Abstract
Nucleosomes in active chromatin are dynamic, but whether they have distinct structural conformations is unknown. To identify nucleosomes with alternative structures genome-wide, we used H4S47C-anchored cleavage mapping, which revealed that 5% of budding yeast (Saccharomyces cerevisiae) nucleosome positions have asymmetric histone-DNA interactions. These asymmetric interactions are enriched at nucleosome positions that flank promoters. Micrococcal nuclease (MNase) sequence-based profiles of asymmetric nucleosome positions revealed a corresponding asymmetry in MNase protection near the dyad axis, suggesting that the loss of DNA contacts around H4S47 is accompanied by protection of the DNA from MNase. Chromatin immunoprecipitation mapping of selected nucleosome remodelers indicated that asymmetric nucleosomes are bound by the RSC chromatin remodeling complex, which is required for maintaining nucleosomes at asymmetric positions. These results imply that the asymmetric nucleosome-RSC complex is a metastable intermediate representing partial unwrapping and protection of nucleosomal DNA on one side of the dyad axis during chromatin remodeling.
Nucleosomes, the fundamental units of chromatin, are dynamic structures characterized by spontaneous conformational fluctuations that lead to reversible loss of histone-DNA and histone-histone contacts. Every nucleosome in the genome has to disassemble at least once during the cell cycle to allow for passage of the DNA replication machinery (Annunziato 2005), and nucleosomes at active regions might turn over several times during each cell cycle (Dion et al. 2007; Deal et al. 2010). Intrinsic to nucleosome dynamics is the formation of nucleosomal intermediates with alternative structures. However, the nature of such intermediate nucleosome structures formed in vivo is not known.
Intermediate nucleosome structures can potentially be identified in vivo using base-pair resolution methods that interrogate histone-DNA contacts genome-wide. The traditional method for high-resolution mapping of nucleosomes is to use micrococcal nuclease (MNase) digestion, which digests away linker regions between nucleosomes (Reeves and Jones 1976). Subjecting MNase-digested DNA fragments to paired-end sequencing (MNase-seq) results in a high-resolution map of nucleosome positions (Hughes and Rando 2014). An alternative method for mapping nucleosomes is H4S47C-anchored cleavage mapping (Brogaard et al. 2012b), which has been used to determine the precise position of nucleosomes in yeast genomes (Brogaard et al. 2012a; Moyle-Heyrman et al. 2013). In this method, histone H4 mutant S47C is derivatized ex vivo with a phenanthroline ligand, converting H4 into a site-specific DNA cleavage agent. Using a modified library preparation and a structural model for H4S47C-anchored cleavage, we extended this method to determine the precise position and orientation of half-nucleosomes (hemisomes) at centromeres (Henikoff et al. 2014), thus showing that alternative nucleosome structures can be probed using this method. In this study, we ask whether alternative nucleosome structures can be found in regions of the yeast genome other than at the centromeres.
