Abstract
The eukaryotic genome has vast intergenic regions containing transposons, pseudogenes, and other repetitive sequences. They produce numerous long noncoding RNAs (lncRNAs) and Piwi-interacting RNAs (piRNAs), yet the functions of the vast intergenic regions remain largely unknown. Mammalian piRNAs are abundantly expressed from the spermatocyte to round spermatid stage, coinciding with the widespread expression of lncRNAs in these cells. Here, we show that piRNAs derived from transposons and pseudogenes mediate the degradation of a large number of mRNAs and lncRNAs in mouse late spermatocytes. In particular, they have a large impact on the lncRNA transcriptome, as a quarter of lncRNAs expressed in late spermatocytes are up-regulated in mice deficient in the piRNA pathway. Furthermore, our genomic and in vivo functional analyses reveal that retrotransposon sequences in the 3′ UTR of mRNAs are targeted by piRNAs for degradation. Similarly, the degradation of spermatogenic cell-specific lncRNAs by piRNAs is mediated by retrotransposon sequences. Moreover, we show that pseudogenes regulate mRNA stability via the piRNA pathway. The degradation of mRNAs and lncRNAs by piRNAs requires PIWIL1 (also known as MIWI) and, at least in part, depends on its slicer activity. Together, these findings reveal the presence of a highly complex and global RNA regulatory network mediated by piRNAs with retrotransposons and pseudogenes as regulatory sequences.
The genomes of complex eukaryotes contain vast intergenic regions that are mostly composed of repetitive DNA with transposons as a major constituent (Taft et al. 2007). Although protein-coding genes have been extensively studied over the past half-century and many genomes have been sequenced in the past decade, the functions of the vast majority of the genome are largely unknown. Although it is now apparent that some transposons and pseudogenes participate in gene regulation (Lunyak et al. 2007; Goodier and Kazazian 2008; Tam et al. 2008; Watanabe et al. 2008; Lynch et al. 2011; Schmidt et al. 2012), the function of the majority of these sequences remains unknown. The recent advent of deep sequencing technology further revealed that a large portion of the mammalian genome is transcribed into an immense number of noncoding RNAs, including more than 10,000 long noncoding RNAs (lncRNAs), thousands of expressed pseudogenes, and millions of Piwi-interacting RNAs (piRNAs)—a class of 21- to 32-nucleotide (nt) small noncoding RNAs predominantly expressed in the germline (Kim et al. 2009; Cabili et al. 2011; Gong and Maquat 2011; Juliano et al. 2011; Ishizu et al. 2012; Kalyana-Sundaram et al. 2012; Kelley and Rinn 2012). The discovery of these new types of RNAs reveals additional levels of genome complexity and presents unprecedented challenges to understanding the function and interrelatedness of the noncoding regions of the genome.
piRNAs bind to Piwi proteins, which represent a subfamily of the Argonaute protein family (Kim et al. 2009; Juliano et al. 2011; Ishizu et al. 2012; Pillai and Chuma 2012; Ross et al. 2014). piRNAs are generated from various portions of longer single-stranded piRNA precursors, which are transcribed from hundreds of genomic loci known as piRNA clusters that are mostly located in the intergenic regions of the genome. Piwi proteins find their target RNAs by using piRNAs as guides and cleave the targets through the RNase (slicer) activity of the PIWI domain (Saito et al. 2006; Reuter et al. 2011). Additionally, they are believed to interact with factors involved in RNA degradation and chromatin modification (Rouget et al. 2010; Gou et al. 2014; Ross et al. 2014). Across the animal kingdom, Piwi proteins and piRNAs suppress retrotransposons in order to prevent excessive mutations in the germline genome (Grimson et al. 2008). Although the regulation of protein-coding genes by piRNAs has been suggested by a few studies in Drosophila and Caenorhabditis elegans (Ishizu et al. 2012), little is known about piRNA function beyond retrotransposon suppression.
During mouse spermatogenesis, the three Piwi proteins, PIWIL1, PIWIL2, and PIWIL4 (also known as MIWI, MILI, and MIWI2), and piRNAs are highly expressed in two phases: the gonocyte stage and the pachytene spermatocyte to round spermatid stage (Pillai and Chuma 2012). PIWIL2 and PIWIL4 are expressed in gonocytes and bind to gonocyte piRNAs (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). PIWIL1 and PIWIL2 are expressed in pachytene spermatocytes and round spermatids, where they bind to pachytene piRNAs (Robine et al. 2009; Reuter et al. 2011; Beyret et al. 2012). Ectopic expression of artificial pachytene piRNAs leads to the degradation of the complementary reporter RNA in pachytene spermatocytes and round spermatids (Yamamoto et al. 2013), suggesting that pachytene piRNAs degrade target RNAs during this period of spermatogenesis. LINE-1 (L1) retrotransposon suppression by PIWIL1 and pachytene piRNAs has been reported (Xu et al. 2008; Reuter et al. 2011); however, other targets of pachytene piRNAs are unknown. Here we report the systematic identification of the target RNAs of pachytene piRNAs and the regulatory sequences in the target RNAs. These analyses, together with functional analyses using several newly generated mouse strains, demonstrate that transposons and pseudogenes regulate mRNAs and lncRNAs via piRNAs.
