Significance
Mitochondria were originally free-living bacteria with their own division machinery, which took up residence within another cell 2 billion y ago. The host cell subsequently tamed mitochondrial division using dynamin, a membrane-pinching protein. We have found that a single ancient dynamin at the root of extant eukaryotes, capable of pinching both mitochondria and vesicles, duplicated independently in plants and animals into specialized mitochondrial and vesicle variants. A “living fossil” of this ancient bifunctional dynamin still survives in scattered eukaryotic species, along with the original bacterial FtsZ division protein. The mitochondria of these organisms, preserved as if in amber, might teach us how the fateful partnership between host and endosymbiont was first established.
Keywords: eukaryote evolution, mitochondria, vesicles, dynamin, FtsZ
Abstract
Eukaryotic cells use dynamins—mechano-chemical GTPases—to drive the division of endosymbiotic organelles. Here we probe early steps of mitochondrial and chloroplast endosymbiosis by tracing the evolution of dynamins. We develop a parsimony-based phylogenetic method for protein sequence reconstruction, with deep time resolution. Using this, we demonstrate that dynamins diversify through the punctuated transformation of sequence segments on the scale of secondary-structural elements. We find examples of segments that have remained essentially unchanged from the 1.8-billion-y-old last eukaryotic common ancestor to the present day. Stitching these together, we reconstruct three ancestral dynamins: The first is nearly identical to the ubiquitous mitochondrial division dynamins of extant eukaryotes, the second is partially preserved in the myxovirus-resistance-like dynamins of metazoans, and the third gives rise to the cytokinetic dynamins of amoebozoans and plants and to chloroplast division dynamins. The reconstructed sequences, combined with evolutionary models and published functional data, suggest that the ancestral mitochondrial division dynamin also mediated vesicle scission. This bifunctional protein duplicated into specialized mitochondrial and vesicle variants at least three independent times—in alveolates, green algae, and the ancestor of fungi and metazoans—accompanied by the loss of the ancient prokaryotic mitochondrial division protein FtsZ. Remarkably, many extant species that retain FtsZ also retain the predicted ancestral bifunctional dynamin. The mitochondrial division apparatus of such organisms, including amoebozoans, red algae, and stramenopiles, seems preserved in a near-primordial form.
Eukaryotes arose through the acquisition of mitochondria by an archaeal host cell about 2 billion y ago (1, 2), a watershed moment in the evolution of the modern compartmentalized cell plan (3). A second transformative endosymbiotic event, the acquisition of a cyanobacterium by a eukaryotic host to form chloroplasts, gave rise to the photosynthetic eukaryotic lineages (4). As the endosymbionts became integrated with their hosts, their growth and division became regulated by host–cellular machinery (5). Proteins of the dynamin superfamily were central to this process: Mitochondria and chloroplasts originally divided using a constricting ring of the prokaryotic cytoskeletal protein FtsZ, but dynamins have been recruited to these roles in all extant eukaryotes (6, 7). By reconstructing the evolutionary history of dynamins, we can probe the process of endosymbiont integration.
The dynamin superfamily is diverse (8, 9), and different dynamin variants remodel membranes at different cellular locations (Table S1 and primary references therein). A major class of dynamins is essential for mitochondrial and peroxisomal division. Another large group drives the scission of clathrin-coated vesicles in organisms such as fungi and alveolates. A related group, the so-called “classical” dynamins that drive clathrin-coated vesicle scission in metazoans and land plants, contains a membrane-targeted pleckstrin homology (PH) domain. Members of the phragmoplastin class of dynamins participate in cell plate formation in land plants. The myxovirus-resistance-like dynamins are implicated in antiviral activity in vertebrates. A truncated dynamin variant is involved in cytokinesis in amoebozoans and plants, as well as in chloroplast fission in photosynthetic lineages; another truncated variant drives mitochondrial inner membrane fusion in fungi and metazoans. Finally, mitofusins and the related bacterial dynamin-like proteins (BDLPs) are potentially ancient members of the dynamin superfamily (10); these are excluded from our study because they are highly diverged at the sequence level.
Here we present the most comprehensive analysis of dynamin evolution yet reported, including thousands of functionally diverse dynamins from hundreds of broadly sampled eukaryotic species. We reconstruct the series of events that led from the primordial dynamins of the 1.8-billion-y-old last eukaryotic common ancestor (LECA) (11) to the great variety of present-day dynamins. The outcome is a nuanced picture of protein diversification, mirroring key events in the evolution of eukaryotes themselves and shedding light on the earliest stages of endosymbiont integration.
Results
Functionally Diverse Dynamins Are Found Across Eukaryotic Supergroups.