Results
H4S47C-anchored cleavage mapping identifies asymmetric nucleosome positions
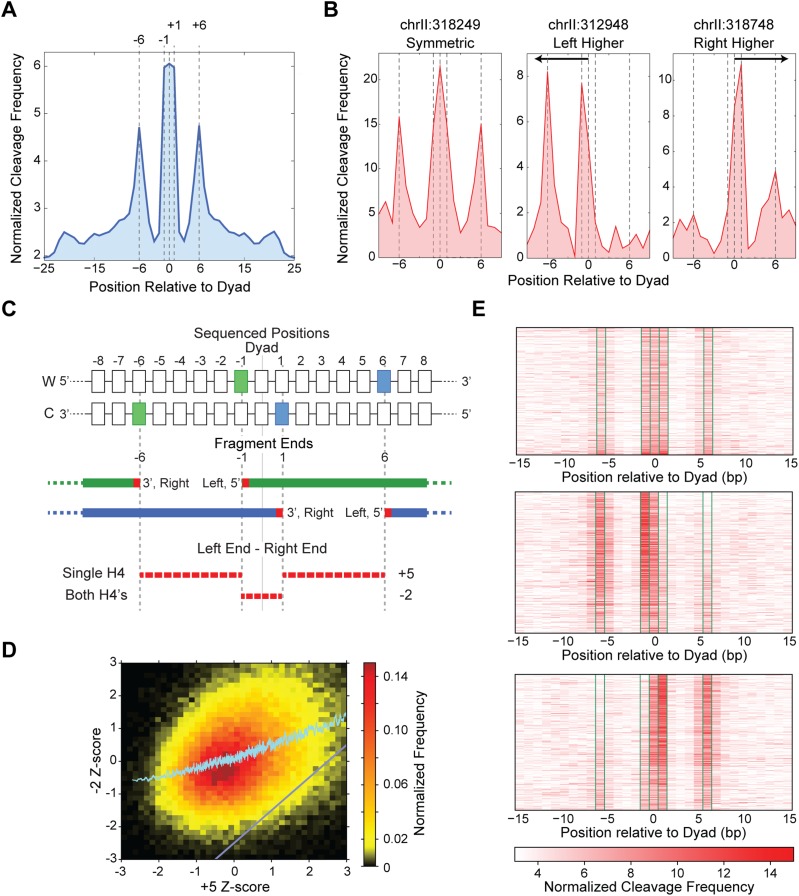
In H4S47C-anchored cleavage mapping, the ends of sequenced fragments reflect cleavage of nucleotide positions that are in close proximity to residue S47C of histone H4. In the map from our recent H4S47C-anchored cleavage data (Henikoff et al. 2014), we observed spikes in cleavage frequency at individual nucleosome positions, as exemplified in a representative 10-kb region (Supplemental Fig. S1A). The genome-wide average of H4S47C-anchored cleavage data around the nucleosome dyad axis reveals specific positions that have high cleavage frequency (−6, −1, 0, +1, and +6 base pairs [bp] relative to the dyad axis; dyad positions obtained from Brogaard et al. 2012a) (Fig. 1A). Figure 1A shows complete concordance between our map of H4S47C-anchored cleavages and the published nucleosome positions (Brogaard et al. 2012a). We used a structural model for H4S47C-anchored cleavage to enable us to distinguish a single H4-contacting DNA from two H4s contacting DNA at a given nucleosome position (Henikoff et al. 2014). We determined that the −6 and −1 cleavages are caused by the H4 upstream of the dyad axis and the +1 and +6 cleavages are caused by the H4 downstream from the dyad axis, whereas the 0 cleavages are caused by either H4. Most positions have two equivalent cleavages, one upstream and one downstream from the dyad axis (Fig. 1B, left), as expected for octameric nucleosomes. However, we also observed nucleosome positions that feature highly asymmetric cleavages, with cleavage frequencies on one side of the dyad axis diminished compared to the other side (Fig. 1B, center and right). The asymmetries represent an excess of nucleosomes at these positions that have lost contact with DNA on one side of the dyad axis.
Figure 1.
H4S47C-anchored cleavage mapping identifies asymmetric nucleosomes. (A) The cleavage data, when averaged over all nucleosomes genome-wide, reveals specific peaks at −6, −1, 0, +1, and +6 bp positions relative to the dyad axis. Dyad axis positions were obtained from Brogaard et al. (2012a). (B) Cleavage signal over single nucleosome dyad axis positions showing a symmetric nucleosome (left), asymmetric nucleosomes with higher signal upstream of the dyad (center) and downstream from the dyad (right). (C) Schematic showing the left and right ends of sequenced fragments that give rise to the specific peaks around the dyad. Cleavages by a single H4 are separated by 5 bp, while cleavages by both the H4s are separated by −2 bp. (D) The −2-ntd Z-score increases monotonically with increasing +5-ntd Z-score. The region below the gray diagonal represents nucleosomes that have a much lower −2-ntd Z-score compared to the +5-ntd Z-score. (E) Heatmap of cleavage mapping data of 1500 symmetric nucleosomes (top), 1654 asymmetric nucleosomes with more cleavages on the left (middle), and 1585 asymmetric nucleosomes with more cleavages on the right side of the dyad (bottom). Significantly more cleavages at −1 and −6 positions compared to +1 and +6 positions in the middle panel and vice versa in the bottom illustrates asymmetry.