Results
mRNAs are negatively regulated by the piRNA pathway
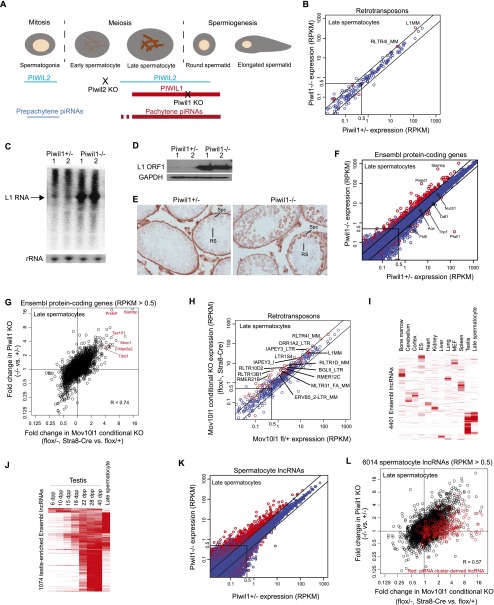
Pachytene piRNAs are expressed from the pachytene spermatocyte to round spermatid stage and bind to PIWIL1 and PIWIL2 (Fig. 1A). In the adult testis, PIWIL1-bound piRNAs, mostly 29–30 nt in length, are more abundant than PIWIL2-bound piRNAs, which are 25–26 nt in length (Supplemental Fig. 1A). Moreover, most of the pachytene piRNAs expressed in late spermatocytes, herein defined as from the mid-pachytene to diakinesis stage, are lost in Piwil1−/− mice (Supplemental Fig. 1B). Together, these results indicate that the expression of a large portion of piRNAs in late spermatocytes is PIWIL1-dependent. To identify endogenous target RNAs of PIWIL1-piRNA complexes, we performed RNA-seq analysis in Piwil1+/− and Piwil1−/− mice (Supplemental Table S1). To minimize the background signal from cells not expressing PIWIL1, we isolated late spermatocytes by fluorescence-activated cell sorting (FACS) from 21- to 23-d post-partum (dpp) mouse testes (Supplemental Fig. 1C,D). Although changes of RNA expression in Piwil1−/− mice were expected to be larger at the round spermatid stage, we selected the late spermatocyte stage, the initial stage of PIWIL1 expression, to minimize secondary effects due to developmental abnormalities (Deng and Lin 2002). We found that the L1 and RLTR4I_MM retrotransposon RNAs were increased in Piwil1−/− late spermatocytes as compared with Piwil1+/− late spermatocytes (Fig. 1B–E). Interestingly, the expression of protein-coding mRNAs was also affected in Piwil1−/− late spermatocytes (Fig. 1F). Here, 172 genes were significantly up-regulated (more than twofold increase, P < 0.025, RPKM > 0.5), and 11 were significantly down-regulated (more than twofold decrease, P < 0.025, RPKM > 0.5). We did not analyze down-regulated mRNAs in this study, because a decrease of these mRNAs is probably secondary, due to developmental abnormalities (Supplemental Results; Supplemental Fig. 1E–G). Of 14 randomly chosen up-regulated genes, 13 were also significantly increased in q-PCR analysis (Supplemental Fig. 1H), confirming that the expression of a subset of mRNAs in spermatocytes is increased in Piwil1−/− mice.
Figure 1.
piRNAs negatively regulate mRNAs in spermatocytes. (A) A schematic drawing of mouse spermatogenesis, with the expression patterns of PIWIL1, PIWIL2, prepachytene piRNAs, and pachytene piRNAs. The stages of developmental arrest in Piwil1−/− and Piwil2−/− mice are shown. (B,F,K) RNA-seq comparison of retrotransposon (B), mRNA (F), and spermatocyte lncRNA (K) expression between Piwil1+/− and Piwil1−/− late spermatocytes. Lines flanking the diagonal represent twofold differences. Significantly changed genes (P < 0.025, Student’s t-test) are indicated in red (n = 3). (RPKM) Reads per kilobase of transcript per million mapped reads. (C,D) Northern blot analysis of L1 full-length RNA (C) and Western blot analysis of L1 ORF1 (D) in 23-dpp testes of Piwil1+/− and Piwil1−/− mice. Two biological replicates from different mice were analyzed. (E) Immunohistochemical analysis of L1 ORF1 in sections of 28-dpp testes from Piwil1+/− and Piwil1−/− mice. Spermatocytes (Spc) and round spermatids (RS) are indicated. (G,L) Piwil1 and Mov10l1 mutations have similar impacts on mRNA (G) and spermatocyte lncRNA (L) expression. lncRNAs derived from the top 50 pachytene piRNA clusters are indicated in red. (H) RNA-seq analysis of retrotransposon expression in late spermatocytes from Mov10l1 CKO and control mice. See B for more information. (I) Approximately 25% of lncRNAs registered in Ensembl are preferentially expressed in the testis. Shown are the expression levels of 4401 Ensembl lncRNAs (rows) across tissues and cells (columns). (J) Expression levels of 1074 testis-enriched Ensembl lncRNAs during spermatogenesis.
To investigate whether mRNA up-regulation could also be observed in other piRNA pathway mutants, we analyzed the transcriptome of Mov10l1 CKO mice (Mov10l1flox/−, Stra8-Cre). MOV10L1 is an RNA helicase essential for piRNA biogenesis (Frost et al. 2010; Zheng et al. 2010), and piRNAs in late spermatocytes were almost entirely absent in Mov10l1 CKO mice (Supplemental Fig. 1I). These mice displayed a Piwil1−/−-like defect with spermatogenesis arrested after meiosis at the early round spermatid stage (Supplemental Fig. 1J,K; Deng and Lin 2002). A set of 81 genes was up-regulated (more than twofold) in both Mov10l1 CKO and Piwil1 KO late spermatocytes (Fig. 1G). No specific GO term in the biological process ontology and the cellular component ontology was enriched among this set, but in the molecular function ontology, “hydrolase activity, acting on glycosyl bond” was frequently observed (six of the 81 genes, P = 2.2 × 10−3). Several genes important for spermatogenesis (Man2a2, Morc1, Tex101, and Tdrd1) were among the set of 81 up-regulated genes. Additionally, several retrotransposons including RLTR4I_MM were up-regulated in Mov10l1 CKO mice (Fig. 1H).