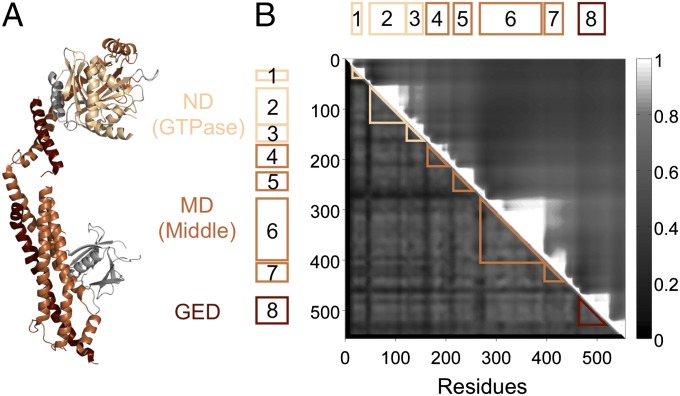
Members of the dynamin superfamily are characterized by three major domains: an N-terminal GTPase domain (ND), a stalk region formed by helices of the middle domain (MD), and a GTPase effector domain (GED) that folds back across the stalk to regulate GTPase activity (Fig. 1A). Most dynamins act by forming homo-oligomeric rings around membrane necks through interactions of the stalk, driving scission via GTP hydrolysis (12). Starting with a curated list of functionally diverse dynamins, we used ND, MD, and GED alignments to query a broadly sampled protein sequence database (Methods, Construction of the Dynamin Database). We thus identified and aligned 3,827 eukaryotic dynamins from 854 species or strains representing all extant eukaryotic supergroups, as well as 14 bacterial dynamins (Datasets S1 and S2; see SI Text for dataset descriptions).
Fig. 1.
Breaking dynamin into evolutionary segments. (A) The crystal structure of human dynamin 1 [Protein Data Bank (PDB) ID: 3SNH] (18). Dynamins have three conserved domains: the N-terminal GTPase domain (ND, light brown), the middle domain (MD, brown), and the GTPase effector domain (GED, dark brown). (B) We assemble a 556-residue concatenated alignment of the ND, MD, and GED. Our analysis relies on finding clusters of proteins with high sequence identity across short segments of this alignment. Two contiguous segments with similar clusters can be merged into a single longer segment with no loss of information. The upper-triangular matrix Rij shows the properties of long segments starting at residue i and ending at residue j; high values indicate that the long segment is made up of multiple subsegments with similar clustering properties (SI Text, Evolutionary Segments). The saw tooth structure shows that dynamin breaks into eight natural evolutionary segments ranging from 22 to 118 residues in length: three in the ND, four in the MD, and one in the GED. Every dynamin is assigned an eight-letter signature, such that two proteins with the same letter at a given segment have high sequence identity across that segment (SI Text, Sequence Clustering).
To examine the diversity within this dataset we used the graphical CLANS tool, which clusters proteins by sequence similarity (SI Text, Sequence Clustering) (13). Applying CLANS to the full alignment, we found that the proteins split into seven major classes (Fig. S1D), each associated with distinct functional annotations (Table S1), whose mutual relationships were not well resolved (Fig. S2). Three of these—which we term superclasses A, B, and C—span multiple eukaryotic supergroups. Given a set of present-day proteins and a backbone tree of eukaryotic species (11, 14), we define the “root species” as the last common ancestor of the species in which those proteins are found and the “root protein” as the last common ancestral protein itself. Class A (blue) consists of 1,891 dynamins, including those involved in mitochondrial and peroxisomal division, vesicle scission, or cell plate formation, with LECA as the root species; class B (orange) consists of 1,390 myxovirus-resistance-like dynamins, with LECA as the root species; class C (green) consists of 136 ND-only dynamins, including those involved in cytokinesis or chloroplast division, with the last common ancestor of amoebozoans and archaeplastids as the root species. The remaining four classes are confined to individual lineages: 16 alveolate-specific proteins related to class C (Fig. S2), 252 fungal (Mgm1) and 142 metazoan (OPA1) mitochondrial fusion dynamins, and 14 bacterial dynamins.
Ancestral Dynamins Can Be Reconstructed by Parsimony.
We are interested in inferring sequence and function of the root proteins of present-day dynamin superclasses. Although evolutionary rates can vary over time and across the protein sequence at billion-year timescales, maximum-likelihood methods incorporating rate heterogeneity can accurately reconstruct protein phylogenies. In effect, conserved regions of a protein similar to the ancestral sequence provide a reliable phylogenetic signal (Fig. S3). However, outside these regions, the reconstructed sequences are often inaccurate. Here we present a sequence reconstruction method based on parsimony, which can outperform maximum likelihood when evolution is heterogeneous (15). This relies on a compact representation of protein sequence that makes clear which regions of an ancestral protein have been reliably reconstructed. We break up the alignment into a small number of contiguous “evolutionary segments” (SI Text, Evolutionary Segments). For a given protein at a given segment, we compress the entire amino acid sequence into a single letter, such that two proteins with the same letter have similar sequences, beyond some threshold identity X% (SI Text, Sequence Clustering). Each present-day protein is thus assigned a short signature consisting of a string of letters: Each position corresponds to a segment, and each letter labels a cluster of proteins with highly similar sequences across the relevant segment. Dollo parsimony then implements the following rule (16): If two present-day proteins have the same letter at a given segment, then their root protein must also have that letter. Ancestral proteins are thus assigned a signature, but with gaps (?) wherever the sequence cannot be reconstructed. If an ancestral signature has a certain letter, it implies the ancestral sequence is >X% identical to present-day proteins with the same letter across the relevant segment.
