Abstract
Though proposed as a promising target antigen for cancer immunotherapy, the prognostic value of Wilms' tumor 1 (WT1) in solid tumors remains inconclusive. Here, we report a systematic review and meta-analysis of the association between WT1 expression and prognosis in solid tumors. PubMed, Web of Science and Google Scholar were searched to identify studies exploring the impact of WT1 on clinical outcomes, including overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS), relapse/recurrence-free survival (RFS) or progression-free survival (PFS), in solid cancer patients. Hazard ratio (HR) and 95% confidence interval (CI) were applied to assess the strength of these associations. Finally, a total of 29 eligible studies with 4090 patients were identified for qualitative analysis, and 22 studies with 3620 patients were enrolled for quantitative synthesis. Overall, positive expression of WT1 was significantly associated with worse OS (metaHR = 1.48, 95% CI = 1.11–1.97) and DFS/RFS/PFS (metaHR = 2.14, 95% CI = 1.42–3.21). Subgroup analyses showed that WT1 positive expression could independently predict unfavorable DFS/RFS/PFS (metaHR = 1.86, 95%CI = 1.04–3.35). In summary, our study suggests that WT1 may be a potential marker to predict DFS/RFS/PFS in solid tumor patients. Further studies are needed to confirm the role of WT1 expression in clinical practice.
Cancer is a major cause of mortality worldwide1. It is well-known that the balance between oncogenes and tumor suppressor genes is a prime determinant of normal development versus tumorigenesis2,3. Therefore, identification and characterization of these genes is of primary importance in understanding the onset and progression of cancer, and ultimately leading to recognition of potential markers and specific targets for prevention and individualized treatment of cancer.
The Wilms' tumor 1 (WT1), located at chromosome 11p13, was first cloned in 1990 as a suppressor in Wilms' tumor4. Subsequent research indicated that WT1 might play an oncologic role in hematologic malignancies and a variety of solid tumors, including leukemia, breast cancer, ovarian cancer, glioblastoma and soft tissue sarcoma, etc5,6,7,8,9. It has been shown that WT1 was overexpressed in a number of cancer cells, and more importantly, knockdown of WT1 by shRNA could induce mitochondrial damage and then inhibit malignant cell growth10. In addition to its prognostic role in a variety cancer types11,12,13,14,15, WT1 was also recognized as a promising target for immunotherapy based on its unique features16,17, and preliminary results from clinical vaccine trials revealed WT1's untapped potential to induce cancer immunity with minimal side effects18,19,20.
Despite the clinical development of WT1-targeted therapies, the prognostic value of WT1 expression in solid tumors remains unclear. It would clearly be desirable to explore whether the tumors manifesting WT1 expression are associated with worse outcome in patients with solid tumors. Therefore, we conducted the present systematic review and meta-analysis in order to appraise the prognostic value of WT1 in solid tumors with the goal of gaining insights into the clinical implications.
Results
Description of Studies
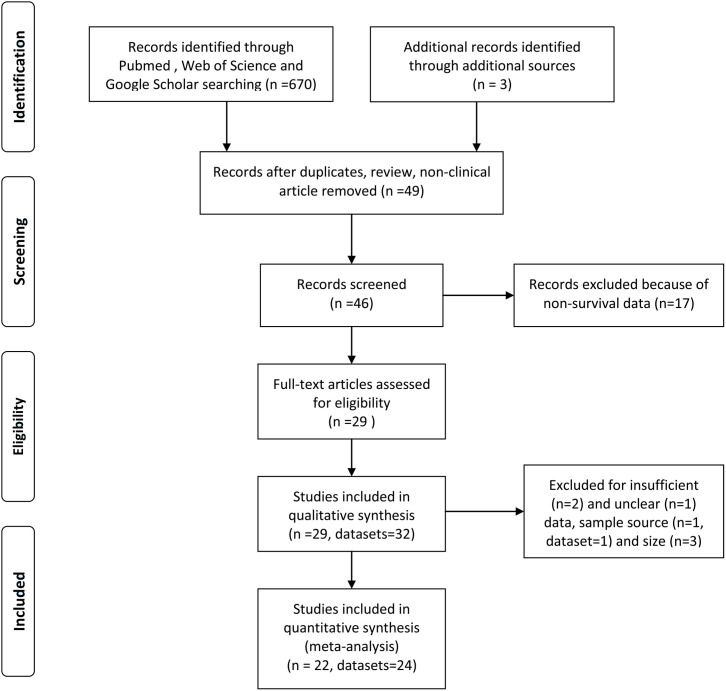
A total of 29 studies9,11,12,13,14,15,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43 with 32 datasets and 4090 patients assessing the relationship between WT1 expression and prognosis of solid tumors were initially identified (Figure 1), and the details of selected studies are shown in Table 1. Twenty-one datasets evaluated carcinoma (9 for ovarian cancer, 5 for breast cancer, 3 for non-small-cell lung cancer [NSCLC], 2 for endometrial cancer, 1 for colorectal cancer and 1 for hepatocellular carcinoma) and 11 datasets evaluated non-carcinoma (2 for soft tissue sarcoma). Twenty-nine datasets reported an adverse impact of WT1 positive expression on prognosis regardless of the statistic power, while, 3 datasets (2 for NSCLC and 1 for soft tissue sarcoma) reported opposite results. Twenty-four datasets used antigen-based methods (21 for immunohistochemistry[IHC], 2 for ELISA and 1 for western blot), and 8 datasets used mRNA-based method (5 for reverse transcription polymerase chain reaction[RT-PCR] and 3 for gene microarray). Thirteen datasets were carried out in Europe, 14 in Asia and 5 in North of America. Twenty datasets had information on overall survival (OS), 5 had information on disease-specific survival (DSS) and 18 had information on disease-free survival (DFS)/relapse or recurrence-free survival (RFS)/progression-free survival (PFS). The median follow-up period of the 12 studies with definite follow-up time was 47 months.
Figure 1. Flow diagram of study identification.