We wanted to determine the prevalence of these asymmetric nucleosome positions genome-wide using a robust and unbiased metric that is independent of the position of the dyad axis. For this metric, we exploited the directionality of cleavages around the dyad axis: The −6 and +1 peaks correspond to right ends of sequenced fragments, whereas the −1 and +6 peaks correspond to left ends (Supplemental Fig. S1B). This arrangement of fragment ends results in the right end of one fragment and the left end of a neighboring fragment separated by distances of +5 and −2 nucleotides (nt) (Fig. 1C). The +5 nucleotide distance (ntd) reflects cleavages by a single H4, and the −2 ntd reflects cleavages by both H4s at the nucleosome position (Fig. 1C). Thus, in asymmetric nucleosome positions, the prevalence of the −2 ntd would be diminished compared to the prevalence of the +5 ntd. As we only need to calculate the distance between fragment ends, we do not explicitly use the position of the dyad axis. We ranked the probability of observing the +5 and the −2 ntd at each nucleosome position using independent Z-scores and plotted the −2-ntd Z-score of each nucleosome position against its +5-ntd Z-score (Fig. 1D). A monotonic increase in the −2-ntd Z-score with respect to the +5-ntd Z-score indicated that for most nucleosome positions, cleavages by both H4s correlated well with cleavages by just one H4, reflecting symmetric nucleosome positions. The nucleosome positions in the lower right quadrant of the plot of −2-ntd vs +5-ntd Z-scores (below the gray line) have +5-ntd Z-scores disproportionately higher than corresponding −2-ntd Z-scores, indicating that cleavages by H4s on one side of the nucleosome position are much higher than the H4s on the other side. We identified 3239 such nucleosome positions (∼5% of the genome-wide total), applying stringent criteria both to pick high asymmetry in probabilities and to filter out nucleosome positions with low read coverage. Cleavages by two H4s also result in fragment ends separated by 12 nt. When we plot the distribution of the difference between the +5-ntd Z-score and the +12-ntd Z-score of each nucleosome position, we observe the distribution for asymmetric nucleosome positions to be right-shifted relative to all nucleosome positions (Supplemental Fig. S2). Thus, in the identified asymmetric nucleosomes, the +5-ntd Z-score is disproportionately higher than the +12-ntd Z-score, validating that in asymmetric nucleosome positions, cleavages by H4s on one side of the nucleosome position are much higher than cleavages by H4s on the other side.
With the 3239 nucleosome positions identified based on disproportionate probabilities of observing the +5 ntd compared to the −2 ntd, we plotted H4S47C cleavage frequencies ±15 bp around the nucleosome dyad axis. As a control, we first displayed the cleavage frequencies for 1500 symmetric nucleosome positions as a heatmap (Fig. 1E, top). As expected from our structural model, we observed peaks at −6, −1, 0, +1, and +6 bp, where 0 is the position of the dyad axis. The peak at −6 bp is comparable to the peak at +6 bp, and the peak at −1 bp is comparable to the peak at +1 bp. We then separated the asymmetric nucleosome positions into two groups based on the direction of asymmetry. For the first group, the peaks at −6 and −1 bp are much higher than the peaks at +1 and +6 bp; that is, the H4S47C cleavages are skewed toward upstream of the dyad axis (Fig. 1E, middle). For the second group, the peaks at +1 and +6 are much higher than the peaks at −1 and −6; that is, the H4S47C cleavages are skewed toward downstream from the dyad axis (Fig. 1E, bottom). Thus, the heatmaps of H4S47C cleavage frequencies imply loss of cleavages on one side of the dyad axis for asymmetric nucleosome positions. The asymmetry in H4S47C cleavage frequencies at the 3239 identified positions is significantly higher than at nucleosome positions genome-wide, although much less than at the 16 centromeric nucleosome positions (Supplemental Fig. S3), all of which are occupied by hemisomes (Henikoff et al. 2014). Since we use a modified Illumina paired-end DNA sequence library preparation that does not include a size-selection step (Henikoff et al. 2011), observation of asymmetric nucleosomes cannot be due to size-related biases. Asymmetric cleavages are highly reproducible, with three independent experiments showing nearly identical profiles (Supplemental Fig. S4), and a highly significant fraction of ±1 asymmetric nucleosome positions could be identified even when data from each individual experiment were analyzed independently (Data set 1: 56%, P-value = 2 × 10−152; Data set 2: 48%, P-value = 2 × 10−197; Data set 3: 48%, P-value = 4.3 × 10−151; P-values calculated using the hypergeometric test). Similar trends were also observed using H4S47C cleavage data from Brogaard et al. (2012a) (Supplemental Fig. S5). Asymmetric cleavages strictly depend on the H4S47C residue, as no asymmetric cleavages were observed using a wild-type H4 strain (Supplemental Fig. S6), which excludes the possibility that they result from phenanthroline-derivatized cysteines on other chromosomal proteins.
