion by the SPREADD.
Using the SPREADD, we searched for chemical compounds that promoted the inclusion of exon 20 in the presence of the FD mutation. We used HeLa cells for the screening because the transfection efficiency was high and our splicing reporters worked similar to their pattern observed in SH-SY5Y cells (Fig. S1). We screened 638 small molecules in our chemical libraries for potential splicing modifiers and pharmaceuticals approved by the US Food and Drug Administration, using kinetin as a positive control. In this screening, we identified a small molecule, which we named RECTAS (Fig. 2A). The potent effect of RECTAS on exon 20 inclusion was confirmed by fluorescence microscopy (Fig. 2B). RECTAS potently promoted exon 20 inclusion, and 2 μM RECTAS had an effect equivalent to kinetin at 50 μM (Fig. 2C).
Fig. 2.

RECTAS, a small molecule that corrects aberrant splicing of FD in the SPREADD. (A) Structure of RECTAS. (B) Microscopic analysis of HeLa cells expressing the IKBKAP-FD reporter treated with the indicated small molecules. (Scale bar: 100 μm.) (C) Quantification of GFP/RFP ratios in HeLa cells expressing the IKBKAP-FD reporter. After 6 h of transfection, cells were treated with each compound at the concentrations described. After a 24-h incubation, the GFP/RFP ratio was quantified. (D) In vitro splicing of 32P-labeled CDC-IKBKAP Ex20 pre-mRNA in HeLa nuclear extract with DMSO or RECTAS (20 μM). (Left) RNAs at each time point were analyzed by denaturing PAGE and autoradiography. (Right) Identities of the pre-mRNA and mRNA are schematically shown. Quantitation of the inclusion ratio to the sum of inclusion and exclusion is normalized with WT DMSO in each experiment. Values are the mean ± SEM (n = 3). P = 0.003438 for FD mutation. (E) In vitro splicing of 32P-labeled IKBKAP Ex20–21 pre-mRNA as in D. (F) Active spliceosomal complex formation on 32P-labeled IKBKAP Ex20–21 pre-mRNA. Complexes at each time point were resolved by native gel electrophoresis. Quantitation of the A-complex ratio to the sum of the A-complex and E-complex is normalized with WT DMSO in each experiment. Values are the mean ± SEM (n = 3). (G) Exon-defining E-complex formation on 32P-labeled IKBKAP int-Ex20-int pre-mRNA. The E-complex at each time point was resolved and quantified as in F. The arrow indicates the E-complex. In D–G, RNA substrates for the assay are illustrated with bold lines and boxes representing IKBKAP sequence (Top).
To test whether RECTAS has a direct role in splicing regulation, it was directly added to HeLa nuclear extract for an in vitro splicing assay with pre-mRNA harboring the FD mutation. We used the chicken δ-crystallin (CDC) pre-mRNA, which has conventionally been used for in vitro splicing (21). The effect of the FD mutation and RECTAS would be recapitulated in this heterologous system if the potential target of RECTAS resides in exon 20 and the flanking intron sequence. As shown in Fig. 2D, exon inclusion of the mutant pre-mRNA was drastically reduced (14/20/15 bands in lanes 10–12) compared with exon inclusion of the WT pre-mRNA (lanes 2–4) when exon 20 with the flanking sequence was placed in the intronic (int) region of the CDC pre-mRNA (CDC-IKBKAP Ex20 pre-mRNA). Preincubation of HeLa nuclear extracts with 20 μM RECTAS before the splicing reaction increased inclusion of the exon upstream of the FD mutation at a rate of approximately twice the rate of the DMSO control [Fig. 2D, compare lane 12 with lane 16, and FD (Right)], whereas the increase of the WT-accompanied exon was smaller and not significant [Fig. 2D, compare lane 4 with 8, and WT (Right)]. These results indicate that RECTAS partially restores the splicing defect through direct regulation of the splicing reaction. Because the mutation affects position +6 of the 5′ splice site (ss) of intron 20, we constructed a pre-mRNA with a sequence flanking this 5′ ss and downstream 3′ ss. These pre-mRNAs (IKBKAP Ex20-21WT and IKBKAP Ex20-21FD), with a shortened intron 20, also showed a partial but direct effect of RECTAS on the mutant when assessed by in vitro splicing [Fig. 2E, compare 20/21 bands in lanes 9 and 12, and FD (Right)]. To elucidate the target of RECTAS, active spliceosomal complexes defined as A, B, and C were analyzed using this pre-mRNA. The A-complex is the early, active prespliceosomal complex, which is formed following the exon-defining E-complex by assembly of U2 snRNP onto the pre-mRNA defined by U1 snRNP and associated factors (22). As shown in Fig. 2F, a short time course showed that RECTAS readily increased the formation of the A-complex, which was attenuated by the mutation [compare the broad bands indicated as A in lanes 9 and 12, and FD (Right)]. Next, we analyzed the effect of RECTAS on E-complex formation using pre-mRNAs of exon 20 with a flanking int sequence (IKBKAP int-Ex20-int WT and IKBKAP int-Ex20-int FD). As shown in Fig. 2G, RECTAS also increased E-complex formation of the mutant pre-mRNA [compare the bands indicated as E in lanes 8 and 9, and FD (Right)], suggesting that RECTAS promotes exon definition in the context of pre-mRNA harboring the FD mutation. It was reported that exon 20 of the IKBKAP gene contains two exonic splicing silencers (ESSs) and that the upstream pyrimidine tract is weak; therefore, exon 20 is poorly recognized (9). Taken together, these results strongly suggest that RECTAS likely affects recognition of exon 20 through promoting U1 snRNP association with the 5′ ss.
RECTAS Promoted Exon 20 Inclusion of Endogenous IKBKAP Pre-mRNA and Increased IKAP Expression in Cells of Patients with FD.
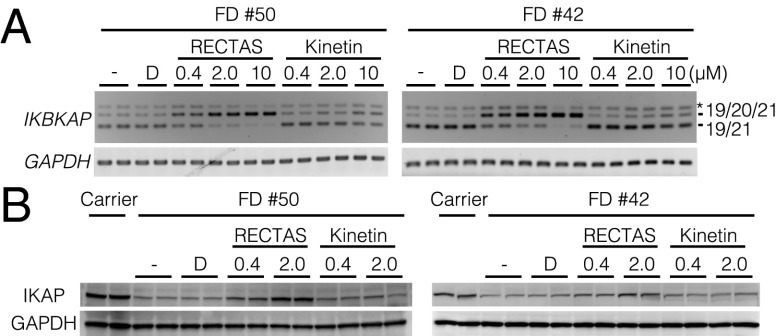
We next examined the effect of RECTAS on splicing of the endogenous IKBKAP pre-mRNA in FD patient fibroblasts (nos. 42 and 50; Coriell Cell Repositories). In cells from both patients, the exon 20-skipped form of IKBKAP mRNA was dominant, as detected by RT-PCR (Fig. 3A, 19/20 in lane D). When RECTAS was added to this cell line, exon 20 inclusion was promoted in a dose-dependent manner at concentrations ranging from 0.4 to 10 μM (19/20/21 in lanes 0.4, 2.0, and 10), whereas the effect of kinetin was seen at concentrations of 10 μM. Even in the carrier cells, RECTAS promoted the inclusion of exon 20 in a dose-dependent manner (Fig. S2). We then examined whether RECTAS increases IKAP expression in the two FD patient cell lines used in Fig. 3A. Immunoblotting with a specific antibody against IKAP demonstrated that treatment with RECTAS increased the level of IKAP in both patient cell lines in a dose-dependent manner. RECTAS at 2 μM recovered the level of IKAP to levels of IKAP in the cell line from the FD carrier who had no clinical symptoms of FD, whereas kinetin showed no effect at 2 μM (Fig. 3B). We then examined the kinetics after oral administration of RECTAS or kinetin to mice, and found that the blood plasma concentration of RECTAS was higher than the blood plasma concentration of kinetin at all of the time points examined (Fig. S3A) and that the concentration in the brain reached the effective dose (∼0.4 μM) required to correct abnormal IKBKAP splicing (Fig. S3B, Bottom). These results indicate that RECTAS is a promising therapeutic compound for patients with FD.