Table 1. Characteristics of studies exploring the relation between WT1 expression and prognosis of solid tumors.
| Author | Year | Patient source | Tumor type | No. of Patients | Stage/Grade | Detection method | No. of WT1 positive expression (%), cutoff value | Follow-up time media(range) | Outcomes, model |
|---|---|---|---|---|---|---|---|---|---|
| Miyoshi Y, et al | 2002 | Japan | Breast cancer | 99 | Operable cancer | RT-PCR | 33/99(33.3%), ≥3.3 | Median 48 m(39–60 m) | DFS, M |
| Dupont J, et al | 2004 | USA | Endometrial carcinoma | 130 | I–IV | IHC | 34/130(26%), score≥ 4 | Median 32 m(1–241 m) | OS, U |
| Hylander B, et al | 2006 | USA | Ovarian cancer | 100 | I–IV | IHC | 78/100(78%), >5% | 1–126 m | OS, U |
| Netinatsunthorn W, et al | 2006 | Thailand | Ovarian cancer | 99 | III–IV | IHC | 50/90(56%), ≥median | 1–168 m | OS/RFS, M |
| Høgdall EV, et al | 2007 | Denmark | Ovarian cancer | 560 | I–IV | IHC | 89/560(16%), >10% | 1–121 m | DSS, M |
| Yamamoto S, et al | 2007 | Japan | Ovarian cancer | 119 | I–IV | IHC | 99/119(83%), >10% | Median 46 m(2–227 m) | DSS, M |
| Sera T, et al | 2008 | Japan | Hepatocellular carcinoma | 42 | I–IV | Western blot | 21/42(50%), ≥1.63 | 1–82 m | DFS, M |
| Kobel, et al | 2008 | Canada | Ovarian carcinoma | 493 | I–III | IHV | 174/493(35%), ≥ 5% | Median 61 m | DSS, M |
| Ohno S, et al | 2009 | Japan | Endometrial cancer | 70 | I–IV | IHC | 31/70(44%), score ≥5 | Median 61 m | OS/RFS, U |
| Oji Y, et ala | 2009 | Japan | NSCLC | 79 | I–III | ELISA | 20/79(25%), median of 8777 WR U | 36 m | DFS, U |
| Bejrananda T, et al | 2010 | Thailand | Colorectal cancer | 157 | I–IV | IHC | 143/157(91%), NA | Median 116 m(77–145 m) | OS, M |
| Camcı, et alb | 2011 | Turkey | Breast cancer | 66 | I–IV | IHC | 32 (48.5%), ≥1+ | NA | RFS, U |
| Vermeij R, et al | 2011 | Netherlands | Ovarian cancer | 229 | I–IV | IHC | 129/229(56%), NA | 1–60 m | DSS/PFS, U |
| Hayashi S, et al | 2012 | Japan | NSCLC | 98 | I–IV | RT-PCR | 60/98(61%), ≥0.0057 | 60 m | OS/DFS, M |
| Qi XW, et al | 2012 | France | Breast cancer | 252 | I–III | Gene microarray | 106/252(42%), ≥2.1 | 1–200 m | DFS, M |
| Qi XW, et al | 2012 | France | Breast cancer | 252 | I–III | Gene microarray | 7/252(3%), ≥3.1 | 1–200 m | DFS, M |
| Choi EJ, et al | 2013 | Korea | Breast cancer | 97 | T1–3 | IHC | 39/97(40), >10% | Median 63 m | DFS, M |
| Wu, et alb | 2013 | China | NSCLC | 159 | I–III | RT-PCR | 79/159(49.7%), ≥3.0013 | 36 | OS/DFS, U |
| Andersson C, et al | 2014 | Sweden | Ovarian carcinoma | 50 | I–IV | IHC | 36/50(72%),>10% | Median 91 m(1–229 m) | OS/PFS, U |
| Andersson C, et ala | 2014 | Sweden | Ovarian carcinoma | 52 | I–IV | ELISA | 26/52(50%), ≥18.8 WRU | Median 91 m(1–229 m) | OS/PFS, U |
| Liu Z, et al | 2014 | Japan | Ovarian cancer | 63 | I–IV | RT-PCR | 18/63(29%), ≥53.94, | Median 14 m(1–66 m) | OS/DFS, U |
| Sotobori T, et al | 2006 | Japan | Soft tissue sarcoma | 52 | NA | RT-PCR | 17/52(33%), ≥0.01 | Median 45 m(9–205 m) | DSS, M |
| Srivastava A, et al | 2006 | USA | Osteogenic sarcoma | 49 | NA | IHC | 12/49(24%), score ≥1 | 4–400 m | OS, U |
| Coosemans A, et al | 2011 | Belgium | Uterine sarcoma | 71 | I–IV | IHC | 49/71(69%), score≥20 | ≥12 m | OS/PFS, M |
| Guntupalli SR, et alc | 2013 | USA | Uterine carcinosarcoma | 64 | I–IV | IHC | 49/64(77%), score >2 | Median 17 m(1–139 m) | OS/PFS, M |
| Kim A, et al | 2014 | Korea | Soft tissue sarcoma | 87 | I–IV | IHC | 47/87(54%), score >21 | Median 29 m(1–187 m) | OS, U |
| Chiba Y, et ald | 2010 | Japan | Glioblastoma | 37 | NA | IHC | 25/37(68%), ≥3score | 1–20 m | OS/PFS, U |
| Garrido-Ruiz MC, et al | 2010 | Spain | Melanoma | 163 | I–IV | IHC | 39.60%, >10% | Median 120 m(1–240 m) | OS, U |
| Scattone A, et ald | 2012 | Italy | Diffuse peritoneal mesothelioma | 31 | I–III | IHC | 10/31(32.3%), ≥25% | 5–60 m | OS, U |
| Cedre's S, et ald | 2013 | Spain | Malignant pleural mesothelioma | 32 | III–IV | IHC | 25/32(78.10%), >1% | 1–40 m | OS, M |
| Rauscher J, et al | 2014 | Germany | Glioma | 328 | Grade II/III | Gene microarray | 96/328(29%), ≥2 | 1–267 m | OS, U |
| Rauscher J, et al | 2014 | Germany | Astrocytoma | 212 | Grade II/III | IHC | 68/212(32%),score ≥1 | 1–150 m | OS, U |
Abbreviations: No. = number; RT-PCR = reverse transcription polymerase chain reaction; IHC = immunohistochemistry; U = univariate; M = multivariate; m = month; OS = overall survival; DFS = disease-free survival; PFS = progression-free survival; DSS = disease-specific survival; RFS = relapse/recurrence-free survival; NSCLC = non-small-cell lung cancer; NA = not available; WRU = WT1-reacting-unit.
aStudy was excluded from quantitative analysis because samples were not form cancer tissue.
bStudy was excluded from quantitative analysis because of insufficient data to estimate HR.
cStudy was excluded from quantitative analysis because of unclear data for survival analysis.
dStudy were excluded from quantitative analysis because of sample size <40.
For quantitative synthesis, 3 studies38,40,41 were excluded due to limited sample size <40, and 2 studies42,43 were rejected for insufficient data, 1 study36 was excluded for unclear data. Finally, 1 study27 and 1 dataset32 were rejected because the samples reported were not tissues-based. Therefore, the consolidated sample for our analysis consisted of 22 studies with 24 datasets. Among the 24 datasets, 14 addressed OS, 5 referred to DSS, and 12 pertained to DFS/RFS/PFS. A total of 3620 patients were enrolled in these studies, and the median sample size was 99, which was used as the criterion for subgroup analysis by sample size.