Asymmetric nucleosomes flank promoters
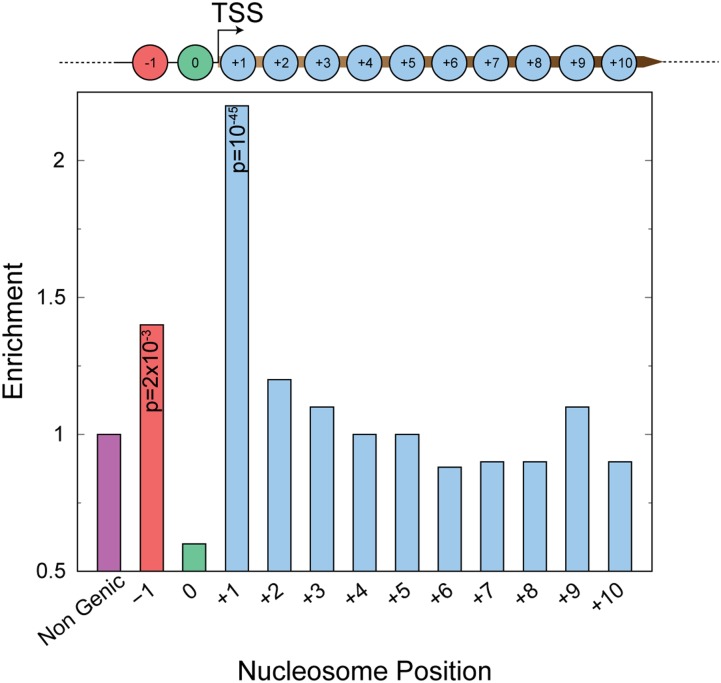
To understand why these nucleosomes are asymmetric, we asked if they are enriched at specific genomic loci. We hypothesized that the asymmetry would be a consequence of nucleosome dynamics, because asymmetric cleavages represent loss of H4-DNA contacts on one side of the dyad axis. Hence, we compared enrichment of asymmetric nucleosome positions at promoters, genic positions, and positions outside of genes and promoters (Fig. 2). Genic nucleosome positions are numbered +1, +2, and so on, based on their position relative to the transcription start site (TSS). If a nucleosome position is found at a promoter, it is numbered 0 and the position upstream of the promoter is numbered −1. We found highly significant enrichment at +1 and significant enrichment at −1 positions compared to other genic and nongenic nucleosome positions. This enrichment of ±1 asymmetric nucleosome positions was observed for asymmetric nucleosomes identified in each of the individual replicates (Supplemental Fig. S7), indicating a reproducible phenomenon. We observed asymmetry on both sides of the dyad axis relative to the TSS (Supplemental Fig. S8), with no evidence for coordination between the +1 and −1 nucleosome position, as only 13 genes had both asymmetric +1 and asymmetric −1 nucleosomes.
Figure 2.
Positional enrichment of asymmetric nucleosomes. Enrichment of asymmetric nucleosomes at genic nucleosome positions and nongenic positions. P-values calculated using a hypergeometric test are shown for significantly enriched positions (+1 and −1).
The −1 and +1 nucleosome positions that flank the promoter represent sites of high nucleosome turnover (Dion et al. 2007). The rate of turnover at asymmetric ±1 positions is as high as all ±1 nucleosome positions (Supplemental Fig. S9), which suggests that asymmetric nucleosomes are formed during a dynamic process.