Fig. 3.
Promotion of exon 20 inclusion with endogenous IKBKAP pre-mRNAs and IKAP expression in cells of patients with FD by treatment with RECTAS. (A, Top) RT-PCR analyses of endogenous IKBKAP mRNAs. (A, Bottom) GAPDH was used as a control in fibroblasts from FD patients #50 (Left) and #42 (Right), respectively. The asterisk indicates a band corresponding to a hybrid of 19/20/21 and 19/20. (B, Top) Immunoblot analyses of endogenous IKAP expression. (B, Bottom) GAPDH was used as a loading control in fibroblasts from FD patients #50 and #42. D, DMSO control; -, medium change only.
Characterization of RECTAS-Responding Exons.
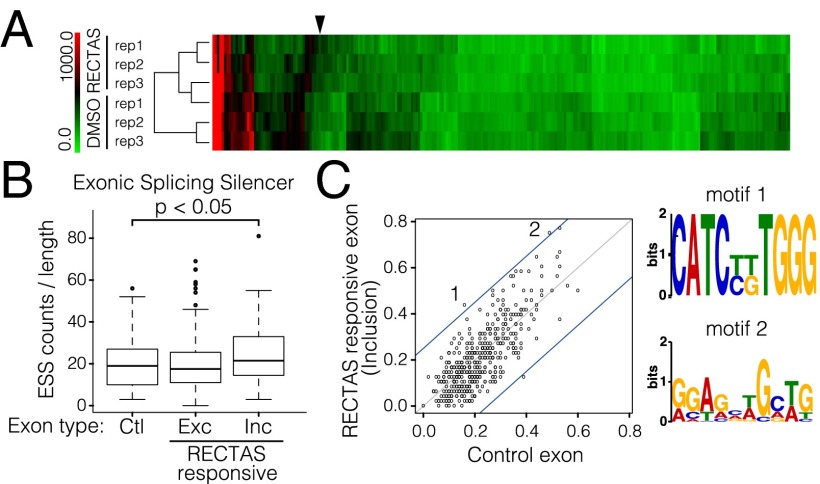
Next, we evaluated the effect of RECTAS on the transcriptome by exon array. To avoid the secondary effect, we extracted total RNAs from the cells of patient no. 42 after a short incubation (6 h) with either DMSO or 2 μM RECTAS. At this time point, the expression level of IKAP was unchanged (Fig. S4A). We found the inclusion-type alteration for 51 exons, including IKBKAP exon 20, and the exclusion-type alteration for 199 exons. The heat map with the relative expression values (SI Materials and Methods) of the exons showed that the magnitude of splicing alteration induced by RECTAS treatment was limited to certain genes (Fig. 4A). Indeed, scatter plots showing the expression values of each exon of RECTAS vs. DMSO treatments indicated that exons affected by RECTAS were a very minor fraction of the entire exon pool (Fig. S4B and detailed in SI Results).
Fig. 4.
Target exons and sequences of RECTAS. (A) Exon array analysis of transcripts in fibroblasts from FD patient no. 42 treated with DMSO or RECTAS. A heat map represents 250 exons differentially expressed in RECTAS-treated cells compared with DMSO-treated cells. The arrowhead indicates IKBKAP exon 20. (B) Tukey box plots show count comparisons of ESS per 100 bases of exon sequences. Each box represents the interquartile range (IQR) from the top (first quartile: Q1) to the bottom (third quartile: Q3), with the whiskers defined as Q1 + 1.5 × IQR to Q3 − 1.5 × IQR. Horizontal lines on the boxes show the median values. Values outside the regions were plotted as individual circles (●). (C) Comparisons of fractions of exons with sequence motifs between inclusion-type exons and control exons. The gray line shows Y = X, whereas the blue lines show Y = X + 0.25 and Y = X − 0.25, where 0.25 is the threshold to define a motif as a differential motif between the datasets. (Right) Sequence logos for motifs enriched in inclusion-type exons are shown.