Quantitative analysis of relationship between WT1 expression and OS
The combined analysis of the 14 datasets showed that WT1 positive expression was significantly associated with worse OS (meta-hazard ratio [metaHR] = 1.48, 95% confidence interval [CI] = 1.11–1.97), and it should be noted that this association only held in univariate model (metaHR = 1.50, 95%CI = 1.11–2.03), but not in multivariate analysis (metaHR = 1.32, 95%CI = 0.59–2.92) (Table 2; Figure 2). However, restricting studies to antigen-based methods, the association between WT1 positive expression and poor OS was indicated in overall (metaHR = 1.67, 95%CI = 1.27–2.21), univariate (metaHR = 1.50, 95%CI = 1.01–2.23) and multivariate (metaHR = 2.06, 95%CI = 1.50–2.83) analyses (Table 3).
Table 2. Meta-analysis of association between WT1 expression and prognosis of solid tumors (WT1 positive versus negative expression).
| OS | DSS | DFS/RFS/PFS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | metaHR(95%CI) | Ph | I2(%) | No. | metaHR(95%CI) | Ph | I2(%) | No. | metaHR(95%CI) | Ph | I2(%) | |
| Total | 14 | 1.48(1.11–1.97) | 0.00 | 65 | 5 | 1.46(0.97–2.20) | 0.01 | 72 | 12 | 2.14(1.42–3.21)a | 0.00 | 72 |
| Cancer type | ||||||||||||
| Carcinoma | 8 | 1.34(0.83–2.15) | 0.00 | 67 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 11 | 2.14(1.37–3.36) | 0.00 | 74 |
| Ovarian | 4 | 1.57(1.10–2.24)b | 0.21 | 34 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 4 | 2.61(1.94–3.52)b | 0.75 | 0 |
| Univariate | 3 | 1.31(0.81–2.10)b | 0.19 | 39 | 1 | 2.10(1.41–3.12) | - | - | 3 | 2.49(1.79–3.44)b | 0.72 | 0 |
| Mutivariate | 1 | 1.98(1.15–3.41) | - | - | 3 | 1.15(0.93–1.43)b | 0.05 | 66 | 1 | 3.36(1.60–7.04) | - | - |
| Breast | 0 | - | - | - | 0 | - | - | - | 4 | 1.43(0.99–2.06)b | 0.10 | 53 |
| Endometrial | 2 | 1.96(1.04–3.72)b | 0.53 | 0 | 0 | - | - | - | 0 | - | - | - |
| Non-carcinoma | 6 | 1.59(1.07–2.37) | 0.01 | 67 | 1 | 2.60(1.00–6.73) | - | - | 1 | 2.17(1.11–4.25) | - | - |
| Uni/Multivariate | ||||||||||||
| Univariate | 10 | 1.50(1.11–2.03) | 0.02 | 53 | 1 | 2.10(1.41–3.12) | - | - | 4 | 2.56(1.87–3.51)b | 0.77 | 0 |
| Mutivariate | 4 | 1.32(0.59–2.92) | 0.00 | 83 | 4 | 1.30(0.83–2.03) | 0.04 | 65 | 8 | 1.86(1.04–3.35) | 0.00 | 78 |
| Detection method | ||||||||||||
| antigen-based | 11 | 1.67(1.27–2.21) | 0.05 | 46 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 7 | 2.61(2.02–3.37)b | 0.26 | 23 |
| mRNA-based | 3 | 0.65(0.14–2.91) | 0.00 | 89 | 1 | 2.60(1.00–6.73) | - | - | 5 | 1.56(0.75–3.24) | 0.00 | 78 |
| Sample size | ||||||||||||
| <99 | 6 | 1.45(0.72–2.91) | 0.00 | 71 | 1 | 2.60(1.00–6.73) | - | - | 5 | 3.16(2.14–4.67)b | 0.56 | 0 |
| ≥99 | 8 | 1.52(1.12–2.08) | 0.01 | 64 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 7 | 1.62(0.94–2.81) | 0.00 | 78 |
| Patient source | ||||||||||||
| Europe | 5 | 1.83(1.52–2.19)b | 0.85 | 0 | 2 | 1.57(0.92–2.66) | 0.03 | 80 | 5 | 1.95(1.52–2.49)b | 0.05 | 57 |
| Asia | 6 | 0.93(0.42–2.05) | 0.00 | 83 | 2 | 2.12(1.23–3.65)b | 0.61 | 0 | 7 | 2.16(1.01–4.62) | 0.00 | 80 |
| North of America | 3 | 1.61(1.06–2.44)b | 0.29 | 20 | 1 | 0.73(0.45–1.18) | - | - | 0 | - | - | - |
| Public bias | ||||||||||||
| Begg's test | 0.38 | 0.81 | 0.45 | |||||||||
| Egger's test | 0.27 | 0.57 | 0.39 | |||||||||
Abbreviations: No. = number; Ph = Pheterogeneity, HR = hazard ratio; CI = confidence interval; OS = overall survival; DSS = disease-specific survival; DFS = disease- free survival; PFS = progression-free survival; RFS = relapse/recurrence-free survival.
Statistically significant associations(P<0.05) are shown in bold if number of studies was equal or more than 2.
aFor DFS, metaHR = 1.41, 95%CI = 1.05–1.88; for RFS, metaHR = 3.47, 95%CI = 1.87–6.45; for PFS, metaHR = 2.38, 95%CI = 1.75–3.23.
bIf number of studies was equal or more than 2, fixed-effects model was used; otherwise, random-effects model was used.
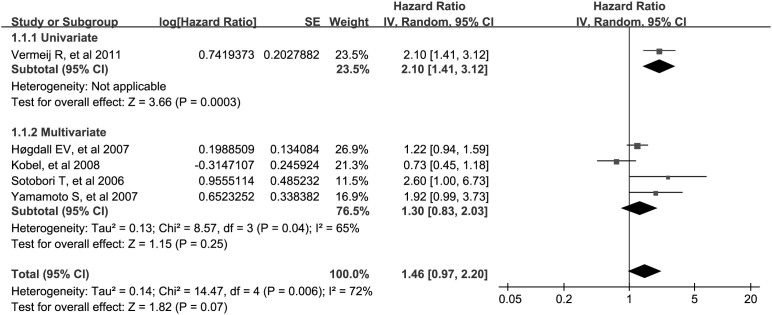
Figure 2. Meta-analysis of impact of WT1 expression on overall survival of patients with solid tumors.
Results are presented as individual and metaHR, and 95% CI.