Asymmetric nucleosomes are characterized by half-nucleosome footprints
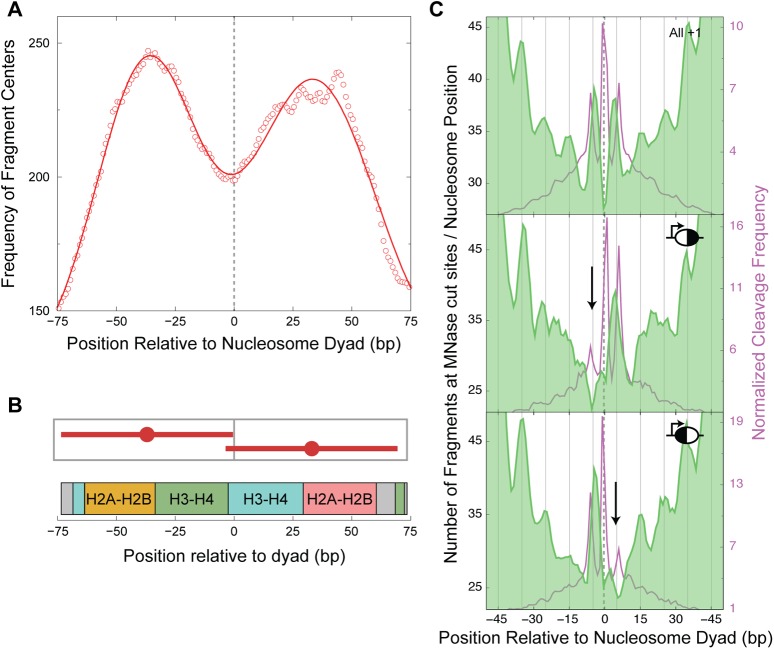
A possible explanation for the lack of H4S47C cleavages on one side of the dyad axis would be the absence of histones on that side, in other words, occupancy of these nucleosome positions by hemisomes. MNase-seq maps chromatin footprints and has been used to uncover subnucleosomal particles (Henikoff et al. 2011). Hence, we used our published paired-end MNase-seq data (Henikoff et al. 2011) to ask if particles smaller than conventional octameric nucleosomes map to the asymmetric positions, which might indicate the presence of nucleosomal intermediates. The MNase-seq data were generated using a modified Illumina paired-end DNA sequence library preparation that does not include a size-selection step, enabling recovery of all fragments between 25 and 500 bp in length (Henikoff et al. 2011). As paired-end data provide both the positions of MNase cleavages and the lengths of the protected fragments, we were able to select shorter fragments (73 ± 20 bp) to map putative nucleosomal intermediates, plotting their distribution around the nucleosomal dyad axis. We oriented nucleosome positions so that the side with the higher H4S47C cleavage frequency would be downstream from the dyad axis. In this orientation, if the lower H4S47C cleavage frequency upstream of the dyad axis is due to the absence of H4 on that side, we would observe decreased MNase-seq protection on the upstream side. However, we observed equivalent protection on both sides of the dyad axis, with a prominent dip in MNase protection centered over the dyad axis, indicating increased MNase accessibility at the dyad axis (Fig. 3A). Furthermore, MNase protection fits a bimodal Gaussian distribution, with mean fragment centers at −37 and +33 bp relative to the dyad axis and a mean fragment length of 73 bp, indicating the presence of half-nucleosomes on both sides of the dyad axis (Fig. 3B). Footprinting of particles that closely correspond to two halves of the nucleosome suggests splitting of the nucleosome at the H3-H3′ interface and excludes the possibility that asymmetric H4S47C cleavages are caused by hemisomes.
Figure 3.
Asymmetric nucleosomes are asymmetrically accessible to MNase. (A) Distribution of the centers of ∼73-bp fragments plotted relative to the +1 dyad axis. A 20-bp running average shows a bimodal distribution with peaks at −37 and +33 bp relative to the dyad, overlaid with its Gaussian fit (solid line). (B) Graphic depicting the fragments that constitute the mean positions obtained from the Gaussian fit, shown in the context of histone-DNA contacts along the path of DNA around the octamer. (C) Distribution of MNase cuts, plotted relative to the nucleosome dyad for all +1 nucleosomes (top, n = 4116) reveals specific cuts at 5 bp on either side of the dyad. A similar distribution of MNase cuts is plotted for asymmetric nucleosomes that have higher cleavages on the downstream side of the dyad with respect to the TSS (middle, n = 171) and for asymmetric nucleosomes that have higher cleavages on the upstream side of the dyad with respect to the TSS (bottom, n = 236). The H4S47C cleavage frequency of each group of nucleosome positions is plotted in the background of their corresponding MNase plots. All nucleosomes are oriented toward the direction of transcription, and the dyad axis position is marked with a dashed line. The downward arrows locate the missing peaks in the asymmetric nucleosomes. The 10-bp periodicity in the MNase peaks can be tracked with the vertical lines that are spaced 10 bp apart.