We next searched for specific sequence features in the exons responsive to RECTAS; 51 for inclusion type and 199 for exclusion type. We used SpliceAid (23) to find known splicing regulatory cis-elements and multiple expectation-maximization for motif elicitation (MEME) (24) algorithm to find the enrichment or depletion of novel sequence motifs compared with a control exon set (SI Materials and Methods). In the analyses with SpliceAid, we found an enrichment of ESSs in a group of inclusion-type RECTAS-responding exons compared with the control set exons (Fig. 4B). In contrast, we did not find clear differences for the exclusion-type exon set (Fig. S4D and detailed in SI Results). These results indicate that the exon recognition of the exons responsive to RECTAS is potentially weakened by ESSs in the exon. For further analysis, we searched for motifs with MEME and identified 450 motif candidates from the inclusion-type, the exclusion-type, and the control exon sets. We counted exons with the motifs for each dataset and compared the frequencies of the motifs among these datasets. We found that three motifs have largely different frequencies (≥0.25) between the inclusion-type and control exon sets, and that two motifs were enriched in the inclusion-type exon set (Fig. 4C). However, we could not find motifs with clear differences in the frequencies between the exclusion-type set and the control set (Fig. S4E). The two motifs of the inclusion-type exons, named motif 1 and 2 in Fig. 4C, were similar to a part of the heterogeneous nuclear ribonucleoprotein (hnRNP) H1 binding motif, GAUCACUGGGGUGGAUCAUCCAGGUGGGGCUUUU, and hnRNP H2 binding motif, GGGGGAGGUGUGGG, respectively. These results suggest that RECTAS specifically restores the inclusion of weak exons whose recognition is suppressed by hnRNP H1/H2, although the detailed molecular mechanism remains to be elucidated.
Hypomodified Wobble Bases in Cytoplasmic tRNAs from FD Patient Fibroblasts.
The Elongator complex has been shown to be required for the formation of mcm5 and ncm5 at wobble uridines in yeast, plants, and worms (4, 5, 25–27). More recently, nucleoside analysis of tRNA derived from IKAP/Elp1 conditional KO mice indicated that mcm5U, ncm5U, and mcm5s2U nucleosides were reduced in total tRNA from the testes of the mice (6). However, tRNA modification at the wobble position of individual tRNAs was not analyzed in Elongator-deficient mammals, and whether tRNA modifications are altered in patients with FD has remained elusive. To determine whether this function of IKAP/Elp1 is conserved in humans, and whether the tRNA modification in wobble uridine is reduced in the cells of patients with FD, we took advantage of the reciprocal circulating chromatography method to isolate individual tRNA species from total RNA (28) (Fig. 5A, Left). Using this method, various cytoplasmic tRNA species were simultaneously isolated from a common source of RNAs under the same conditions (Fig. S5A). After the isolation of individual tRNAs, each tRNA was digested with RNase T1, and the fragments were subsequently analyzed by liquid chromatography/MS measurement to determine the modification status. We isolated tRNAs containing a wobble uridine, including tRNAVal(UAC), tRNAArg(UCU), and tRNAGly(UCC), from cells of patients with FD as well as from FD carrier cells. As shown in Fig. 5B, we found that human tRNAVal(UAC) from FD carrier cells mainly contained ncm5U at the wobble position, which was confirmed by collision-induced dissociation of the anticodon-containing fragment (Fig. S5B). In cells of patients with FD, however, the wobble uridine in tRNAVal(UAC) was hypomodified (Fig. 5B). Similarly, we found that tRNAArg(UCU) and tRNAGly(UCC) from FD carrier cells were fully modified with mcm5s2 and mchm5 at their wobble uridine, respectively, but the wobble uridine in these tRNAs from cells of patients with FD was hypomodified (Fig. 5 C and D and detailed in SI Results). We then tested whether the expression level of these tRNAs was altered in cells of patients