Table 3. Meta-analysis of association between WT1 expression and prognosis of solid tumors (WT1 positive versus negative expression) restricted to studies with antigen-based methods.
| OS | DSS | DFS/RFS/PFS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | metaHR(95%CI) | Ph | I2(%) | No. | metaHR(95%CI) | Ph | I2(%) | No. | metaHR(95%CI) | Ph | I2(%) | |
| Total | 11 | 1.67(1.27–2.21) | 0.05 | 46 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 7 | 2.61(2.02–3.37)a | 0.26 | 23 |
| Cancer type | ||||||||||||
| Carcinoma | 6 | 1.83(1.41–2.38)a | 0.66 | 0 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 6 | 2.69(2.04–3.55)a | 0.19 | 33 |
| Ovarian | 3 | 1.69(1.16–2.44)a | 0.29 | 19 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 3 | 2.58(1.89–3.52)a | 0.57 | 0 |
| Univariate | 2 | 1.46(0.88–2.43)a | 0.18 | 45 | 1 | 2.10(1.41–3.12) | - | - | 2 | 2.44(1.73–3.44)a | 0.46 | 0 |
| Mutivariate | 1 | 1.98(1.15–3.41) | - | - | 3 | 1.15(0.93–1.43)a | 0.05 | 66 | 1 | 3.36(1.60–7.04) | - | - |
| Endometrial | 2 | 1.96(1.04–3.72)a | 0.53 | 0 | 0 | - | - | - | 1 | 3.74(1.20–11.66) | - | - |
| Non-carcinoma | 5 | 1.52(0.83–2.79) | 0.01 | 72 | 0 | - | - | - | 1 | 2.17(1.11–4.25) | - | - |
| Uni/Multivariate | ||||||||||||
| Univariate | 8 | 1.50(1.01–2.23) | 0.03 | 56 | 1 | 2.10(1.41–3.12) | - | - | 3 | 2.53(1.82–3.51)a | 0.59 | 0 |
| Mutivariate | 3 | 2.06(1.50–2.83)a | 0.89 | 0 | 3 | 1.15(0.93–1.43)a | 0.05 | 66 | 4 | 2.73(1.81–4.12)a | 0.09 | 55 |
| Sample size | ||||||||||||
| <99 | 5 | 1.65(0.77–3.54) | 0.00 | 74 | 0 | - | - | - | 4 | 3.20(2.10–4.87)a | 0.40 | 0 |
| ≥99 | 6 | 1.72(1.39–2.12)a | 0.70 | 0 | 4 | 1.36(0.88–2.10) | 0.01 | 76 | 3 | 2.31(1.67–3.19)a | 0.19 | 40 |
| Patient source | ||||||||||||
| Europe | 4 | 1.81(1.38–2.37)a | 0.71 | 0 | 2 | 1.57(0.92–2.66) | 0.03 | 80 | 3 | 2.38(1.75–3.23)a | 0.73 | 0 |
| Asia | 4 | 1.41(0.66–3.04) | 0.00 | 79 | 1 | 1.92(0.99–3.73) | - | - | 4 | 3.23(2.02–5.18)a | 0.11 | 50 |
| North of America | 3 | 1.61(1.06–2.44)a | 0.29 | 20 | 1 | 0.73(0.45–1.18) | - | - | 0 | - | - | - |
| Public bias | ||||||||||||
| Begg's test | 0.44 | 1.00 | 0.76 | |||||||||
| Egger's test | 0.92 | 0.86 | 0.65 | |||||||||
Abbreviations: No. = number; Ph = Pheterogeneity, HR = hazard ratio; CI = confidence interval; OS = overall survival; DSS = disease-specific survival; DFS = disease- free survival; PFS = progression-free survival; RFS = relapse/recurrence-free survival.
Statistically significant associations(P<0.05) are shown in bold if number of studies was equal or more than 2.
a If number of studies was equal or more than 2, fixed-effects model was used; otherwise, random-effects model was used.
Subgroup analyses by cancer type showed that WT1 positive expression had an unfavorable impact on OS for patients with ovarian cancer (metaHR = 1.57, 95%CI = 1.10–2.24), endometrial cancer (metaHR = 1.96, 95%CI = 1.04–3.72) and non-carcinoma malignancies (metaHR = 1.59, 95%CI = 1.07–2.37) (Table 2). For ovarian cancer, the prognostic value of WT1 was evaluated only in univariate model (metaHR = 1.31, 95%CI = 0.81–2.10), because only 1 study reported multivariate HR (Table 2). Subgroup analyses also suggested that the correlation of WT1 positive expression with worse OS was statistically significant among studies with using antigen-based methods (metaHR = 1.67, 95%CI = 1.27–2.21), with sample size ≥99 (metaHR = 1.52, 95%CI = 1.12–2.08), and in Europe (metaHR = 1.83, 95%CI = 1.52–2.19) as well as North of America (metaHR = 1.61, 95%CI = 1.06–2.44) (Table 2).
Quantitative analysis of relationship between WT1 expression and DSS
The combined analysis of the 5 datasets showed that WT1 positive expression had borderline association with worse DSS (metaHR = 1.46, 95%CI = 0.97–2.20), and this association was also not significant in multivariate model (metaHR = 1.30, 95%CI = 0.83–2.03) (Table 2; Figure 3). In addition, restricting studies to antigen-based methods did not alter the results (for overall analysis, metaHR = 1.36, 95%CI = 0.88–2.10; for multivariate analysis, metaHR = 1.15, 95%CI = 0.93–1.43) (Table 3). Subgroup analyses also did not reveal any significant associations, except for studies carried out in Asia (metaHR = 2.12, 95%CI = 1.23–3.65) (Table 2).
Figure 3. Meta-analysis of impact of WT1 expression on disease-specific survival of patients with solid tumors.
Results are presented as individual and metaHR, and 95% CI.
Quantitative analysis of relationship between WT1 expression and DFS/RFS/PFS
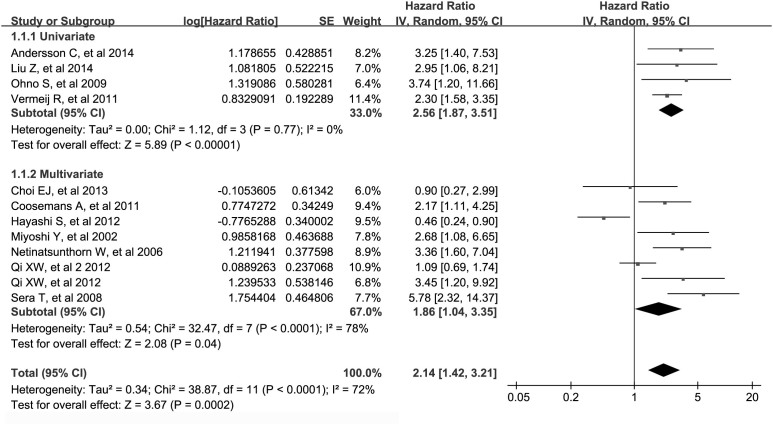
The combined analysis of the 12 datasets showed that WT1 positive expression was associated with worse DFS/RFS/PFS (metaHR = 2.14, 95%CI = 1.42–3.21), and these associations were clearly evident both in univariate (metaHR = 2.56, 95%CI = 1.87–3.51) and multivariate models (metaHR = 1.86, 95%CI = 1.04–3.35) (Table 2; Figure 4). In addition, after restricting studies to antigen-based methods, the associations were still significant in overall (metaHR = 2.61, 95%CI = 2.02–3.37), univariate (metaHR = 2.53, 95%CI = 1.82–3.51) and multivariate (metaHR = 2.73, 95%CI = 1.81–4.12) analyses (Table 3).