Asymmetric nucleosomes are asymmetrically accessible to MNase
To further characterize asymmetric nucleosomes, we pooled several yeast MNase-seq data sets, for a total of ∼500 million paired-end fragments of lengths between 25 bp and 500 bp, to probe the relative MNase accessibility of nucleosomal DNA. By plotting the MNase cut density relative to the +1 dyad axis, we determined that the overall frequency of MNase cut sites gradually increased going from the dyad axis toward the entry/exit positions of the nucleosome (−73 bp and +73 bp relative to the dyad axis) (Supplemental Fig. S10). We also observed that peaks in the MNase cutting frequency showed a striking 10-bp periodicity (Supplemental Fig. S10), which reflects exposure of DNA to MNase at the surface of the nucleosome every 10 bp. We also observed that preferred MNase cut sites internal to the nucleosome map at ±5 bp relative to the dyad axis (Fig. 3C, top). Thus, at high sequencing depth, MNase cut site frequencies map the relative accessibility of nucleosomal DNA in vivo. This method of probing structure from outside the nucleosome is independent of H4S47C-anchored cleavage mapping, which probes structure from inside the nucleosome.
We next asked if accessibility to MNase digestion is altered at asymmetric +1 nucleosomes. We oriented asymmetric +1 nucleosome positions in the direction of transcription. Asymmetric nucleosome positions were split into two groups: those with a lower frequency of H4S47C cleavages upstream of the dyad axis (Fig. 3C, middle) and vice versa (Fig. 3C, bottom). For positions with fewer H4S47C cleavages upstream of the dyad axis, MNase cut sites upstream of the dyad axis (at dyad axis − 5 bp) disappeared (Fig. 3C, middle). For positions with fewer H4S47C cleavages downstream from the dyad axis, MNase cut sites downstream from the dyad axis (at dyad axis + 5 bp) disappeared (Fig. 3C, bottom). In other words, asymmetric nucleosome positions become less MNase accessible on the side of the dyad axis that has fewer H4S47C cleavages. Thus, the process that causes loss of H4-DNA contacts on one side of the dyad axis protects the DNA on that side from MNase cutting.
RSC is enriched over asymmetric nucleosome positions
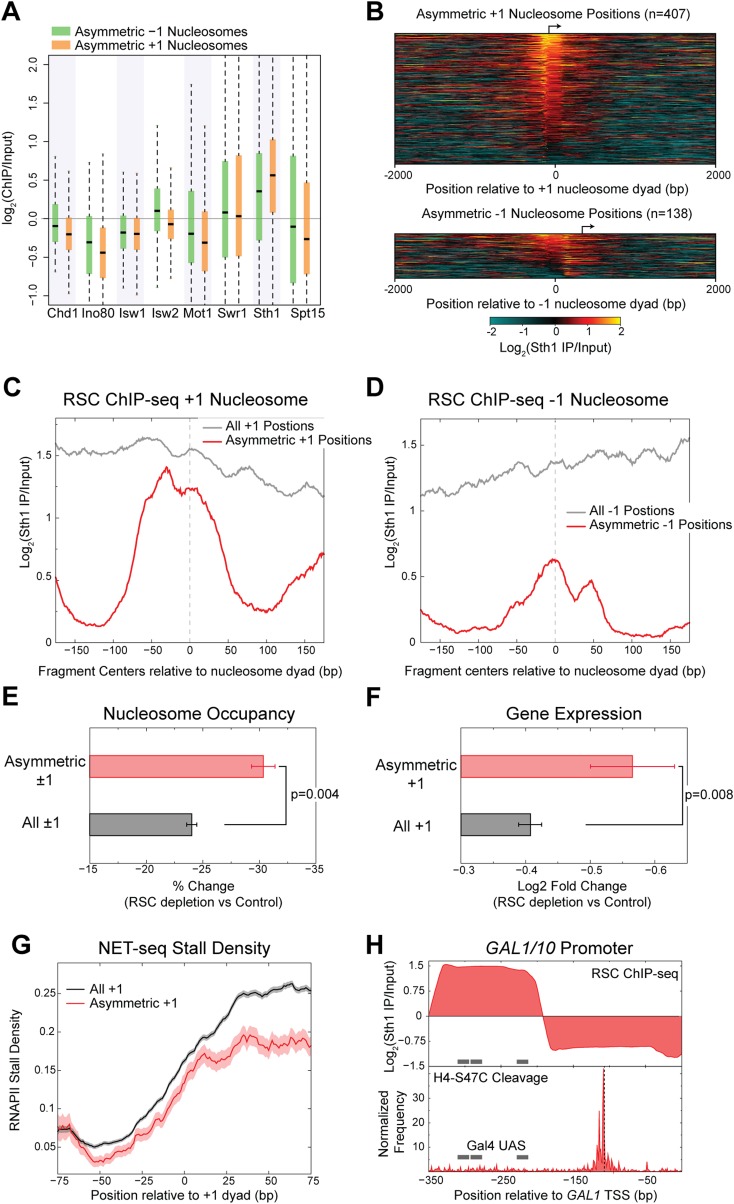
Partial protection of nucleosomal DNA from MNase digestion suggests that a protein or complex is bound to the outside of the nucleosome. To also reduce H4S47C cleavage on the MNase-protected side of the dyad axis, the bound protein or complex must distort the nucleosome structure so that some histone-DNA contacts are lost while others are maintained. Likely candidates for causing both loss of H4-DNA contacts and protection of DNA at asymmetric nucleosomes are SWI/SNF family chromatin remodelers, which