Testis-enriched lncRNAs are negatively regulated by the piRNA pathway
Recent transcriptome analyses in mammals revealed that a large and diverse set of lncRNAs is specifically expressed in the testis (Cabili et al. 2011; Soumillon et al. 2013). We consistently found that a large number of lncRNAs (24.4%; 1074/4401) registered in Ensembl were most highly expressed in testes as compared to other tissues and cells (Fig. 1I; Flicek et al. 2014). Many of the testis-enriched Ensembl lncRNAs were initially expressed at 15−22 dpp, when the first wave of spermatogenesis reaches the pachytene spermatocyte to early round spermatid stages (Fig. 1J). Thus, their expression overlaps with that of PIWIL1 and pachytene piRNAs. To analyze the impact of the piRNA pathway on lncRNA expression, we first identified 6014 lncRNAs (RPKM > 0.5 either in Piwil1+/− or in Piwil−/−) from intergenic regions using our late spermatocyte RNA-seq data. Many of these lncRNAs are preferentially expressed in the testis, with expression first detected at 15–18 dpp (Supplemental Fig. 1L,M); this suggests that these lncRNAs are expressed from the late spermatocyte stage. However, very few (2.4%) were derived from pachytene piRNA clusters, suggesting that most of them are not piRNA precursors. Remarkably, 485 of the 6014 identified lncRNAs were significantly increased in Piwil1−/− mice (more than twofold increase, P < 0.025), while only 32 were significantly decreased (more than twofold decrease, P < 0.025) (Fig. 1K). In Mov10l1 CKO mice, 473 of 4872 lncRNAs (RPKM > 0.5 either in Mov10l1 CKO or in control) were significantly increased (more than twofold increase, P < 0.025), and many of the increased lncRNAs were also up-regulated in Piwil1−/− mice (Fig. 1L). In addition, 20.4% (1229/6014) and 21.5% (1051/4872) of lncRNAs were up-regulated by more than 1.5-fold in Piwil1 KO and Mov10l1 CKO mice, respectively (Fig. 2B). When comparing RNA levels between Piwil1 and Mov10l1 mutant mice, a lower correlation of fold-change in lncRNA levels (R = 0.57) was observed as compared with mRNA levels (R = 0.74). This is at least partly due to the relatively lower expression level of lncRNAs (Supplemental Fig. 1N), because the expression levels of lowly expressed RNAs tend to be variable. Additionally, many of the piRNA precursors are up-regulated in Mov10l1 CKO mice but only mildly affected in Piwil1 KO mice (see below) (Fig. 1L).
Figure 2.
The extent of mRNA and lncRNA up-regulation is strongly correlated to the number of piRNA matches. (A,B) piRNAs are preferentially mapped to the up-regulated mRNAs (A) and lncRNAs (B). The y-axis represents the numbers (RPM) of piRNAs mapped in the antisense orientation by allowing up to four mismatches including up to two indels. Protein-coding genes and lncRNAs on the x-axis are sorted based on the extent of changes in the RNA-seq analysis in Piwil1 KO. (RPM) Reads per million sequenced reads. (C,D) A box plot analyzing the number of piRNA matches in the antisense orientation. Protein-coding genes (C) and lncRNAs (D) are classified into five groups based on the extent of up-regulation in Piwil1−/− mice. When the RPM value was <1, the value was treated as 1. (E) MA plot analysis of mRNA (left) and lncRNA (right) expression in Piwil1 KO and Mov10l1 CKO. The top 200 mRNAs and lncRNAs generating many sense unique piRNAs are highlighted in red. The top 200 mRNAs and lncRNAs having many antisense piRNA hits (four mismatch condition) are highlighted in blue. (F) piRNAs are preferentially generated from the up-regulated mRNAs (top) and lncRNAs (bottom) in Mov10l1 CKO. Protein-coding genes and lncRNAs on the x-axis are sorted based on the extent of changes in the RNA-seq analysis in Mov10l1 CKO. (G) Box plots analyzing the number of unique sense piRNAs generated from mRNAs (top) and lncRNAs (bottom). mRNAs and lncRNAs are classified into five groups based on the extent of up-regulation in Mov10l1 CKO (left) and Piwil1 KO (right) mice. When the RPM value was <0.1, the value was treated as 0.1.
The regulation of mRNAs and lncRNAs by PIWIL1 is piRNA-dependent
To determine if PIWIL1 regulates the potential target RNAs in a piRNA-sequence-dependent manner, we mapped the published PIWIL1-associated piRNAs (Robine et al. 2009) against mRNAs and lncRNAs expressed in late spermatocytes, and we counted the number of piRNA reads that were matched to each mRNA and lncRNA in the antisense orientation (see Supplemental Results and Supplemental Fig. 2 for details on mapping and counting). Notably, piRNAs were preferentially matched to the up-regulated mRNAs and lncRNAs (Fig. 2A–D), suggesting that piRNAs guide PIWIL1 to target RNAs. In addition, the average number of piRNA matches to the moderately up-regulated (one- to twofold) transcripts was much higher than that to the down-regulated (more than onefold) transcripts. This result suggests that some moderately up-regulated RNAs are also direct targets of PIWIL1 and piRNAs. We attempted to pull down PIWIL1 target RNAs by immunoprecipitation using the anti-PIWIL1 antibody. However, the target RNAs were not enriched in the immunoprecipitated RNAs, possibly due to a transient association between PIWIL1 and the target RNAs that are degraded (Supplemental Results; Supplemental Fig. 3).
piRNA precursors were up-regulated in Mov10l1 CKO mice but only mildly affected or unaffected in Piwil1−/− mice (Fig. 1L). Consistent with this, there was a positive correlation between the extent of mRNA and lncRNA up-regulation in Mov10l1 CKO mice and the number of unique sense piRNAs (Fig. 2E–G). This correlation was modest or almost absent in Piwil1−/− mice (Fig. 2E,G). These results suggest that the expression of a subset of mRNAs and lncRNAs up-regulated in Mov10l1 CKO mice is regulated by the piRNA precursor processing machinery upstream of PIWIL1.
Pseudogene-derived piRNAs regulate their cognate functional gene
The most up-regulated gene in Piwil1−/− late spermatocyte RNA-seq was Stambp, which encodes a deubiquitinase required for neuronal cell survival (Ishii et al. 2001). PIWIL1-associated piRNAs were mapped to several regions within this mRNA in the antisense orientation (Fig. 3A). Specifically, most of these piRNAs were mapped to Stambp mRNA with mismatches but were perfectly and uniquely matched to its processed pseudogene (Stambp-ps1) in a piRNA cluster. This suggests that piRNAs from Stambp-ps1 regulate Stambp mRNA. To explore this hypothesis, we generated two mouse strains from ES lines with gene-trap insertions just upstream of Stambp-ps1 in the piRNA cluster (Fig. 3B). In spermatocytes from mice homozygous for the insertion (Stambp-ps1Gt1/Gt1 and Stambp-ps1Gt2/Gt2), both putative piRNA precursor transcripts and piRNAs from the Stambp-ps1 region were dramatically decreased (Fig. 3B–D). As predicted, Stambp mRNA was significantly increased in late spermatocytes from Stambp-ps1Gt1/Gt1 and Stambp-ps1Gt2/Gt2 mice, but Stambp pre-mRNA was not changed (Fig. 3E). Although we did not find any phenotype in Stambp-ps1Gt1/Gt1 mice (Supplemental Fig. 4A–D), a marked increase in Stambp protein was observed in Stambp-ps1Gt1/Gt1 late spermatocytes and round spermatids (Fig. 3F,G). Together, these results indicate that piRNAs from the Stambp pseudogene regulate the Stambp gene at the post-transcriptional level. Inspired by this example, we aligned all mRNAs to piRNA clusters expressed in spermatocytes and identified six other pseudogenes that potentially targeted their functional cognate mRNAs. Of the six cognate mRNAs, three were significantly up-regulated in both Piwil1−/− and Mov10l1 CKO spermatocytes (Supplemental Fig. 4E).