Crucially, the results of Dollo parsimony are guaranteed to be valid independent of the choice of segments and thresholds, as long as convergence can be ruled out. To prevent convergence, we must use some combination of long segments and high thresholds, so the chance of two unrelated proteins accidentally having >X% identity across the entire segment is negligible. However, if the segments are too long or the threshold is too high, it is likely that only recently diverged protein pairs have >X% identity across whole segments. We would then be able to reconstruct recent ancestral proteins, but the signatures of ancient proteins would consist mostly of gaps. Also, if the threshold were so low that any pair of homologous proteins would have >X% identity across all segments, all proteins both present day and ancestral would trivially have the same signature. This would prevent us from inferring the function of ancestral variants. The details of how segment boundaries are chosen and how sequence stretches at each segment are grouped into clusters corresponding to letter labels are given in SI Text, Evolutionary Segments and SI Text, Sequence Clustering. To understand our results, only the following properties of segments and letter clusters are relevant. First, we find that dynamins split into eight well-defined evolutionary segments ranging in length from 22 to 118 residues, smaller than domains but significantly correlated with secondary-structural elements (Fig. 1B) (17, 18). In practice, segments 20 residues or longer are sufficient to prevent accidental convergence and can retain nearly as much phylogenetic information as an entire protein (Fig. S3D). Our segments are therefore sufficiently long for parsimony to be valid. Second, when we examine the clusters of sequences corresponding to each segment-wise letter label, almost all clusters (223 of 238 clusters across segments; Dataset S1, sheet 2) have a median pairwise identity >X = 35%. This is comparable to the level of conservation between pairs of proteins with similar function (19). Therefore, if we do find present-day proteins with signatures nearly identical to those of some reconstructed ancestor, we can infer the function of the ancestor based on the function of its extant descendants.
Ancient Segments, Unchanged for Billions of Years, Are “Living Fossils.”
It is illuminating to benchmark our segment-based approach against a maximum-likelihood phylogenetic analysis; further validation steps are described in Fig. S3 and SI Text, Benchmarking Segment-Based Parsimony. Each letter label at each segment is associated with a cluster of proteins. For each such cluster, we define three attributes (Fig. 2 and Dataset S1, sheet 2):
Attribute 1: The pairwise identity between sequences within a cluster. This allows us to estimate the sequence of root proteins: If convergence is ruled out, the root protein of a pair of present-day proteins is at least as similar to each of them as they are to one another. Of 238 clusters, 223 have a median intracluster pairwise identity >35%. If the phylogeny within such clusters is balanced, this implies that the root protein is >35% identical to at least one present-day protein over the relevant segment.
Attribute 2: The root species of a cluster. This allows us to estimate the age of root proteins: Assuming no horizontal transfer, if a cluster contains proteins from multiple eukaryotic supergroups, its root species and root protein are necessarily ancient, over 1.5 billion y old (11). In contrast, the root proteins of supergroup-specific clusters are potentially more recent. We find few ancient clusters: Class A dynamins have an ancient cluster at each of segments 1–8, class B dynamins have an ancient cluster at each of segments 1–4 and 6, and the ND-only class C dynamins have an ancient cluster at each of segments 1–3.
Attribute 3: Whether the cluster is monophyletic. This establishes that the root protein of one cluster is not ancestral to that of any other cluster. To be monophyletic, member sequences of a cluster must repeatedly form a well-defined clade over 1,000 bootstrap phylogenetic trees calculated for a given segment (Methods, Maximum-Likelihood Analysis). The majority of supergroup-specific clusters were strongly monophyletic (median monophyly support: class A, 800/1,000; class B, 567/1,000; and class C, 759/1,000; Fig. 2C, Left). However, we found that the monophyly support levels of ancient clusters were significantly different from those of supergroup-specific clusters (P = 5E-6, Kolmogorov–Smirnov test; Fig. 2C, Right). This is striking, given that both ancient and supergroup-specific clusters had sequence identities in the same range of 50–90%. Whereas ancient class C clusters were monophyletic (median monophyly support: 356/1,000; Fig. 2C, Center), ancient class A and B clusters were not (median monophyly support: class A, 0/1,000; class B, 0/1,000; Fig. 2C, Center).
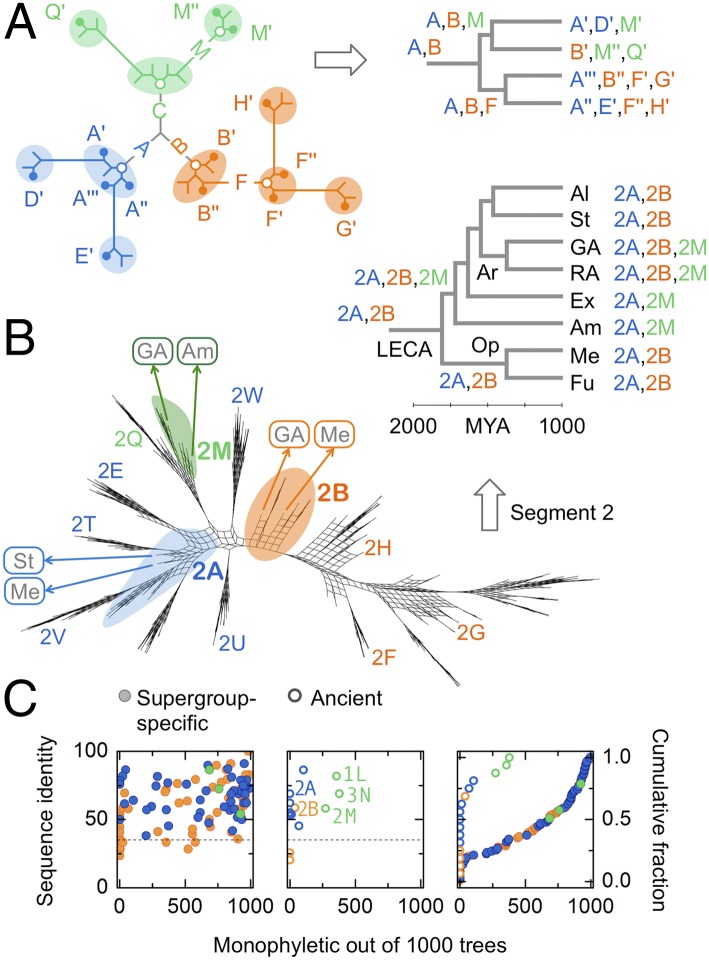
Fig. 2.