Figure 4. Meta-analysis of impact of WT1 expression on disease-free survival/progression-free survival/relapse or recurrence-free survival of patients with solid tumors.
Results are presented as individual and metaHR, and 95% CI.
Subgroup analyses by cancer type showed that WT1 positive expression had an unfavorable impact on DFS/RFS/PFS for patients with carcinoma (metaHR = 2.14, 95%CI = 1.37–3.36), including ovarian cancer (metaHR = 2.61, 95% CI = 1.94–3.52). For ovarian cancer, the prognostic value of WT1 was demonstrated only in univariate model (metaHR = 2.49, 95%CI = 1.79–3.44) (Table 2). Subgroup analyses also found that the correlation of WT1 positive expression with worse DFS/RFS/PFS was significantly evident among studies using antigen-based methods (metaHR = 2.61, 95%CI = 2.02–3.37), with sample size<99 (metaHR = 3.16, 95%CI = 2.14–4.67), and in Europe (metaHR = 1.95, 95%CI = 1.52–2.49) as well as Asia (metaHR = 2.16, 95%CI = 1.01–4.62) (Table 2).
Publication bias
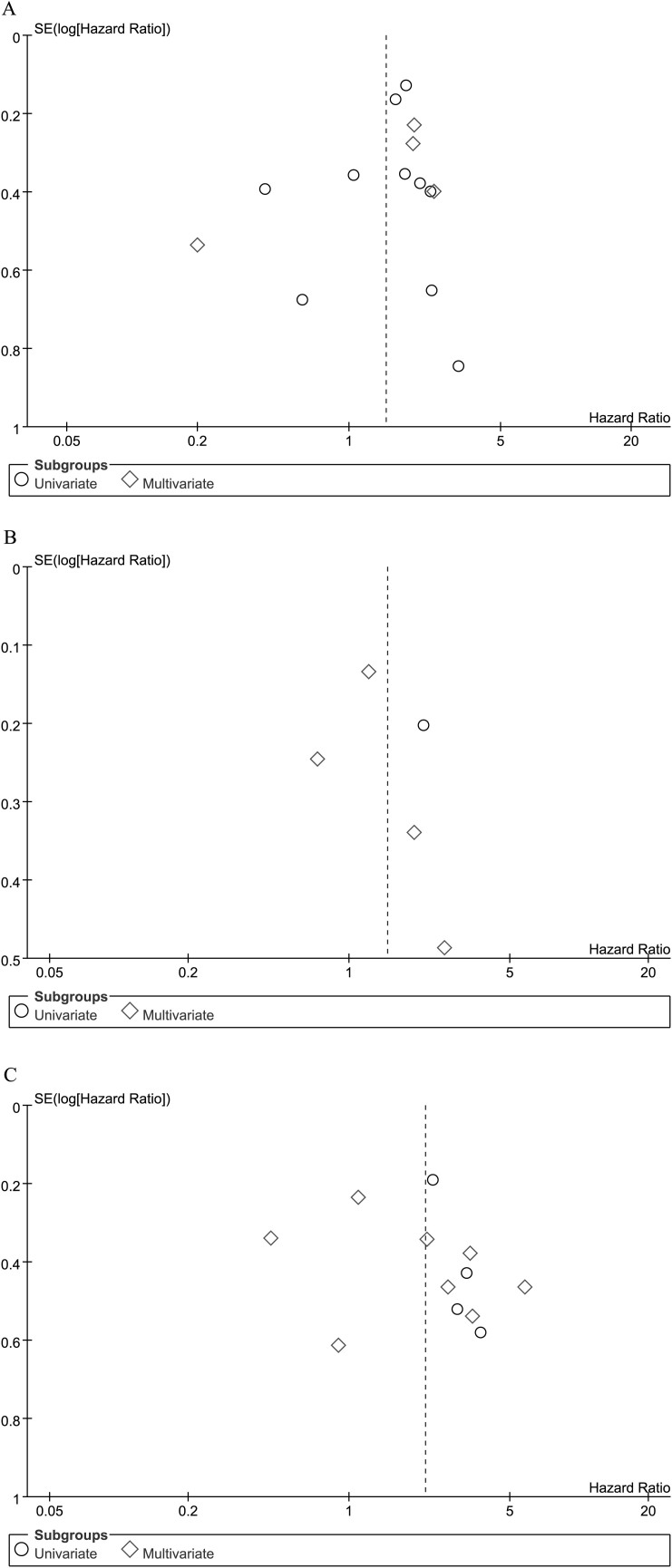
The shapes of the funnel plots did not reveal evidence of obvious asymmetry for OS (Figure 5A), DSS (Figure 5B) and DFS/RFS/PFS (Figure 5C) analyses, which was confirmed by the Begg's and Egger's tests (Table 2). The results of publication bias for studies using antigen-based methods to detect WT1 expression are shown in Table 3.
Figure 5. Funnel plot for the evaluation of potential publication bias in the impact of WT1 on overall survival (A), disease-specific survival (B) and disease-free survival/progression-free survival/relapse or recurrence-free survival (C) of patients with solid tumors.
The funnel graph plots the log of HR against the standard error of the log of the HR (an indicator of sample size). The circles indicate the individual studies in the meta-analysis. The line in the center represents the metaHR.
Sensitivity analysis
Sensitivity analysis was performed by sequential removal of individual studies to assess the influence of each study on the metaHRs. Results showed that metaHRs were not statistically altered (data not shown).
Discussions
The WT1 gene, which encodes a protein consisting of four zinc finger domains at the C terminus and an glutamine and proline-rich domain at the N terminus, plays an important role in cell proliferation, differentiation, apoptosis, organ development and the maintenance of several adult tissues44,45. Though recognized as a classic tumor suppressor gene in Wilms' tumor, there is a growing body of evidence demonstrating that wild-type WT1 is expressed in a variety of tumors arising from different tissues that normally do not express WT146,47,48. It is possible that WT1 could inhibit cell apoptosis by transcriptional activation and/or upregulation of proto-oncogenes10. Recent work has also revealed that WT1 is a key regulator in overcoming senescence downstream of KRAS signaling49, and is involved in the apoptotic response to cytotoxic stress50, further demonstrating its oncogenic effects. Moreover, WT1 could also promote invasion, migration and metastasis43,51,52, facilitate angiogenesis53,54 and confer drug resistance to cancer cells55,56.