Figure 3.
The Stambp pseudogene regulates its cognate mRNA via the piRNA pathway. (A) Distribution of PIWIL1-associated piRNAs matched to Stambp mRNA in the antisense orientation. CDS is represented by the hatched region in mRNA. (B) Structure of the Stambp-ps1 piRNA cluster. The Stambp pseudogene (Stambp-ps1) is indicated in green. The insertion sites of the two gene trap lines (Gt1 and Gt2) are indicated by arrows and brown bars. piRNAs generated from this locus and Chr 17 piRNA cluster (control) in Stambp-ps1+/Gt1 and Stambp-psGt1/Gt1 mice are shown. Watson (Crick) strand piRNAs are shown in red (blue). (C) Northern blotting analysis of a piRNA derived from Stambp-ps1. The location of the probe used is shown in B. (D) q-PCR analysis of the putative piRNA precursor in Stambp-ps1Gt1/Gt1 (left) and Stambp-ps1Gt2/Gt2 (right) late spermatocytes. The locations of the primers used are shown in B. Error bars represent the SD (n = 4). (*) P < 0.01 (Student’s t-test). (E) q-PCR analysis of Stambp mRNA and pre-mRNA in Stambp-ps1Gt1/Gt1, Stambp-ps1Gt2/Gt2, Piwil1−/−, and Mov10l1 CKO late spermatocytes. Error bars represent the SE (n = 3 for Mov10l1 CKO and n = 4 for others). (*) P < 0.01 (Student’s t-test). (F) Immunofluorescence analysis of Stambp protein. Round spermatids (RS) are located at the center of seminiferous tubules and indicated. (G) Western blot analysis of Stambp protein. Late spermatocytes and round spermatids were FACS-isolated from 26-dpp mouse testes.
Transcripts containing retrotransposon sequences are enriched in up-regulated RNAs
To examine the piRNA target sequences within the regulated RNAs, we identified piRNA-complementary and, therefore, potential target sequences within mRNAs. Almost all of the piRNA-complementary sequences were located within the 3′ UTR rather than 5′ UTR and coding DNA sequence (CDS) of mRNAs (Fig. 4A). The piRNA-complementary sequences were much more densely observed in the 3′ UTR of the up-regulated mRNAs than the down-regulated mRNAs, consistent with the results of preferential mapping of piRNAs to the up-regulated RNAs (Fig. 2). Many (68.5%) of the piRNA-complementary sequences within the up-regulated mRNAs corresponded to retrotransposon sequences (Fig. 4B), of which 83.2% corresponded to SINE, nonautonomous retrotransposons with a length of 150–300 nt (Goodier and Kazazian 2008). When repeat sequence-derived piRNAs and nonrepeat sequence-derived piRNAs were separately mapped to mRNAs, strong preferential mapping to the up-regulated mRNAs was observed only for the repeat sequence-derived piRNAs, and these were mostly retrotransposon-derived (Fig. 4C). A similar result was obtained for lncRNAs (Fig. 4D). These results raise the possibility that retrotransposon sequences in mRNAs and lncRNAs are targeted by PIWIL1 and piRNAs.
Figure 4.
Retrotransposon sequences are enriched in the up-regulated mRNAs and lncRNAs. (A) Most piRNA-complementary sequences were observed within the 3′ UTR. The average number (RPM) of antisense piRNA matches at each position was calculated using a moving window of 100 bases (step size = 100 bases). (TSS) Transcription start site. (B) Many of the piRNA-complementary sequences in mRNAs were SINE sequences. Each position in piRNA-complementary sequences within the up-regulated mRNAs was annotated. (C,D) The number of matches to repeat-derived piRNAs was correlated with the extent of up-regulation. Box plots analyzing the number of matches to nonrepeat-derived piRNAs (left) and repeat-derived piRNAs (right) are shown. When the RPM value was <0.1, the value was treated as 0.1. (E,F) The up-regulated mRNAs (E) and lncRNAs (F) frequently contain retrotransposon sequences. The number of genes and lncRNAs in each class is shown above the bar on the left graph. P-values between the <1 and >2 groups were calculated by the R function prop.test. (G,H) High density of retrotransposon sequences in the 3′ UTR of the up-regulated mRNAs (G) and across the entire region of the up-regulated lncRNAs (H). Regions examined are divided into 0.1-kb bins (window size = 100 bases, step size = 100 bases).
To evaluate the importance of retrotransposon sequences in piRNA-mediated mRNA and lncRNA regulation, we classified mRNAs and lncRNAs based on the extent of up-regulation in the Piwil1−/− mice. Remarkably, the frequencies of mRNAs and lncRNAs containing retrotransposon sequences were significantly higher among the up-regulated RNAs than the down-regulated RNAs for all retrotransposon classes analyzed (Fig. 4E,F). For example, 46.4% of the up-regulated mRNAs, but only 12.9% of the down-regulated mRNAs, contained SINE sequences. In addition, the density of retrotransposon sequences was considerably higher in the 3′ UTR of the up-regulated mRNAs (Fig. 4G). For lncRNAs, a higher density was observed across the entire region of the up-regulated lncRNAs (Fig. 4H). This correlation implies that many of the retrotransposon sequences in the up-regulated mRNAs and lncRNAs are targeted by PIWIL1 for degradation.