with FD by Northern blotting analyses of individual tRNAs. We found that the level of these tRNAs was indeed up-regulated to various extents in FD patient cells (Fig. S5C), implicating the occurrence of dosage compensation to overcome the low fidelity of codon recognition in FD cells. Additionally, we analyzed tRNAThr(UGU) and found that most of the wobble-modified uridine of tRNAThr(UGU) seemed to be ncm5U. Consistent with Val, Arg, and Gly, the frequency of this modification is reduced in the cells of patients with FD (Fig. S5E and detailed in SI Results). These results demonstrate that human IKAP/Elp1 is involved in the biogenesis of the wobble uridine modification (ncm5U, mcm5s2U, and mchm5U) of cytoplasmic tRNAs. In addition, the hypomodification at the wobble positions was observed, at least in tRNAs for Val, Arg, Gly, and Thr prepared from the cells of patients with FD, which express reduced levels of IKAP/Elp1.
Fig. 5.
Analysis of modifications at the first nucleotide of the anticodons in human tRNAVal(UAC), tRNAArg(UCU), and tRNAGly(UCC) from both FD carrier and patient cells. (A, Left) Scheme for tRNA modification analysis of the anticodon loop using reciprocal circulating chromatography (RCC) and liquid chromatography/mass spectrometry (LC/MS) measurements. AEC, anion exchange chromatography. (A, Right) ELPs are likely involved in the step for ncm5U synthesis. Modified residues are highlighted by the colors consistent with those colors used for chromatograms and histograms shown in B–D. (B–D, Top) Secondary structures and sequences at the anticodon stem–loop region of indicated tRNAs are shown. The wobble uridine residue of each tRNA is indicated with possible modifications. The reported modifications m3C and t6A near the anticodon are also indicated. The sequences indicated in bold letters are the RNase T1 fragments harboring the anticodons. (B–D, Middle) LC/MS analyses of the RNase T1 fragments of each tRNA from FD carrier (Carrier) and patient (#42 and #50) cells are shown. Each arrow with the number corresponding in Table S2 indicates the position of the peak of the mass chromatogram of each RNase T1 fragment. All sequences of RNase T1 fragments analyzed by LC/MS are listed in Table S2. (B–D, Bottom) Histograms show the percentage of modifications in total uridine residues at the wobble position for each tRNA.
RECTAS Restores tRNA Modification at the Wobble Position in Cells of Patients with FD.
The observation that hypomodification at the wobble positions of four cytoplasmic tRNAs led us to examine whether RECTAS treatment could restore the frequency of the tRNA modifications in cells of patients with FD. For these experiments, cells from patient no. 42 were cultured for 1 d after plating and then treated with either 0.02% DMSO as a control or 2 µM RECTAS for 4 d before the extraction of total RNA. Those four species of tRNA were isolated and analyzed (Fig. 6 and Fig. S6). Under this culture condition, the frequency of wobble modifications in cells from patient no. 42 cultivated in the presence of DMSO was lower than the frequency of wobble modifications of the carrier cells, as observed in Fig. 5 B–D (i.e., 20% of ncm5U in tRNAVal, 47% of mcm5s2U in tRNAArg, and 68% of mchm5U in tRNAGly). When this cell line was treated with RECTAS, the frequency of wobble modifications for these tRNA species was restored remarkably (Fig. 6 and Fig. S6 B–E). Dosage compensation of this tRNA was also observed (Fig. S6F). These results demonstrate that RECTAS treatment increased the steady-state level of IKAP/ELP1 through rectifying the aberrant splicing of IKBKAP pre-mRNAs harboring an FD-causing mutation, and then recovered the reduced wobble modifications of cytoplasmic tRNAs. Finally, we examined the effect of RECTAS on the growth of FD cells because cells from patient no. 42 exhibited lower cell growth than carrier cells. When RECTAS was added to the culture medium at 10 μM, the growth of cells from patient no. 42 improved to a level similar to the level of the carrier cells (Fig. 6B).