Because of its implications in the etiology of cancer, clinical values of WT1 expression are the subject of increasing scrutiny. To date, the role of WT1 expression as a potential prognostic marker has been extensively explored in a number of solid tumors as listed in Table 1. In addition, considering its unique features16,18, much attention has been given to the use of WT1 peptides in eliciting an immune response. In phase I and II clinical trials for solid tumors, two patients obtained a complete remission, three had a partial remission, and five showed a decrease in tumor marker or tumor size18. Nevertheless, the clinical implications of WT1 expression are still largely unknown, and the prognostic value of WT1 expression in solid tumors remains controversial. Therefore, we conducted the present systemic review and meta-analysis to clarify this issue and to provide more evidence for facilitating the development of anti-WT1 target research.
The present study included a total of 29 eligible studies with 32 datasets and 4090 patients for qualitative analysis. Ovarian cancer ranked first among the reported cancer types, and IHC was most commonly used for detection. For most of these studies, positive expression of WT1 was linked with an unfavorable impact on the prognosis. However, two of three studies for NSCLC (one using ELISA, and the other one using RT-PCR) revealed that patients with high WT1 expression had a better prognosis than controls, which might be attributed to differences in detection methods as well as cellular context49. For meta-analysis, we excluded studies using ELISA as a detection method because the source WT1 IgG antibody in plasma was largely unclear and controversial27,32. Finally, 22 studies with 24 datasets and 3620 patients were subjected to quantitative analysis. Overall, WT1 positive expression showed significant association with worse OS and DFS/RFS/PFS, and borderline association with poor DSS, in solid cancer patients. Multivariate analyses indicated that WT1 positive expression was an independent unfavorable predictor for DFS/RFS/PFS in solid cancer patients, and the correlation became more prominent if we only focused on WT1 protein expression. Taking together, we can infer that WT1 may be of limited prognostic value for all-caused and disease-specific mortality in solid tumor patients. However, it could be a potentially useful marker to predict the risk of relapse and progression for these patients.
Since ovarian cancer was the most common cancer type reported in the current meta-analysis, we conducted overall and subgroup (uni/multivariate) analyses to explore the correlations of WT1 expression with prognosis in these patients. Results showed that WT1 positive expression could only predict poor outcomes for ovarian cancer in univariate model, but not in multivariate model. Restricting studies to antigen-based methods did not alter the results, which indicated that WT1 was not an independent prognostic marker for ovarian cancer. In fact, two large scale studies with a sample size ≥50011,24 also found that WT1 might be of limited prognostic value in ovarian cancer patients under multivariable model. There are several distinct subtypes of ovarian cancer, which develop differently and respond differently to chemotherapy11. At the same time, WT1 was expressed differently among these subtypes, mostly in serous carcinoma11,22,24,29,33. Therefore, the negative prognostic significance of WT1 in the entire cohort might reflect subtype differences in expression. It is obviously desirable to investigate the prognostic value of WT1 in each subtype individually rather than treating ovarian cancer as a homogeneous malignancy; however, since it is not feasible at present to secure the specific data regarding WT1 expression status and patients' survival data in each ovarian cancer subtype, we were unable to do this potentially important individualized analysis. Therefore, studies attempting to clarify the specific association between WT1 expression and prognosis in subtypes of ovarian cancer are strongly encouraged, as they offer the distinct possibility of advancing our insights into the clinical implications of WT1 expression in ovarian cancer.
It should be noted that there are some limitations to the analyses presented here. First, publication bias is a concern, because more positive results tended to be published, thus potentially exaggerating the association between WT1 expression and poor outcomes. Second, despite intensive effort, it was not possible to communicate with some investigators as we attempted to indentify all relevant date. In addition, because of limited number of studies, we were unable to conduct analyses for certain types of cancer, such as non-carcinoma cancer, and subtypes of specific cancer, such as ovarian cancer; in addition, we were also not allowed to do subgroup analyses specific to DFS/RFS/PFS. Third, some HRs were estimated indirectly as previously reported57, however, these data were less reliable than direct data from the original literature. Finally, though subgroup analysis by detection method was done, we were unable to carry out stratified analysis according to cutoff values of WT1 expression owing to great methodological variation among studies we selected here. Clearly, it is urgent to establish a uniform and validated method for assessment of WT1 expression to reduce the substantial heterogeneity currently present in clinical reporting protocols.
In summary, our study suggests that WT1 may be a potential marker to predict the risk of relapse and progression in solid cancer patients, but the indicative value of WT1 for all-caused and disease-specific mortality in these patients might be limited. Further data from large multicenter prospective studies are needed to validate the clinical importance of WT1.
Methods
This study was performed according to the guidelines and recommendations for improving the quality of reporting of medical research such as REMARK and PRISMA58,59.
Search strategy
A systematic literature search was performed to identify articles regarding WT1 and prognosis of solid tumors. The Pubmed, Web of Science, Google Scholar were used simultaneously, with the combination of terms “WT1 or Wilms' tumor 1 or Wilms' tumor gene 1 or Wilms' tumor protein 1 or Wilms' tumor suppressor gene 1” and “survival or prognosis or mortality or death” and ‘‘neoplasm or cancer or tumor or carcinoma’’ (up to 8 August 2014). All articles were initially reviewed by abstract and title browsing to select the relevant reports, which were subjected to further screening.
Study selection criteria
Studies that were included in the systemic review had to meet all of the following criteria: 1) the publication explored the relation between WT1 expression and solid tumor prognosis, such as OS, DSS, DFS, RFS and PFS, 2) if one study reported multi-datasets based on different detection methods, cutoff values or populations, datasets would been recognized individually, and 3) publication language was confined to English. For qualitative meta-analysis, additional criteria were needed: 1) there were sufficient, clear, and available data to extract or estimate57 the individual HR and 95% CI, 2) sample size was 40 or more, 3) WT1 expression was detected in cancer tissues.
Data extraction
Three authors (X.W. Qi, F. Zhang and H. Wu) independently extracted information using predefined criteria. The following information of each study was collected: first author, reference year, patient source, cancer type, number of patients, detection method, number and percentage of WT1 positive expression and its cutoff value, median and range of follow-up time, outcome endpoint, univariate or multivariate HR and 95%CI for WT1 positive expression (exposed group) versus WT1 negative expression (unexposed group). If univariate and multivariate HR and 95%CI were both reported, we preferred the multivariate result. For the studies with inadequate or unclear information, the authors were contacted by E-mail if possible. However, information from those authors was not forthcoming.