PIWIL1 degrades target mRNAs and lncRNAs in a slicer-dependent fashion
Among the 13 known piRNA pathway genes (Pillai and Chuma 2012), Tdrd1, Piwil2, and Piwil1 mRNAs contain SINE sequences potentially targeted by piRNAs (Fig. 5A; Supplemental Fig. 5A). Moreover, these mRNAs were up-regulated in Piwil1−/− or Mov10l1 CKO mice (Supplemental Fig. 5B). In the 3′ UTR of Tdrd1 mRNA, two SINE sequences (PB1D10 and ID2) were found (Fig. 5A). Interestingly, we observed a sharp increase in RNA-seq reads at one site in the PB1D10 sequence of Tdrd1 mRNA (Fig. 5B, arrow). The reason for this sharp increase can be inferred from our RNA-seq method: We prepared strand-specific RNA-seq libraries by fragmenting RNAs and ligating the 5′ ends of the resulting RNA fragments to an RNA linker. The library was then sequenced from the linker. Therefore, if there were many RNA ends at a particular site generated by RNase cleavage in vivo in addition to in vitro fragmentation, more reads would be produced from the ends at that site. Because the sharp increase in RNA-seq reads at the PB1D10 sequence was observed only in Piwil1+/− but not Piwil1−/− mice (Fig. 5B), it likely resulted from the PIWIL1-mediated cleavage. In support of this, PIWIL1-associated piRNAs were mapped predominantly to one region encompassing the site of sharp increase in the antisense orientation. Furthermore, the site of sharp increase occurred between the 10th and 11th positions of the mapped piRNAs (Fig. 5A,C), which exactly corresponded to the cleavage site of PIWIL1 and these piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007; Reuter et al. 2011). Additionally, the 5′ end of one of the piRNAs generated from Tdrd1 mRNA exactly corresponds to the site of sharp increase (Fig. 5B,C), showing characteristics of secondary piRNAs, which are generated from 5′ ends of RNAs cleaved by Piwi-piRNAs (Juliano et al. 2011; Ishizu et al. 2012; Pillai and Chuma 2012). This further supports the PIWIL1-mediated cleavage at the site of sharp increase. In addition, expression of Tdrd1 and other target RNAs were increased in late spermatocytes from Piwil1 slicer mutant (Piwil1−/ADH) mice (Fig. 5D; Supplemental Results; Reuter et al. 2011), indicating that slicer activity contributes to the target RNA degradation.
Figure 5.

PIWIL1 slicer activity contributes to the degradation of target RNAs. (A) PIWIL1-associated piRNAs are predominantly mapped to PB1D10 SINE sequences (gray box) in the 3′ UTR of Tdrd1 mRNA. (B) The sharp increase in RNA-seq reads at the PB1D10 region. Shown are RNA-seq reads and PIWIL1-associated piRNAs derived from this region (below). (C) A magnified view of the site of the sharp increase. The location is indicated by the blue line in A. (D) q-PCR analysis of several target mRNAs (top) and lncRNAs (bottom) in Piwil1 KO (blue) and in Piwil1 slicer mutant (red) mice. The locations of PCR amplicons are shown in Figures 3A (Stambp), 5A (Tdrd1), and 7A (Prelid1). Error bars represent the SE (n = 3).
PIWIL1 promotes the decay of a reporter mRNA containing a B1 or B2 SINE sequence at its 3′ UTR
To further demonstrate that retrotransposon sequences in a target mRNA indeed mediate the degradation of the target mRNA by the PIWIL1-piRNA pathway, we generated mice with a single-copy transgene that expresses mCherry mRNA with a retrotransposon sequence in its 3′ UTR (Fig. 6A; Supplemental Fig. 7). The retrotransposon sequence in mCherry mRNA was flanked by LoxP sites, and therefore can be removed in the presence of Cre protein. We chose B1 and B2 SINE retrotransposons because they are frequently found in the up-regulated mRNAs and lncRNAs in Piwil1−/− mice (Fig. 4E,F; Supplemental Fig. 7). Upon introduction of Stra8-Cre, B1 and B2 sequences in the transgenes were deleted in late spermatocytes (Fig. 6B,C), and mCherry fluorescence signal in late spermatocytes increased by 2.2-fold (mCherry-B1flox/0 vs. mCherry-B1flox/0, Stra8-CreTg/0) and 3.2-fold (mCherry-B2flox/0 vs. mCherry-B2flox/0, Stra8-CreTg/0) (Fig. 6D,E, left). A similar level of increase (2.8-fold in mCherry-Bflox/0 and 2.7-fold in mCherry-B1flox/0) was also observed at the mRNA level; however, no change was observed at the pre-mRNA level (Fig. 6E, center and right). These results indicate that B1 and B2 sequences confer instability to mature mRNAs in late spermatocytes.
Figure 6.
B1 and B2 SINE sequences confer instability to mRNAs. (A) A schematic representation of a transgene construct. The PEST motif was fused to the C terminus of the mCherry protein, which results in a shorter half-life of the protein. (B) FACS analysis of testicular cells from 28-dpp mice using Hoechst 33342. (C) Deletion of B1 (top) and B2 (bottom) sequences from transgenes in late spermatocytes from Stra8-Cre mice. A set of PCR primers spanning LoxP sites was used for PCR. (D) The increase of mCherry fluorescence upon deletion of retrotransposon sequences. (E) mCherry fluorescence (left), mCherry mRNA (center), and mCherry pre-mRNA (right) levels in the absence (blue) or the presence (red) of Stra8-Cre allele in late spermatocytes are shown. RNA levels were measured by q-PCR. Error bars represent the SE (n = 3). (*) P < 0.05 (Student’s t-test). (F,G) PIWIL1 targets B1 and B2 sequences for degradation. mCherry fluorescence (left) and mCherry mRNA (right) levels in Piwil1+/− (blue) and Piwil1−/− (red) late spermatocytes are shown. Error bars represent the SE (n = 3). (*) P < 0.05 (Student’s t-test).