Fig. 6.
Recovery of uridine modification of tRNAs by RECTAS in cells of patients with FD. (A) LC/MS analyses of RNase T1 fragments of tRNAs obtained from the cells of FD patient #42 treated with either DMSO (−) or RECTAS (+).The histograms for modification percentages obtained from the mass chromatograms of each tRNA are presented. (B) Cell growth of carrier cells and cells of FD patient #42 with or without RECTAS treatment. Shown is a representative experiment repeated two times. Values are the mean ± SEM (n = 6).
Discussion
In this study, we used the dual-color splicing reporter of mutated IKBKAP to identify modulators of the aberrant splicing that causes the hereditary disease FD. Our dual-color reporter system, the SPREADD, identified a small molecule named RECTAS that promotes exon 20 inclusion of IKBKAP pre-mRNA and expression of IKAP. A plant cytokinin, kinetin (6-furfurylaminopurine) was shown to rescue the splicing abnormality and restore normal IKAP levels in FD fibroblasts and transformed lymphoblast lines (15). Animal studies showed that kinetin is well absorbed orally and is distributed into plasma and the CNS, but the therapeutic daily dose was estimated to be over 1 g for humans (29). The newly identified RECTAS was shown to be ∼25-fold more potent than kinetin for the promotion of the exon 20 inclusion of IKBKAP in FD cell lines (Fig. 3A). RECTAS directly affected mRNA splicing (Fig. 2 D–G), and our transcriptome analyses revealed that RECTAS affected the splicing of only a limited set of genes, suggesting it has high specificity and a direct role in correcting IKBKAP aberrant splicing (Fig. 4A and Fig. S4). Sequence analysis of the exons responsive to RECTAS suggests that RECTAS promotes exon recognition through either inhibiting splicing suppressors, such as hnRNPs, or activating splicing enhancers. Because we do not yet know the direct target of RECTAS in splicing, we cannot distinguish between these possibilities. Further experiments, including identification of the direct target of RECTAS, are required. RECTAS was rapidly absorbed after oral administration, was more stable in the blood plasma than kinetin (Fig. S3A), and was transported into the brain at sufficient levels to reach the effective dose (Fig. S3B), suggesting that RECTAS has therapeutic potential in FD. Artificial splicing rectification with RECTAS may offer an unexplored direction for pharmacological intervention in other hereditary diseases, such as Duchenne muscular dystrophy (30).
tRNAs from all organisms contain modified nucleosides, and a uridine present in the wobble position (U34) is almost universally modified (31, 32). Accumulating evidence indicates that the Elongator complex, including ELP1, also plays a pivotal role in the regulation of translation, in addition to participating in transcriptional regulation, through the formation of ncm5 and mcm5 side chains on the wobble uridines of tRNAs (5, 25–27). These modified wobble uridines stabilize codon–anticodon interactions and increase the efficiency of decoding of A- and G-ending codons during translation, as reported in S. cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, and Arabidopsis thaliana (5, 25, 26, 33), and uridine modifications served by ELPs are required for translation of specific mRNAs, which are highly expressed upon stress conditions (34). Several studies have shown that uridine modification is involved in some neurodegenerative diseases, such as ALS and mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (reviewed in refs. 35, 36). A direct link between FD pathogenesis and tRNA modification remains elusive, although a reduction in tRNA modifications was observed in the testes of IKBKAP KO mice, which led to male infertility (6). We showed here for the first time, to our knowledge, that the levels of ncm5, mcm5s2, and mchm5 side-chain formation of tRNAs were lower in cells of patients with FD than in control cells (Fig. 5 B–D and Fig. S5E), implying that the reduced expression of IKAP in patients who have FD results in the down-regulation of tRNA modification. Furthermore, we succeeded in recovering tRNA modification and cell growth by treatment with RECTAS (Fig. 6), which rectifies the aberrant splicing of IKBKAP pre-mRNA. Continued therapeutic research on RECTAS using FD model mice and FD-induced pluripotent stem cells should confirm the role of tRNA modification in the pathological phenotype of FD and pave the way to the first clinical drug for patients who have FD.