Living fossils: tight, ancient, paraphyletic clusters. (A) (Left) Schematic protein tree. Proteins are grouped into clusters of high pairwise identity corresponding to letter labels (colored ovals and circles). For each cluster we show extant proteins (solid dots) and their last common ancestor (root proteins: open dots). If descendants of a root protein are confined to the cluster itself, the cluster is monophyletic; otherwise it is paraphyletic, with other clusters branching out of it. (Right) Extant proteins are mapped to the leaves of a species tree. If a cluster contains proteins from multiple species, their last common ancestor is its root species. Three superclasses (blue, orange, and green) emerge from three root proteins (“A,” “B,” and “C”). Proteins similar to A and B survive to the present day, and no proteins similar to C survive. (B) Maximum-likelihood trees for one trial of segment 2. We show a consensus split network (48) representing 100 bootstrap replicates; nodes with strong bootstrap support appear as thin stems. Ancient clusters (2A, 2B, and 2M) are shown with colored overlays and the rest with letters; not all clusters are labeled. Paraphyletic clusters 2A and 2B form “galls” from which other class A (blue) and class B (orange) clusters emerge; monophyletic clusters like 2M have no outward branches. Clusters 2U and 2W map to the center of the tree due to long-branch attraction (Fig. S3D). We map ancestral proteins to a species tree (Mya: millions of years ago); species group labels are as in Fig. 3. Ancient clusters contain proteins from multiple supergroups (labels in boxes). (C) To find the phylogenetic attributes of clusters corresponding to each letter label, the analysis shown in Fig. 2B for segment 2 is repeated for each segment separately. We show the attributes of all 110 clusters in our dataset with more than 10 members. Attribute 1: pairwise identity (y axis, Left and Center; dashed line, 35% identity). Attribute 2: root species (open circles, ancient, root species >1.5 billion y old; solid circles, supergroup-specific, root species <1.5 billion y old). Attribute 3: monophyly (x axis, Left, Center, and Right, monophyly support out of 1,000 trees; Right, cumulative distribution of monophyly support). If a cluster is tight (pairwise identity >35%), ancient (root species >1.5 billion y old), and paraphyletic (monophyly support <∼100/1,000), its members are living fossils: extant proteins similar in sequence to the ancient root proteins of functional superclasses. All ancient class A and class B clusters (e.g., 2A and 2B, Center) are paraphyletic and therefore ancestral. The ancient class C clusters (1L, 2M, and 3N) are monophyletic.
There are two reasons a cluster might not be monophyletic: Either its phylogenetic support is weak (e.g., cluster 2E in Fig. 2B) or it is paraphyletic, meaning its root protein is ancestral to members of multiple clusters. As evident in Fig. 2B (and also Fig. S3A) the ancient class A and class B clusters at each segment are truly paraphyletic: They are not only ancient, but also ancestral to other supergroup-specific class A and class B clusters. To reflect their important status, we label the unique ancestral cluster at each segment after the class itself (e.g., 2A or 2B at segment 2; for ancestral clusters, the same letter indicates distinct sequences at each segment; e.g., 1A and 2A are distinct). These clusters are living fossils: sets of extant proteins similar in sequence (over the relevant segment) to the ancient root proteins of functional superclasses. Although sequence remnants of the root class C dynamin are lost, those of the root class A and class B dynamins persist to the present day.
Entire Ancestral Dynamin Variants Are Preserved in Extant Species.
Every protein in our dataset is assigned an eight-letter signature (Table S1 and Dataset S1, sheet 1; the segment value is redundant and therefore omitted in signatures; e.g., 1A, 2A, … , 8A becomes AAAAAAAA; class C dynamins have three-letter signatures). By carrying out the parsimony analysis independently for each segment, we can reconstruct the full signatures of ancestral protein variants. We find that LECA had a class A dynamin identical in signature to that of mitochondrial and peroxisomal division dynamins of many extant species (AAAAAAAA; Fig. 3A). Vesicle scission dynamins in fungi and metazoans retain similarity to the ancestral variant only in their N-terminal GTPase domains; those of land plants and alveolates are even more diverged. A class A variant of unknown function in green algae (A:A::XYZ) appears to have duplicated in land plants, into a vesicle dynamin (:W:-:XYZ) and a phragmoplastin (AVAA:XYZ) responsible for cell plate formation (20) (Fig. S3). LECA also had a class B dynamin whose closest extant variants are found in the metazoans (BBBB:B::; Fig. 3B). These dynamins have a patchy present-day distribution and appear to have undergone a recent radiation in fungi; in vertebrates these myxovirus-resistance-like proteins have antiviral functions (21).