bind nucleosomes and alter histone-DNA contacts (Narlikar et al. 2013). To identify remodelers that might be enriched over +1 and −1 nucleosome positions, we performed native ChIP-seq of Ino80, Swr1, and Sth1. We also examined our published ChIP-seq data for Chd1, Isw1, and Isw2 (Zentner et al. 2013), using TATA-binding protein (Spt15) and the SWI/SNF family Spt15 regulator Mot1 (Zentner and Henikoff 2013) as negative controls. We observed that only RSC was enriched over asymmetric ±1 nucleosome positions relative to the input, whereas other SWI/SNF family proteins and Spt15 were depleted over these nucleosome positions (Fig. 4A; Supplemental Table S4).
Figure 4.
Asymmetric nucleosomes are stabilized by RSC. (A) ChIP enrichment of chromatin remodelers and Spt15 over the dyad axis ±70 bp of asymmetric +1 and −1 nucleosome positions is shown as box-plots. (B) Heatmap of the log2 enrichment of the RSC catalytic subunit, Sth1 over input, plotted relative to the dyad axis of asymmetric +1 nucleosome positions (top) and asymmetric −1 nucleosome positions (bottom). (C) ChIP-seq of the RSC catalytic subunit, Sth1, shows enrichment of RSC specifically over the nucleosome at asymmetric +1 positions compared to enrichment at the upstream NDR for all +1 positions. Log enrichment of the normalized density of fragment centers is plotted. The fragment centers were obtained from paired-end sequencing data and correspond to fragments of length 200 ± 20 bp. Data are averaged over a 20-bp moving window. (D) Same as C for asymmetric −1 nucleosome positions. (E) Depletion of the RSC component Sth1 results in a significantly greater decrease in nucleosome occupancy at asymmetric ±1 positions compared to all ±1 positions. P = 0.004 (Kolmogorov-Smirnov test). (F) Depletion of the RSC component Sth1 results in a significantly greater decrease in the expression of well-expressed genes with asymmetric +1 nucleosome position compared to all well-expressed genes with a +1 nucleosome position. P = 0.008 (Kolmogorov-Smirnov test). For E and F, mean and SEM are plotted. (G) Well-expressed genes with asymmetric +1 nucleosome positions feature decreased stalling by RNA polymerase II at the +1 nucleosome compared to well-expressed genes with a +1 nucleosome position. (H) (Top) Plot showing the log2 enrichment of Sth1 ChIP-seq at the GAL1/10 promoter, for fragment lengths between 120 and 160 bp, with the Gal4 binding sites indicated with gray bars. (Bottom) H4-S47C cleavage frequency is plotted for the GAL1/10 promoter, showing a highly asymmetric nucleosome position downstream from the Gal4 binding sites. The dyad position of the asymmetric nucleosome is indicated with a dashed black line, showing that the frequency of cleavages is higher upstream of the dyad.