To examine if the observed instability caused by B1 and B2 sequences is PIWIL1-dependent, mCherry-B1flox/0 and mCherry-B2flox/0 mice were crossed with Piwil1 mutant mice. Elevated levels of mCherry mRNA were observed in Piwil1−/− late spermatocytes as compared with the Piwil1+/− control in both mCherry-B1flox/0 and mCherry-B2flox/0 mice (1.8-fold for mCherry-B1flox/0 and 3.4-fold for mCherry-B2flox/0) (Fig. 6F, right), suggesting that PIWIL1 is involved in the degradation of B1- and B2-containing mCherry mRNAs. The larger increase observed for B2-containing mCherry mRNA as compared to B1-containing mCherry mRNA in Piwil1−/− mice is consistent with the preferential enrichment of the B2 sequence in highly up-regulated RNAs in the RNA-seq analysis (Figs. 4E,F, 6F). When B1 and B2 sequences were removed (mCherry-B1Δlox/0 and mCherry-B2Δlox/0), no increase was observed in Piwil1−/− mice compared with Piwil1+/− mice (Fig. 6G). These observations indicate that PIWIL1 targets B1 and B2 sequences in the mCherry mRNAs for degradation.
PIWIL1 promotes the decay of an endogenous mRNA containing conserved retrotransposon sequence in its 3′ UTR
To directly validate that retrotransposon sequences regulate the stability of endogenous mRNAs via a PIWIL1-piRNA-mediated mechanism, we deleted such retrotransposon sequences in the 3′ UTR of an endogenous mRNA, Prelid1 mRNA, and examined its expression in wild-type versus Piwil1 mutant backgrounds. The two most frequently observed retrotransposon sequences in pachytene piRNAs were MIR and LINE2 (L2) repeats (Supplemental Fig. 8A), inactive retrotransposons that were propagated before the mammalian radiation and widely exist in mammalian genomes (Silva et al. 2003). Their sequences are often found in mRNAs, as 6.3% and 2.5% of mouse RefSeq mRNAs contain MIR and L2a sequences, respectively (Faulkner et al. 2009). Prelid1 mRNA contains both MIR and L2a sequences in the 3′ UTR, and PIWIL1-associated piRNAs were mapped predominantly to the MIR and L2a sequences in the antisense orientation (Fig. 7A). Up-regulation of this mRNA in Piwil1−/− mice as compared to Piwil1+/− mice (approximately fourfold) was observed only at the mRNA level, but not at the pre-mRNA level (Supplemental Fig. 8B), indicating that PIWIL1 degrades this mRNA.
Figure 7.
The mammalian conserved retrotransposon sequence in Prelid1 mRNA regulates its stability in late spermatocytes. (A) The distribution of PIWIL1-associated piRNAs mapped to Prelid1 mRNA in the antisense orientation. PIWIL1-associated piRNAs are mapped to the MIR and L2 retrotransposon sequences (gray boxes) in the antisense orientation. (B) 3′RACE analysis of Prelid1 mRNA in Prelid1pA/pA late spermatocytes. The arrows indicate 3′RACE PCR products amplified from the 3′ ends of wild-type and truncated Prelid1 mRNAs. The insertion site of the polyadenylation signal sequence and the location of the primer used are shown in A. (C) q-PCR analysis of Prelid1 mRNA and pre-mRNA in Prelid1pA/pA late spermatocytes (left) and livers (right). Error bars represent the SE (n = 3). (*) P < 0.05 (Student’s t-test). The locations of the primers used for the analyses of the mRNA are shown in A. (D) Published microarray data (1431420_a_at probe) are shown (Fallahi et al. 2010). (E) Clustering analysis of the relative expression levels of 12,077 Ensembl protein-coding genes among early spermatocytes (Espc), late spermatocytes (Lspc), and round spermatids (RS) is shown. Expression levels were determined by RNA-seq in individual cell types. (F) The read number (RPM) of PIWIL1-associated piRNAs complementary to mRNA(s) of each gene is shown. (G) Retrotransposons are less frequently found in round spermatid (RS) mRNAs than early spermatocyte (Espc) mRNAs. The number of genes in each class is shown above the bar on the left graph. P-values between the <1 and >2 groups were calculated using the R function prop.test. (H) RS mRNAs have a short 3′ UTR. Box plots analyzing the length of 5′ UTR, CDS, and 3′ UTR of all Ensembl mRNAs, Espc mRNAs, and RS mRNAs are shown. (I) PIWIL1 and piRNAs derived from pseudogenes and retrotransposons in piRNA clusters degrade spermatocyte-expressed mRNAs and lncRNAs at the mature RNA level via piRNA-complementary sequences in the RNAs. The PIWIL1 and piRNA-induced degradation is mostly dependent on the slicer activity of PIWIL1.
To generate mice expressing Prelid1 mRNA without the retrotransposon sequences, we inserted a synthetic polyadenylation signal sequence just upstream of the two retrotransposon sequences in the 3′ UTR using the CRISPR-Cas9 system, because the polyadenylation signal sequence of this mRNA is in the L2a retrotransposon sequence, which is located immediately downstream from the MIR retrotransposon sequence (Fig. 7A). Mice homozygous for the mutant allele (Prelid1pA/pA) generated truncated Prelid1 mRNA lacking the retrotransposon sequences (Fig. 7B). Although we did not find any phenotype in Prelid1pA/pA mice, Prelid1 mRNA, but not its pre-mRNA, was increased approximately sixfold in late spermatocytes from Prelid1pA/pA mice as compared to wild-type (Prelid1+/+) control mice (Fig. 7C). Such an increase was not observed in the liver, where PIWIL1 is not expressed (Fig. 7C). Furthermore, in contrast to the full-length Prelid1 mRNA, the truncated Prelid1 mRNA was not increased in Piwil1−/− late spermatocytes (Supplemental Fig. 8C), indicating that PIWIL1 targets the retrotransposon sequences in the 3′ UTR of Prelid1 mRNA.