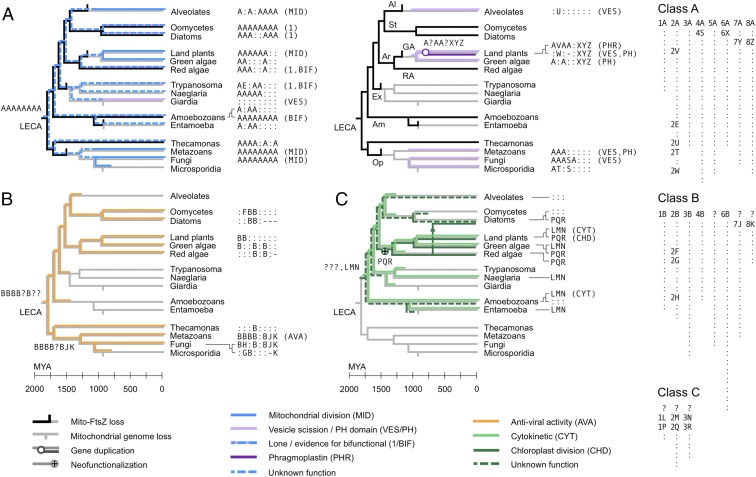
Fig. 3.
Punctuated diversification of dynamins across 1.8 billion y. The heterogeneous nature of dynamin evolution is highlighted, over time and across the protein sequence. The evolutions of different dynamin superclasses (colored) and of mitochondrial FtsZ (black) are overlaid on the eukaryotic tree (gray). The timing of speciation events, from 2,000 million years ago (Mya) to the present day, is approximately as reported in Parfrey et al. (11); the FtsZ loss data are derived from Fig. S6. Certain events (mitochondrial genome loss, FtsZ loss, other gene loss and duplication events, and horizontal transfers) are shown on the correct branches but not necessarily at the correct times. Species group labels: Al, alveolates; St, stramenopiles; Ar, archaeplastids; GA, green algae (including land plants); RA, red algae; Ex, excavates; Am, amoebozoans; Op, opisthokonts; Me, metazoans; Fu, fungi. We represent dynamins with 8-letter signatures (Dataset S1, sheets 1 and 2; class C dynamins span only segments 1–3). Each segment-wise letter labels a cluster of similar sequences; two proteins with the same letter at a given segment thus have high sequence identity across that segment. The key (Right) shows the full set of 238 letter clusters across segments and superclasses, sorted as in Dataset S1, sheet 2. The letters A and B represent distinct clusters at each segment, corresponding to the reconstructed root proteins of classes A and B; the root protein of class C cannot be reconstructed. Most supergroup-specific clusters are labeled with the shorthand symbol “:” but are assigned full cluster labels in Dataset S1; no two instances of : in this figure correspond to the same full label. A subset of clusters, including all those spanning multiple supergroups, is labeled by segment-wise letters for ease of reference. Gaps are labeled “-”; segments of ancestral proteins that cannot be reconstructed are labeled “?.” We show functional annotations [mitochondrial division (MID), vesicle scission (VES), etc.] where they have been experimentally verified (Table S1). (A) (Left) Class A dynamins specialized for mitochondrial division (MID; solid blue), or lone/bifunctional (1/BIF; dashed blue and lavender; “lone” indicates a single copy per genome). (Right) Derived class A dynamins specialized for vesicle scission (VES; lavender) or phragmoplast/cell plate formation [phragmoplastin (PHR); purple]. (B) Class B dynamins involved in antiviral activity (AVA) (orange). (C) Class C dynamins specialized for cytokinesis (CYT) (green) or chloroplast division (CHD) (dark green). The cytokinetic (LMN) and plastid division dynamins (PQR) are sister groups, descended from an ancestral dynamin of unknown sequence (???) (Fig. S2). The vertical arrow shows horizontal gene transfer by secondary endosymbiosis of red algae by diatoms (Bacillariophyta). Some rootings of the eukaryotic tree would place the ancestral class C dynamin at LECA itself (Fig. S4).
Because the ancestral class C dynamin appears lost and therefore cannot be reconstructed by parsimony, we carried out a phylogenetic analysis of GTPase domains (concatenated segments 1, 2, and 3) for every class C dynamin in our dataset (Fig. S2). This revealed that the cytokinetic dynamins were monophyletic (LMN: 88% bootstrap support), and represented a sister group to the chloroplast division dynamins (PQR: 43% bootstrap support). The last common ancestor of amoebozoans and archaeplastids therefore had a class C cytokinetic dynamin (LMN) (22) that persists essentially unchanged in many lineages, as well as another class C dynamin of unknown sequence (???) (Fig. 3C). Descendants of this unknown ancestor are specialized for chloroplast division in archaeplastids and plastid-bearing stramenopiles such as diatoms (PQR); other diverged descendants are present but uncharacterized in alveolates, amoebozoans, one rhizarian (Bigelowiella natans), and one diatom (Thalassiosira oceanica). Because diatom plastids are derived from the secondary endosymbiosis of red algae (23) and the chloroplast dynamins of these lineages branch close to one another (Fig. S2), we attribute the occurrence of the PQR variant in stramenopiles to horizontal transfer.
Extant Eukaryotes Have Separate Mitochondrial and Vesicle Scission Dynamins or a Potentially Bifunctional Dynamin.