Statistical analysis
MetaHR and 95% CI were applied to assess the association between WT1 expression and outcomes of solid cancer patients. Outcome endpoints were divided into three groups, OS, DSS and DFS/RFS/PFS, based on the data acquired in the current study and previous report60. An observed metaHR >1 implied increased hazard of all-caused mortality for OS, disease-specific mortality for DSS, and relapse/progression for DFS/RFS/PFS, respectively, in patients with WT1 positive expression. Subgroup analyses were done according to cancer type (carcinoma and non-carcinoma; for carcinoma, ovarian cancer, breast cancer, endometrial cancer and others), uni/multivariate model, detection method (antigen-based and mRNA-based), sample size (<median value and ≥median value) and patient source (Europe, Asia and North of America). Because it was the most common method used to detect WT1 expression, we conducted overall and subgroup analyses restricted to studies with antigen-based methods. In addition, we also did subgroup analysis by uni/multivariate approaches to explore whether WT1 was an independent prognostic factor for patients with ovarian cancer, since it was the most extensively studied type of solid cancer in the current meta-analysis. Heterogeneity assumption was assessed by chi-based Q-test and I2 test. The heterogeneity was considered statistically significant if P < 0.05. If heterogeneity across studies was not identified, the fixed-effects model (Mantel-Haenszel) was used. Otherwise, the random effects model (DerSimonian and Laird) was used. Publication bias was analyzed by performing funnel plots qualitatively, and estimated by Begg's and Egger's test quantitatively. Sensitivity analysis was carried out by removing each study at a time to evaluate the stability of the results. Two-sided P < 0.05 was considered statistically significant. The metaHRs and funnel plots were generated by Review Manager Version 5.3 (The Nordic Cochrane Centre, Copenhagen, Denmark). The analysis of publication bias and sensitivity was performed using STATA version 11.0 (StataCorp, College Station, TX, USA).
Author Contributions
Conceived and designed the experiments: J.J., X.W.Q. Performed the experiments: X.W.Q., F.Z. and H.W. Analyzed the data: X.W.Q., F.Z., H.W. Contributed reagents/materials/analysis tools: X.W.Q., J.L.L., B.G.Z. and C.X. Wrote the paper: X.W.Q., F.Z. and J.J. All authors reviewed and approved the final manuscript.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 81102030), and Key Laboratory of Tumor Immunopathology, Ministry of Education of China (No. 2012JSZ101). We would like to extend our sincere gratitude and appreciation to Steve Ward, Professor Emeritus of Department of Anatomy and Neurobiology, Northeastern Ohio Medical University, Rootstown, OH, USA, for language editing, and Ying Zhang from University of San Francisco, San Francisco, CA,USA, for proof-reading.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–74 (2011). [DOI] [PubMed] [Google Scholar]
- Essafi A. & Hastie N. D. WT1 the oncogene: a tale of death and HtrA. Mol Cell 37, 153–5 (2010). [DOI] [PubMed] [Google Scholar]
- Call K. M. et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell 60, 509–20 (1990). [DOI] [PubMed] [Google Scholar]
- Miwa H., Beran M. & Saunders G. F. Expression of the Wilms' tumor gene (WT1) in human leukemias. Leukemia 6, 405–9 (1992). [PubMed] [Google Scholar]
- Oji Y. et al. Expression of the Wilms' tumor gene WT1 in solid tumors and its involvement in tumor cell growth. Jpn J Cancer Res 90, 194–204 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen H. D. et al. Wilms' tumor gene (WT1) expression in lung cancer, colon cancer and glioblastoma cell lines compared to freshly isolated tumor specimens. J Cancer Res Clin Oncol 126, 226–32 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka S. et al. Immunohistochemical detection of WT1 protein in a variety of cancer cells. Mod Pathol 19, 804–14 (2006). [DOI] [PubMed] [Google Scholar]
- Miyoshi Y. et al. High expression of Wilms' tumor suppressor gene predicts poor prognosis in breast cancer patients. Clin Cancer Res 8, 1167–71 (2002). [PubMed] [Google Scholar]
- Tatsumi N. et al. Wilms' tumor gene WT1-shRNA as a potent apoptosis-inducing agent for solid tumors. Int J Oncol 32, 701–11 (2008). [PubMed] [Google Scholar]
- Kobel M. et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 5, e232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera T. et al. Wilms' tumour 1 gene expression is increased in hepatocellular carcinoma and associated with poor prognosis. Eur J Cancer 44, 600–8 (2008). [DOI] [PubMed] [Google Scholar]
- Qi X. W. et al. High Wilms' tumor 1 mRNA expression correlates with basal-like and ERBB2 molecular subtypes and poor prognosis of breast cancer. Oncol Rep 28, 1231–6 (2012). [DOI] [PubMed] [Google Scholar]
- Sotobori T. et al. Prognostic significance of Wilms tumor gene (WT1) mRNA expression in soft tissue sarcoma. Cancer 106, 2233–40 (2006). [DOI] [PubMed] [Google Scholar]
- Rauscher J. et al. WT1 expression increases with malignancy and indicates unfavourable outcome in astrocytoma. J Clin Pathol 67, 556–61 (2014). [DOI] [PubMed] [Google Scholar]
- Cheever M. A. et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res 15, 5323–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T. et al. Targeting the intracellular WT1 oncogene product with a therapeutic human antibody. Sci Transl Med 5, 176ra33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche A., Berneman Z. N. & Van Tendeloo V. F. Active specific immunotherapy targeting the Wilms' tumor protein 1 (WT1) for patients with hematological malignancies and solid tumors: lessons from early clinical trials. Oncologist 17, 250–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H. et al. Impact of dendritic cell vaccines pulsed with Wilms' tumour-1 peptide antigen on the survival of patients with advanced non-small cell lung cancers. Eur J Cancer 49, 852–9 (2013). [DOI] [PubMed] [Google Scholar]
- Koido S. et al. Treatment with chemotherapy and dendritic cells pulsed with multiple Wilms' tumor 1 (WT1)-specific MHC class I/II-restricted epitopes for pancreatic cancer. Clin Cancer Res 20, 4228–39 (2014). [DOI] [PubMed] [Google Scholar]
- Dupont J. et al. Wilms Tumor Gene (WT1) and p53 expression in endometrial carcinomas: a study of 130 cases using a tissue microarray. Gynecol Oncol 94, 449–55 (2004). [DOI] [PubMed] [Google Scholar]
- Hylander B. et al. Expression of Wilms tumor gene (WT1) in epithelial ovarian cancer. Gynecol Oncol 101, 12–7 (2006). [DOI] [PubMed] [Google Scholar]