Materials and Methods
Reporter Construction.
We constructed the IKBKAP splicing reporter vectors as described in Fig. 1. A human IKBKAP genomic DNA fragment spanning exon 19 to exon 21 was amplified and cloned into the Gateway Destination vector (Invitrogen), carrying both EGFP (Clontech) and mRFP with different reading frames under the control of the cytomegalovirus enhancer/chicken β-actin hybrid (CAGGS) promoter, as previously reported (18). To adjust the reading frame to the reading frame of RFP/GFP, we introduced a single nucleotide deletion into exon 20 in the reporter. We introduced the FD mutation using the QuikChange method (Stratagene). Sequences of primers used in the construction are listed in Table S1.
Cell-Based Screening of Small Chemical Compounds.
We reversely transfected HeLa cells with the hIKBKAP-FD reporter and plated them on a 96-well plate. At ∼4–6 h after transfection, chemical compounds were added to the cells. After 24 h of incubation, the cells were fixed and stained with Hoechst, as described in SI Materials and Methods (Analysis of Transfected Cells). After staining the nucleus, cells were kept in PBS and visualized using a Cellomics ArrayScan VTi (Thermo Fisher Scientific). Fluorescence quantification in cell-based screening of small chemical compounds is described in SI Materials and Methods.
Liquid Chromatography/MS Analysis of tRNAs.
Details about total RNA preparation and isolation of each tRNA are described in SI Materials and Methods. One picomole of isolated tRNA was digested with RNase T1, and the digest (0.5 pmol) was analyzed using a linear iontrap–orbitrap hybrid mass spectrometer equipped with a custom-made nanospray ion source, a splitless nano-HPLC system, a C18 trap cartridge, and a C18 capillary column. More information is provided in SI Materials and Methods.
Cells from patients with FD, cell culture, splicing substrate construction, semiquantitative RT-PCR, small chemical compounds, in vitro splicing and complex formation assay, single-dose oral administration, immunoblot, expression analysis, sequence analysis, Northern blotting, and cell viability assay are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the members of the M.H. laboratory for experimental assistance, helpful discussions, and comments on this manuscript. Chemical screening was performed at the Medical Research Support Center (Kyoto University). We are grateful to Maki Sakuma for critical reviews prior to submission, as well as to anonymous reviewers whose comments helped improve the final manuscript. We also thank Dr. Akira Kakizuka for support and encouragement. This work was supported by Grants-in-Aid for Scientific Research (Grant 21249013 to M.H. and Grant 23112706 to N.K.); the Research Program of Innovative Cell Biology (Grant 231006 to M.H.); the Platform for Drug Discovery (M.H.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; the National Institute of Biomedical Innovation (Grant 241011 to M.H.); and Create Revolutionary Technological Seeds for Science and Technology Innovation (CREST) of the Japan Science and Technology Agency (Grant 231038 to M.H.).
Footnotes
Conflict of interest statement: S.Y., T.H., and M.H. are coinventors on pending patent applications based on this work (Japanese patent application no. 2013-146891).
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession no. GSE58038 (transcriptome analysis of the effect of RECTAS on fibroblast cells derived from a familial dysautonomia patient)].
See Commentary on page 2637.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415525112/-/DCSupplemental.