Alveolates, land plants, metazoans, and fungi each have distinct class A dynamins specialized for mitochondrial division (the ancestral variant; Fig. 3A, Left) and vesicle scission (a derived variant; Fig. 3A, Right). In amitochondriate eukaryotes (24) such as microsporidia and the Entamoebidae, the ancestral class A variant appears diverged or deleted (although microsporidia retain the diverged fungal vesicle dynamin). Surprisingly, several eukaryotic lineages encode just a single class A dynamin; these include both the oomycete and diatom lineages of stramenopiles (25/30 species or strains, median protein repertoire 17,368), red algae (6/7 species or strains, median protein repertoire 7,176), and excavates (30/36 species or strains, median protein repertoire 9,858). Assuming that dynamins are essential for mitochondrial division as well as for clathrin-coated vesicle scission, we predict these lone dynamins to drive both processes. There is partial although not conclusive experimental support for this idea. The class A dynamin of the excavate Trypanosoma brucei localizes to mitochondria (25) but appears to be required for mitochondrial fission as well as endocytosis (26); that of the amitochondriate excavate Giardia lamblia colocalizes with clathrin-coated vesicles (27). The class A dynamin of the red alga Cyanidioschyzon merolae has a punctate distribution on the cell surface between mitochondrial division events, suggesting it might also participate in vesicle scission (28, 29). The amoebozoan Dictyostelium discoideum has two class A variants, of which DymA is ancestral; this protein is not essential for mitochondrial fission but influences mitochondrial morphology (30) and participates in both mitochondrial and vesicle activities (31).
Discussion
LECA Had a Bifunctional Dynamin That Duplicated Multiple Independent Times.
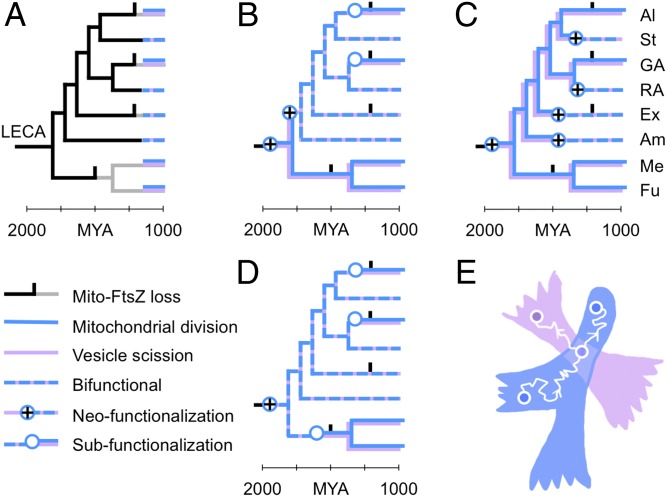
Eukaryotic lineages fall into two groups: those that use two distinct dynamins for mitochondrial and vesicle scission and those for which there is support for a single bifunctional dynamin (Fig. 4A). Three independent arguments suggest that the ancestral protein itself was bifunctional. First, we find no evidence of a distinct ancestral vesicle scission dynamin (Fig. 3A, Right): Features shared across vesicle dynamins are also shared with mitochondrial dynamins. Second, the scenario in which LECA had a single bifunctional dynamin is more parsimonious than scenarios in which LECA contained specialized mitochondrial and vesicle dynamins (Fig. 4; Fig. S4 shows the same result for alternative rootings of the eukaryotic tree). Third, given extant bifunctional and specialized variants, a population-genetic analysis strongly supports a bifunctional ancestor (Fig. 4E; Fig. S5; and SI Text, Routes of Gene Duplication) (32). Together, these observations imply that the bifunctional ancestral dynamin duplicated and specialized into mitochondrial and vesicle variants at least three independent times (Fig. 4D and Fig. S4): in green algae, in alveolates, and in the opisthokonts. (Opisthokont vesicle dynamins do not form a robust clade, so we cannot rule out separate duplications in fungi and metazoans.) Independent observations support this multiple-duplication scenario: The PH domains of plant and metazoan classical vesicle dynamins appear to have independent origins (33); the mitochondrial and vesicle dynamins of alveolates are suggested to be related (34); and the mitochondrial and vesicle dynamins in yeast (Dnm1 and Vps1) share a role in peroxisomal division (35), indicating a recent divergence. Following duplication, the mitochondrial variant retains most of its ancestral signature (AAAAAAAA; Fig. 3A, Left). Conversely, all specialized vesicle scission dynamins have diverged from the ancestral state in segments 6, 7, and 8 (Fig. 3A, Right and Table S1), where the oligomerization interface is found (17, 18). This suggests that hetero-oligomerization must be suppressed before a duplicate dynamin is free to take on new roles.
Fig. 4.
Duplication and specialization of a bifunctional ancestral dynamin. (A) Present-day lineages have either a bifunctional dynamin with both mitochondrial and vesicle roles (dashed blue and lavender) or two dynamins specialized for mitochondrial division and vesicle scission (stacked blue and lavender). We want to find the most parsimonious explanation for this distribution. Scenarios using alternative rootings of the eukaryotic tree are shown in Fig. S4. Species group labels are as in Fig. 3. (B and C) Scenarios in which LECA had two specialized dynamins require multiple gain-of-function events. (D) The scenario in which LECA had a single bifunctional dynamin predicts that specialized dynamins emerge from at least three independent subfunctionalization events, all coupled with the loss of FtsZ. The causal relationship between gene duplication and FtsZ loss is not clear. (E) A 2D schematic of protein sequence space. A single protein sequence is represented as a point (circles); as its evolves by random mutation, the corresponding point traces a curve through the space (white wiggly arrows represent the passage of time). Some amino acid sequences do not encode functional proteins (white background); other regions encode mitochondrial dynamins (X; blue) and vesicle dynamins (Y; lavender). If such regions overlap, it implies the possibility of a bifunctional dynamin. A protein starts in one of the monofunctional regions (bottom left circle). As it diffuses through sequence space (white wiggly arrow) it is confined to this region by purifying selection for function X. Eventually, it discovers a bifunctional region XY, in the overlap of the blue and lavender regions (middle circle). If the protein then duplicates (branch), the two new copies subfunctionalize by rapidly exiting the bifunctional region, back into the blue and lavender regions (white wiggly arrows moving toward the top left and right circles; each arrow represents a distinct protein copy). The dynamics of this process are derived in SI Text, Routes of Gene Duplication.