During spermatogenesis, Prelid1 mRNA is expressed at high levels until the early-pachytene stage, and it is down-regulated after the expression of pachytene piRNAs at the mid-pachytene stage, reaching a very low level at the round spermatid stage (Fig. 7D). Although the expression of many of the mRNAs that are down-regulated from the early spermatocyte stage to the late spermatocyte stage were not affected in Piwil1−/− late spermatocytes (data not shown), a subset of these mRNAs may be cleared or repressed by pachytene piRNAs in late spermatocytes and round spermatids, as observed for Prelid1 mRNA (Fig. 7D). This low level of Prelid1 expression in round spermatids, together with strong derepression of Stambp expression in Piwil1−/− round spermatids (Fig. 3F), suggests that genes that function in round spermatids are not repressed by the piRNA pathway. Consistent with this idea, PIWIL1-associated piRNAs were less preferentially matched to mRNAs expressed in round spermatids (Rspd mRNAs) compared with mRNAs expressed in early spermatocytes (Espc mRNAs) (Fig. 7E,F). Furthermore, retrotransposon sequences were found less often in Rspd mRNAs than in Espc mRNAs (Fig. 7G). This low frequency is likely due to the shorter 3′ UTRs of Rspd mRNAs, which have a median 3′ UTR length of 328 nucleotides (nt) compared to 729 nt and 973 nt for all mRNAs and Espc mRNAs, respectively (Fig. 7H). Therefore, it is tempting to speculate that round spermatid genes might have evolved to possess a short 3′ UTR in order to escape retrotransposon insertions that result in deleterious effects on these important reproductive genes.
Discussion
We have identified a global RNA regulatory mechanism mediated by retrotransposons and pseudogenes that regulate mRNAs and lncRNAs through our systematic search for the potential target RNAs of pachytene piRNAs. Pachytene piRNAs are abundantly expressed in mouse late spermatocytes and round spermatids, and their deficiency leads to spermatogenic arrest at the early round spermatid stage. Therefore, they have been proposed to play an important role during spermatogenesis; however, the mechanisms and targets of their regulation were largely unknown. The present study reveals that pachytene piRNAs and PIWIL1 negatively regulate protein-coding mRNAs and lncRNAs as well as L1 retrotransposons in late spermatocytes, with piRNAs acting as guide molecules to identify target RNAs via sequence complementarity. The degradation of target RNAs by PIWIL1 and piRNAs is mostly dependent on the slicer activity of PIWIL1. Furthermore, our results reveal two mechanisms for the piRNA guidance (Fig. 7I). First, piRNAs generated from pseudogene sequences in piRNA clusters target mRNAs transcribed from their functional cognate genes. Second, piRNAs from retrotransposon sequences target RNAs that contain retrotransposon sequences.
Our results reveal that PIWIL1 and piRNAs have a large impact on the expression of lncRNAs. Many of them are probably regulated via retrotransposon sequences. Given that 66% of mouse lncRNAs contain transposable elements (Kelley and Rinn 2012), the large effect on the lncRNA transcriptome is not surprising. In late spermatocytes and round spermatids, standard histones are replaced with several histone variants associated with an open chromatin structure, and a genome-wide low level of H3K9me2 and high level of H3K4me2 are observed (Kimmins and Sassone-Corsi 2005; Tachibana et al. 2007; Soumillon et al. 2013). The open chromatin structure presumably facilitates the replacement of the histones with transition proteins (Doenecke et al. 1997). Although we cannot rule out the possibility that widespread transcription of lncRNAs in late spermatocytes and early round spermatids have some functional purposes such as producing a diverse and large set of pachytene piRNAs (Ha et al. 2014), most of these intergenic RNAs could be a “by-product” of extensive chromatin remodeling occurring in these cells (Soumillon et al. 2013). Nonfunctional RNAs generated from intergenic regions often contain retrotransposon sequences, since ∼40% of the mouse genome is derived from retrotransposon sequences. Some of them might have harmful effects on spermatogenesis. The piRNA pathway would be ideal for regulating such lncRNAs, as it mainly targets retrotransposon sequences. Because the expression of PIWIL1 and pachytene piRNAs occurs at the same time as the chromatin remodeling and the resultant pervasive transcription, it is possible that piRNAs are abundantly expressed in late spermatocytes and round spermatids in part to degrade “leaked” intergenic transcripts that may cause harmful effects.
Our results reveal that piRNAs derived from inactive as well as active retrotransposons regulate mRNAs, including Tdrd1 and Prelid1 mRNA. piRNAs are generated from two biogenesis pathways: the primary and secondary pathways (Kim et al. 2009; Juliano et al. 2011; Ishizu et al. 2012; Pillai and Chuma 2012). The secondary pathway specifically amplifies piRNAs derived from active retrotransposons. Given that the amplification depends on high similarity among active retrotransposon copies, inactive retrotransposon-derived piRNAs are unlikely to be efficiently amplified by the secondary pathway. Mouse pachytene piRNAs are known to be generated mostly through the primary pathway (Reuter et al. 2011; Beyret et al. 2012; Pillai and Chuma 2012). Similarly, it has just been reported that adult testicular piRNAs in the common marmoset are generated mainly via the primary pathway, and both young and old retrotransposon sequences equally contribute to the production of these piRNAs (Hirano et al. 2014). In addition, this study also identified pseudogene-derived piRNAs that potentially regulate parental mRNAs. Thus, generation of piRNAs from inactive retrotransposons and pseudogenes via the primary pathway in adult testes seems to be a conserved phenomenon in mammals. Although it is unclear whether this is due to the predominant presence of cellular mechanisms activating the primary pathway and/or the involvement of only a small subset of pachytene piRNAs in silencing active retrotransposons, it is possible that pachytene piRNA generation highly depends on the primary pathway in order to generate sufficient unamplifiable piRNAs, such as inactive retrotransposon-derived piRNAs, to regulate mRNAs and lncRNAs.
In summary, our genome-wide analysis of pachytene piRNA targets in late spermatocytes reveals a regulatory network in which the stability of many lncRNAs and mRNAs are regulated by retrotransposons and pseudogenes via their derivative piRNAs. In addition, retrotransposons auto-regulate the turnover of their own RNAs also via the piRNA pathway. Interestingly, the expression of an unusually large number of lncRNAs as well as pachytene-like piRNAs in the testis is a well-conserved phenomenon among mammals and birds (Li et al. 2013; Soumillon et al. 2013; Hirano et al. 2014). Therefore the genome-wide interregulatory mechanisms reported here for piRNAs, lncRNAs, mRNAs, pseudogene RNAs, and transposon RNAs may be well conserved in the testes of vertebrates.