References
- 1.Hawkes NA, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277(4):3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 2.Svejstrup JQ. Elongator complex: How many roles does it play? Curr Opin Cell Biol. 2007;19(3):331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen L, Humbert S, Saudou F, Chariot A. Elongator—An emerging role in neurological disorders. Trends Mol Med. 2010;16(1):1–6. doi: 10.1016/j.molmed.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Esberg A, Huang B, Johansson MJ, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24(1):139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 5.Huang B, Johansson MJ, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11(4):424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin FJ, Shen L, Jang CW, Falnes PO, Zhang Y. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 2013;9(5):e1003516. doi: 10.1371/journal.pgen.1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axelrod FB. A world without pain or tears. Clin Auton Res. 2006;16(2):90–97. doi: 10.1007/s10286-006-0326-7. [DOI] [PubMed] [Google Scholar]
- 8.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10(5):828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim EC, et al. Weak definition of IKBKAP exon 20 leads to aberrant splicing in familial dysautonomia. Hum Mutat. 2007;28(1):41–53. doi: 10.1002/humu.20401. [DOI] [PubMed] [Google Scholar]
- 10.Slaugenhaupt SA, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68(3):598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson SL, Qiu J, Rubin BY. EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem Biophys Res Commun. 2003;310(2):627–633. doi: 10.1016/j.bbrc.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Keren H, et al. Phosphatidylserine increases IKBKAP levels in familial dysautonomia cells. PLoS ONE. 2010;5(12):e15884. doi: 10.1371/journal.pone.0015884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochner R, et al. Phosphatidylserine increases IKBKAP levels in a humanized knock-in IKBKAP mouse model. Hum Mol Genet. 2013;22(14):2785–2794. doi: 10.1093/hmg/ddt126. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Anderson SL, Qiu J, Rubin BY. Cardiac glycosides correct aberrant splicing of IKBKAP-encoded mRNA in familial dysautonomia derived cells by suppressing expression of SRSF3. FEBS J. 2013;280(15):3632–3646. doi: 10.1111/febs.12355. [DOI] [PubMed] [Google Scholar]
- 15.Slaugenhaupt SA, et al. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum Mol Genet. 2004;13(4):429–436. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 16.Lee G, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25(12):1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 17.Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods. 2006;3(11):909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi A, Hosokawa M, Nojima T, Hagiwara M. Splicing reporter mice revealed the evolutionally conserved switching mechanism of tissue-specific alternative exon selection. PLoS ONE. 2010;5(6):e10946. doi: 10.1371/journal.pone.0010946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orengo JP, Bundman D, Cooper TA. A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic Acids Res. 2006;34(22):e148. doi: 10.1093/nar/gkl967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci USA. 2008;105(32):11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataoka N, et al. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6(3):673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 22.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15(2):108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piva F, Giulietti M, Nocchi L, Principato G. SpliceAid: A database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics. 2009;25(9):1211–1213. doi: 10.1093/bioinformatics/btp124. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 25.Mehlgarten C, et al. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol. 2010;76(5):1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Tuck S, Byström AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5(7):e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer F, et al. Translational control of cell division by Elongator. Cell Reports. 2012;1(5):424–433. doi: 10.1016/j.celrep.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyauchi K, Ohara T, Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35(4):e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Axelrod FB, et al. Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr Res. 2011;70(5):480–483. doi: 10.1203/PDR.0b013e31822e1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishida A, et al. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat Commun. 2011;2:308. doi: 10.1038/ncomms1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366(1):1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T. 2005. Biosynthesis and function of tRNA wobble modifications. Fine-Tuning of RNA Functions by Modification and Editing, Topics in Current Genetics, ed Grosjean H (Springer, Heidelberg), Vol 12, pp 23–69.
- 33.Johansson MJ, Esberg A, Huang B, Björk GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28(10):3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernández-Vázquez J, et al. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013;9(7):e1003647. doi: 10.1371/journal.pgen.1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20(6):306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45(45):299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.