The Primordial Mitochondrial Division Apparatus Is Preserved in Many Extant Lineages.
Most present-day chloroplasts and plastids are simultaneously squeezed from the outside by dynamin and pulled from the inside by an FtsZ ring during division (7). However, most present-day mitochondria have lost their prokaryotic FtsZ apparatus entirely and rely on dynamin to drive division (6). All known lineages that use specialized mitochondrial and vesicle dynamins have independently lost mitochondrial FtsZ (Fig. 3A), although the order of specialization and FtsZ loss is not clear. The excavates, which use a potentially bifunctional dynamin, have also lost FtsZ. However, mitochondrial FtsZ persists in amoebozoans, red algae, glaucophytes, haptophytes, and stramenopiles (36, 37), as well as in the apusozoan Thecamonas trahens (a member of a sister group to the opisthokonts; Fig. S6) (38). LECA therefore must have used FtsZ, along with a bifunctional dynamin, to divide its mitochondria. The dynamins of glaucophytes, haptophytes, and Thecamonas are currently uncharacterized; by determining their roles we might elucidate the correlation between specialization and FtsZ loss. Remarkably, the remaining FtsZ-containing species appear to retain LECA’s mitochondrial division apparatus in its entirety: Stramenopiles, red algae, and amoebozoans all encode an ancestral class A dynamin known or predicted to be bifunctional, as well as mitochondrial FtsZ. A detailed characterization of mitochondrial division in these organisms would provide a glimpse of an early period of eukaryote evolution in which dynamin interacted both with the clathrin-coated vesicles of the host and with the FtsZ ring of the endosymbiont. Frozen in time, the persistence of ancestors converts a problem of evolutionary speculation into a problem of experimental measurement, allowing us to probe the cell biology of ancient organisms across a span of billions of years.
Methods
Construction of the Dynamin Database.
We built a nonredundant protein database combining UniProtKB (May 2014) (39) with proteomes inferred from 65 protist genomes (Dataset S1, sheet 3). We assembled a curated list of dynamins from seven model organisms (Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, D. discoideum, and Arabidopsis thaliana) (8). We used HMMER (40) to annotate protein domains according to the Pfam database of hidden Markov models (41) and used MUSCLE (42) to build multiple-sequence alignments of the ND (PF00350), the MD (PF01031), and the GED (PF02212). For each domain, we performed PSI-BLAST (43) against the protein database, with the most lenient E-value cutoff consistent with convergence within 50 iterations. We checked whether functional dynamin subtypes were able to pick up members of other subtypes; the highly diverged mitofusins and BDLPs failed this cross-validation step and were excluded. From the resulting dataset, we used IsoSVM (44) to detect splice isoforms and retained the longest isoform per gene. We used HMMER and DomainFinder3 (45) to annotate the ND (168 residues), MD (296 residues) and GED (92 residues) of these sequences, using an E-value cutoff of 1, and retained sequences that contained at least one of these domains. For 387 proteins we found multiple hits to the same HMM profile; in these cases we retained the most C-terminal hit for the alignment, but retained all hits for clustering. This produced a final alignment of 3,841 sequences across 556 residues (Dataset S2). A total of 3,827 sequences were from 854 eukaryotic species or strains according to NCBI Taxonomy IDs (asterisks indicate accepted eukaryotic supergroups): 646 opisthokonts* (280 fungi, 2 mesomycetozoans, 1 nucleariid, 2 choanoflagellates, and 361 metazoans), 1 apusozoan, 16 amoebozoans*, 36 exacavates*, 69 archaeplastids* (1 glaucophyte, 7 red algae, 14 green algae, and 47 land plants), 2 hacrobians (1 haptophyte and 1 cryptophyte), and 84 species from the SAR supergroup* (30 stramenopiles, 51 alveolates, and 3 rhizarians). Fourteen additional dynamins mapped to bacteria.
Maximum-Likelihood Analysis.
We computed all maximum-likelihood trees using RAxML (46), with an LG amino acid substitution model and gamma-distributed rate heterogeneity. The protein evolution model was selected using ProtTest (47). Because running RAxML on the full dataset is computationally expensive, we assessed the monophyly of clusters, using a sampling strategy. For each segment, we ran 100 bootstrap replicates on a dataset consisting of 10 random representatives from each cluster (excluding clusters with fewer than 10 members). We repeated this process for 10 randomly subsampled datasets, thus generating 1,000 trees for each segment. We scored each cluster by asking how many times it was recovered as precisely monophyletic from these 1,000 trees (Dataset S1, sheet 2). Consensus split networks were generated using SplitsTree4 (48) with default parameters, with the “Use weights” and “Convex hull” options unchecked.