Methods
RNA-seq library construction and sequencing
Total RNAs were isolated from FACS-isolated cells or testes using TRIzol (Invitrogen). RNA-seq data from several tissues and cell types that were sequenced in Bing Ren’s laboratory were obtained from the Gene Expression Omnibus (GEO) database under accession number GSE36026. See Supplemental Methods for details on RNA-seq library preparation, mapping, calculation of RPKM values, and lncRNA identification.
Flow cytometry and sorting of spermatogenic cells
To prepare single-cell suspensions, decapsulated testes were treated with a digestion solution (0.5 mg/mL collagenase type IV, 0.05% trypsin-EDTA, and 25 μg/mL DNaseI in DMEM) at 35°C for 10 min. See Supplemental Methods for details.
Generation of knock-in, gene-trap, and transgenic mice
An oligo DNA for homologous recombination (50-nt homologous sequence−polyadenylation signal sequence−50-nt homologous sequence), Cas9 mRNA, and chimeric guide RNA were mixed and then injected into the cytoplasm of one-cell embryos of B6;SJL F2 mice. See Supplemental Methods for details about CRISPR-mediated knock-in mouse, gene-trap mouse, and transgenic mouse generation.
Mapping of PIWIL1-associated piRNAs to mRNAs and lncRNAs
PIWIL1-associated small RNA reads (Sequence Read Archive [SRA] accession number SRR033657) were annotated as previously described (Watanabe et al. 2011), and miRNAs and degradation products of abundant noncoding RNAs (rRNA, tRNA, snRNA, snoRNA, scRNA, srpRNA, and RNA) were removed. The remaining small RNAs were mapped to the masked mRNAs and lncRNAs using SeqMap software (Jiang and Wong 2008; http://www-personal.umich.edu/∼jianghui/seqmap/) allowing up to four mismatches, including two indels. See Supplemental Methods for details about piRNA mapping analyses.
Small RNA analysis in Stambp pseudogene mutant mice
Small RNA libraries were constructed using total RNAs isolated from 23-dpp Stambp-ps1+/Gt1 and Stambp-ps1Gt1/Gt1 mouse testes. See Supplemental Methods for details.
Analyses of retrotransposon sequences in mRNAs and lncRNAs
The mRNA (lncRNA) sequences with 5′ and 3′ flanking intergenic sequences were obtained from the UCSC Table Browser (http://genome.ucsc.edu/cgi-bin/hgTables). Retrotransposon sequences in the downloaded sequences were identified using RepeatMasker (http://www.repeatmasker.org). See Supplemental Methods for details about retrotransposon analyses.
Identification of pseudogenes in piRNA clusters
Pseudogenes in piRNA clusters were identified by aligning mRNA sequences to piRNA cluster sequences. See Supplemental Methods for details.
Data access
The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE42004.
Supplementary Material
Acknowledgments
We thank Timothy Nottoli and Carole Pease for generating transgenic and CRISPR-Cas9-mediated KO mice, Michael Reuter and Ramesh Pillai for Piwil1 slicer mutant mice, Robert Frost and Eric Olson for Mov10l1 mutant mice, Sandra Martin for anti-L1 ORF1, Celina Juliano, Robert Ross, Molly Weiner, and Anastasios Vourekas for comments on the manuscript, Yoshiaki Tanaka and Mengjie Chen for discussion about bioinformatics analysis, and Chia-ling Hsieh for help with sample preparation. This work was supported by NIH grant R37HD42012 and a Mathers Foundation award to H.L. T.W. was a fellow of the Japan Society for the Promotion of Science (JSPS) and the Uehara Memorial Foundation during part of this work. E.C. was supported by the Connecticut Stem Cell Research Grants-in-Aid Program (grant 10SCA13). T.W. performed the experiments and conducted bioinformatics analysis. E.C. was involved in FACS sorting. M.Z. performed the sequencing. T.W. and H.L. designed the study and wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.180802.114.
References
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H. 2012. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res 22: 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. [DOI] [PubMed] [Google Scholar]
- Doenecke D, Drabent B, Bode C, Bramlage B, Franke K, Gavenis K, Kosciessa U, Witt O. 1997. Histone gene expression and chromatin structure during spermatogenesis. Adv Exp Med Biol 424: 37–48. [DOI] [PubMed] [Google Scholar]
- Fallahi M, Getun IV, Wu ZK, Bois PRJ. 2010. A global expression switch marks pachytene initiation during mouse male meiosis. Genes 1: 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. 2010. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci 107: 11847–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35. [DOI] [PubMed] [Google Scholar]
- Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y, Kang JY, Wang X, Li H, Hua MM, et al. 2014. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24: 680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Ha H, Song J, Wang S, Kapusta A, Feschotte C, Chen KC, Xing J. 2014. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 15: 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Iwasaki YW, Lin ZY, Imamura M, Seki NM, Sasaki E, Saito K, Okano H, Siomi MC, Siomi H. 2014. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 20: 1223–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Owada Y, Yamada M, Miura S, Murata K, Asao H, Kondo H, Sugamura K. 2001. Loss of neurons in the hippocampus and cerebral cortex of AMSH-deficient mice. Mol Cell Biol 21: 8626–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC. 2012. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 26: 2361–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH. 2008. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24: 2395–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. 2011. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 45: 447–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, Asangani IA, Kothari V, Prensner JR, Lonigro RJ, et al. 2012. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149: 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. 2007. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317: 248–251. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1159. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Chuma S. 2012. piRNAs and their involvement in male germline development in mice. Dev Growth Differ 54: 78–92. [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Current Biol 19: 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RJ, Weiner MM, Lin H. 2014. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. 2010. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Shabalina SA, Harris DG, Spouge JL, Kondrashovi AS. 2003. Conserved fragments of transposable elements in intergenic regions: evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet Res 82: 1–18. [DOI] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3: 2179–2190. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N, Shinkai Y. 2007. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J 26: 3346–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 29: 288–299. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB. 2008. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod 79: 51–57. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Watanabe T, Hoki Y, Shirane K, Li Y, Ichiiyanagi K, Kuramochi-Miyagawa S, Toyoda A, Fujiyama A, Oginuma M, et al. 2013. Targeted gene silencing in mouse germ cells by insertion of a homologous DNA into a piRNA generating locus. Genome Res 23: 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci 107: 11841–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.