- Netinatsunthorn W., Hanprasertpong J., Dechsukhum C., Leetanaporn R. & Geater A. WT1 gene expression as a prognostic marker in advanced serous epithelial ovarian carcinoma: an immunohistochemical study. BMC Cancer 6, 90 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogdall E. V. et al. Expression level of Wilms tumor 1 (WT1) protein has limited prognostic value in epithelial ovarian cancer: from the Danish "MALOVA" ovarian cancer study. Gynecol Oncol 106, 318–24 (2007). [DOI] [PubMed] [Google Scholar]
- Yamamoto S. et al. Clinicopathological significance of WT1 expression in ovarian cancer: a possible accelerator of tumor progression in serous adenocarcinoma. Virchows Arch 451, 27–35 (2007). [DOI] [PubMed] [Google Scholar]
- Ohno S. et al. Immunohistochemical detection of WT1 protein in endometrial cancer. Anticancer Res 29, 1691–5 (2009). [PubMed] [Google Scholar]
- Oji Y. et al. WT1 IgG antibody for early detection of nonsmall cell lung cancer and as its prognostic factor. Int J Cancer 125, 381–7 (2009). [DOI] [PubMed] [Google Scholar]
- Bejrananda T. et al. WT1 expression as an independent marker of poor prognosis in colorectal cancers. Cancer Biomark 8, 35–42 (2010). [DOI] [PubMed] [Google Scholar]
- Vermeij R. et al. Tumor-infiltrating cytotoxic T lymphocytes as independent prognostic factor in epithelial ovarian cancer with wilms tumor protein 1 overexpression. J Immunother 34, 516–23 (2011). [DOI] [PubMed] [Google Scholar]
- Hayashi S. et al. Low Wilms' tumor gene expression in tumor tissues predicts poor prognosis in patients with non-small-cell lung cancer. Cancer Invest 30, 165–71 (2012). [DOI] [PubMed] [Google Scholar]
- Choi E. J. et al. Prognostic significance of RSPO1, WNT1, P16, WT1, and SDC1 expressions in invasive ductal carcinoma of the breast. World J Surg Oncol 11, 314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson C. et al. Prognostic significance of specific anti-WT1 IgG antibody level in plasma in patients with ovarian carcinoma. Cancer Med 3, 909–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. High levels of Wilms' tumor 1 (WT1) expression were associated with aggressive clinical features in ovarian cancer. Anticancer Res 34, 2331–40 (2014). [PubMed] [Google Scholar]
- Srivastava A. et al. High WT1 expression is associated with very poor survival of patients with osteogenic sarcoma metastasis. Clin Cancer Res 12, 4237–43 (2006). [DOI] [PubMed] [Google Scholar]
- Coosemans A. et al. Wilms tumor gene 1 (WT1) is a prognostic marker in high-grade uterine sarcoma. Int J Gynecol Cancer 21, 302–8 (2011). [DOI] [PubMed] [Google Scholar]
- Guntupalli S. R. et al. Wilms' tumor 1 protein and estrogen receptor beta expression are associated with poor outcomes in uterine carcinosarcoma. Ann Surg Oncol 20, 2373–9 (2013). [DOI] [PubMed] [Google Scholar]
- Kim A. et al. Prognostic significance of WT1 expression in soft tissue sarcoma. World J Surg Oncol 12, 214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y. et al. Prognostic value of WT1 protein expression level and MIB-1 staining index as predictor of response to WT1 immunotherapy in glioblastoma patients. Brain Tumor Pathol 27, 29–34 (2010). [DOI] [PubMed] [Google Scholar]
- Garrido-Ruiz M. C., Rodriguez-Pinilla S. M., Perez-Gomez B. & Rodriguez-Peralto J. L. WT 1 expression in nevi and melanomas: a marker of melanocytic invasion into the dermis. J Cutan Pathol 37, 542–8 (2010). [DOI] [PubMed] [Google Scholar]
- Scattone A. et al. High Wilms' tumour gene (WT1) expression and low mitotic count are independent predictors of survival in diffuse peritoneal mesothelioma. Histopathology 60, 472–81 (2012). [DOI] [PubMed] [Google Scholar]
- Cedres S. et al. Expression of Wilms' tumor gene (WT1) is associated with survival in malignant pleural mesothelioma. Clin Transl Oncol 16, 776–82 (2014). [DOI] [PubMed] [Google Scholar]
- Camcý C. et al. Prognostic significance of Wilms Tumor 1 (WT1) protein expression in breast cancer. Gaziantep Med J 17, 67–72 (2011). [Google Scholar]
- Wu C. et al. WT1 Promotes Invasion of NSCLC via Suppression of CDH1. J Thorac Oncol 8, 1163–9 (2013). [DOI] [PubMed] [Google Scholar]
- Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 11, 111–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y. Y. & Hastie N. D. The role of Wt1 in regulating mesenchyme in cancer, development, and tissue homeostasis. Trends Genet 28, 515–24 (2012). [DOI] [PubMed] [Google Scholar]
- Hohenstein P. & Hastie N. D. The many facets of the Wilms' tumour gene, WT1. Hum Mol Genet 15 Spec No 2, R196–201 (2006). [DOI] [PubMed] [Google Scholar]
- Yang L., Han Y., Suarez Saiz F. & Minden M. D. A tumor suppressor and oncogene: the WT1 story. Leukemia 21, 868–76 (2007). [DOI] [PubMed] [Google Scholar]
- Sugiyama H. WT1 (Wilms' tumor gene 1): biology and cancer immunotherapy. Jpn J Clin Oncol 40, 377–87 (2010). [DOI] [PubMed] [Google Scholar]
- Vicent S. et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest 120, 3940–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkamp J., Carpenter B. & Roberts S. G. The Wilms' tumor suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol Cell 37, 159–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbolina M. V., Adley B. P., Shea L. D. & Stack M. S. Wilms tumor gene protein 1 is associated with ovarian cancer metastasis and modulates cell invasion. Cancer 112, 1632–41 (2008). [DOI] [PubMed] [Google Scholar]
- Brett A., Pandey S. & Fraizer G. The Wilms' tumor gene (WT1) regulates E-cadherin expression and migration of prostate cancer cells. Mol Cancer 12, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty G., Awad O. & Loeb D. M. WT1 protein directly regulates expression of vascular endothelial growth factor and is a mediator of tumor response to hypoxia. J Biol Chem 286, 43634–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katuri V. et al. WT1 regulates angiogenesis in Ewing Sarcoma. Oncotarget 5, 2436–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. et al. Wilms' tumor 1 suppressor gene mediates antiestrogen resistance via down-regulation of estrogen receptor-alpha expression in breast cancer cells. Mol Cancer Res 6, 1347–55 (2008). [DOI] [PubMed] [Google Scholar]
- Perugorria M. J. et al. Wilms' tumor 1 gene expression in hepatocellular carcinoma promotes cell dedifferentiation and resistance to chemotherapy. Cancer Res 69, 1358–67 (2009). [DOI] [PubMed] [Google Scholar]
- Tierney J. F., Stewart L. A., Ghersi D., Burdett S. & Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane L. M. et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 93, 387–91 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J. & Altman D. G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6, e1000097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti A. et al. Pathologic Complete Response As a Potential Surrogate for the Clinical Outcome in Patients With Breast Cancer After Neoadjuvant Therapy: A Meta-Regression of 29 Randomized Prospective Studies. J Clin Oncol 32, 3883–3891 (2014). [DOI] [PubMed] [Google Scholar]