Supplementary Material
Acknowledgments
We thank Jitu Mayor, Anjali Jaiman, Madan Rao, Iñaki Ruiz-Trillo, and Madhu Venkadesan for useful discussions. We are indebted to K. S. Krishnan, who showed us how science should be done. M.T. was partly funded by a Wellcome Trust/Department of Biotechnology India Alliance Intermediate Fellowship 500103/Z/09/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.W.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407163112/-/DCSupplemental.
References
- 1.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504(7479):231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 2.Baum DA, Baum B. An inside-out origin for the eukaryotic cell. BMC Biol. 2014;12:76. doi: 10.1186/s12915-014-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramadas R, Thattai M. New organelles by gene duplication in a biophysical model of eukaryote endomembrane evolution. Biophys J. 2013;104(11):2553–2563. doi: 10.1016/j.bpj.2013.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 2004;91(10):1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- 5.Osteryoung KW, Nunnari J. The division of endosymbiotic organelles. Science. 2003;302(5651):1698–1704. doi: 10.1126/science.1082192. [DOI] [PubMed] [Google Scholar]
- 6.Kuroiwa T, et al. Structure, function and evolution of the mitochondrial division apparatus. Biochim Biophys Acta. 2006;1763(5-6):510–521. doi: 10.1016/j.bbamcr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Miyagishima SY. Mechanism of plastid division: From a bacterium to an organelle. Plant Physiol. 2011;155(4):1533–1544. doi: 10.1104/pp.110.170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Praefcke GJK, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5(2):133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13(2):75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444(7120):766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 11.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108(33):13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morlot S, Roux A. Mechanics of dynamin-mediated membrane fission. Annu Rev Biophys. 2013;42:629–649. doi: 10.1146/annurev-biophys-050511-102247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frickey T, Lupas A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20(18):3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 14.Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59(5):429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolaczkowski B, Thornton JW. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431(7011):980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- 16.Farris JS. Phylogenetic analysis under Dollo’s law. Syst Zool. 1977;26(1):77–88. [Google Scholar]
- 17.Ford MGJ, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477(7366):561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faelber K, et al. Crystal structure of nucleotide-free dynamin. Nature. 2011;477(7366):556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 19.Orengo CA, et al. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5(8):1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 20.Gu X, Verma DP. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 1996;15(4):695–704. [PMC free article] [PubMed] [Google Scholar]
- 21.Horisberger MA, Staeheli P, Haller O. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc Natl Acad Sci USA. 1983;80(7):1910–1914. doi: 10.1073/pnas.80.7.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc Natl Acad Sci USA. 2008;105(39):15202–15207. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 24.Shiflett AM, Johnson PJ. Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol. 2010;64:409–429. doi: 10.1146/annurev.micro.62.081307.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan GW, Goulding D, Field MC. The single dynamin-like protein of Trypanosoma brucei regulates mitochondrial division and is not required for endocytosis. J Biol Chem. 2004;279(11):10692–10701. doi: 10.1074/jbc.M312178200. [DOI] [PubMed] [Google Scholar]
- 26.Chanez AL, Hehl AB, Engstler M, Schneider A. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J Cell Sci. 2006;119(Pt 14):2968–2974. doi: 10.1242/jcs.03023. [DOI] [PubMed] [Google Scholar]
- 27.Gaechter V, Schraner E, Wild P, Hehl AB. The single dynamin family protein in the primitive protozoan Giardia lamblia is essential for stage conversion and endocytic transport. Traffic. 2008;9(1):57–71. doi: 10.1111/j.1600-0854.2007.00657.x. [DOI] [PubMed] [Google Scholar]
- 28.Nishida K, et al. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA. 2003;100(4):2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imoto Y, et al. Single-membrane-bounded peroxisome division revealed by isolation of dynamin-based machinery. Proc Natl Acad Sci USA. 2013;110(23):9583–9588. doi: 10.1073/pnas.1303483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schimmel BG, Berbusse GW, Naylor K. Mitochondrial fission and fusion in Dictyostelium discoideum: A search for proteins involved in membrane dynamics. BMC Res Notes. 2012;5:505. doi: 10.1186/1756-0500-5-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wienke DC, Knetsch ML, Neuhaus EM, Reedy MC, Manstein DJ. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol Biol Cell. 1999;10(1):225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154(1):459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YW, Su AI, Schmid SL. The evolution of dynamin to regulate clathrin-mediated endocytosis: Speculations on the evolutionarily late appearance of dynamin relative to clathrin-mediated endocytosis. BioEssays. 2012;34(8):643–647. doi: 10.1002/bies.201200033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. Elucidation of clathrin-mediated endocytosis in tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 2005;1(5):e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuravi K, et al. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 19):3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 36.Beech PL, et al. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287(5456):1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- 37.Kiefel BR, Gilson PR, Beech PL. Diverse eukaryotes have retained mitochondrial homologues of the bacterial division protein FtsZ. Protist. 2004;155(1):105–115. doi: 10.1078/1434461000168. [DOI] [PubMed] [Google Scholar]
- 38.Torruella G, et al. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29(2):531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UniProt Consortium Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42(Database issue):D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spitzer M, Lorkowski S, Cullen P, Sczyrba A, Fuellen G. IsoSVM—distinguishing isoforms and paralogs on the protein level. BMC Bioinformatics. 2006;7:110. doi: 10.1186/1471-2105-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeats C, Redfern OC, Orengo C. A fast and automated solution for accurately resolving protein domain architectures. Bioinformatics. 2010;26(6):745–751. doi: 10.1093/bioinformatics/btq034. [DOI] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 48.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.