RSC is an essential remodeling complex (Cairns et al. 1996) that maintains nucleosome depletion at promoters and positioning of nucleosomes genome-wide (Hartley and Madhani 2009). To determine if RSC acts at asymmetric nucleosome positions, we analyzed the high-resolution enrichment of the Sth1 catalytic subunit of RSC relative to the nucleosome dyad axis over ±1 nucleosome positions. We found that RSC is enriched over most of asymmetric ±1 nucleosome positions (Fig. 4B), regardless of the direction of asymmetry relative to the TSS (Supplemental Fig. S11). At +1 nucleosome positions, we observed enrichment of RSC both over the upstream nucleosome-depleted region (NDR) and over the +1 nucleosome position (Fig. 4C). This enrichment agrees with the known function of RSC in evicting nucleosomes over NDRs and positioning the +1 nucleosome (Hartley and Madhani 2009). To determine if RSC is bound to asymmetric nucleosomes, we compared the ChIP enrichment of RSC at asymmetric nucleosome positions to all +1 nucleosome positions. In stark contrast to the average for all +1 nucleosomes, asymmetric +1 nucleosomes featured RSC enrichment directly over the nucleosome position and not over the upstream NDR. In other words, RSC is excluded from the linkers belonging to asymmetric +1 nucleosome positions, whereas enrichment directly over the asymmetric +1 nucleosome positions is similar to all +1 nucleosome positions. A similar trend was observed for asymmetric −1 nucleosome positions (Fig. 4D). The binding of RSC directly over asymmetric nucleosome positions and not upstream indicates that a RSC-nucleosome complex occupies asymmetric nucleosome positions.
Asymmetric nucleosomes are sites of RSC action
Our mapping of RSC directly over asymmetric nucleosome positions argues for direct action of RSC on the nucleosome but does not rule out the alternative possibility that RSC and the nucleosome independently occupy the same position in different cells in the population. To test for an interaction between RSC and asymmetric nucleosomes, we analyzed the genome-wide nucleosome landscape upon partial RSC depletion (Van de Vosse et al. 2013). We observed that twofold RSC depletion resulted in an overall reduction in nucleosome occupancy at asymmetric positions (Fig. 4E; Supplemental Fig. S12), which strongly argues that direct action of RSC on the nucleosome causes it to be asymmetric.
Remodeler action at the +1 nucleosome position is associated with RNA polymerase II (RNAPII) transit across nucleosomal DNA, both in facilitating RNAPII elongation (Soutourina et al. 2006; Schwabish and Struhl 2007) and in repositioning nucleosomes in the wake of RNAPII transit (Smolle et al. 2012). If RSC is acting at the asymmetric +1 position to facilitate transcription, its depletion should result in expression changes at genes with asymmetric +1 nucleosome positions. To test for expression changes of genes with asymmetric +1 nucleosomes, we analyzed published expression data after complete RSC depletion using a degron strain (Hartley and Madhani 2009). Indeed, we observed that for well-expressed genes, the presence of an asymmetric nucleosome position correlates with a reduction in expression upon depletion of RSC (Fig. 4F). Thus, RSC is required for the robust expression of genes with asymmetric +1 positions. To determine if RSC facilitates RNAPII transit, we calculated the extent of RNAPII stalling at +1 nucleosome positions using published NET-seq data (Churchman and Weissman 2011). We observed decreased stalling of RNAPII across the +1 nucleosome of well-expressed genes with an asymmetric +1 nucleosome position compared to all well-expressed genes (Fig. 4E).
A previous study had reported a partially unwrapped nucleosome bound by RSC at the Gal4 binding sites of the GAL1/10 promoter (Floer et al. 2010). RSC action at this site is required for rapid induction of GAL1/10 upon shift from glucose to galactose as the carbon source (Floer et al. 2010). Our RSC ChIP-seq shows high enrichment of RSC at the Gal4 binding sites (Fig. 4F, top), reproducing the published RSC profile (Floer et al. 2010). We also observed an absence of H4-S47C cleavage at the Gal4 binding sites (Fig. 4F, bottom), indicating that if histones were present at this site, H4 was not contacting DNA. However, adjacent to the Gal4 sites and closer to the GAL1 TSS, we observed a highly asymmetric nucleosome position. Thus, at the GAL1/10 promoter, which is a well-characterized site of RSC action, an asymmetric nucleosome position is found downstream from the RSC and Gal4 binding site.
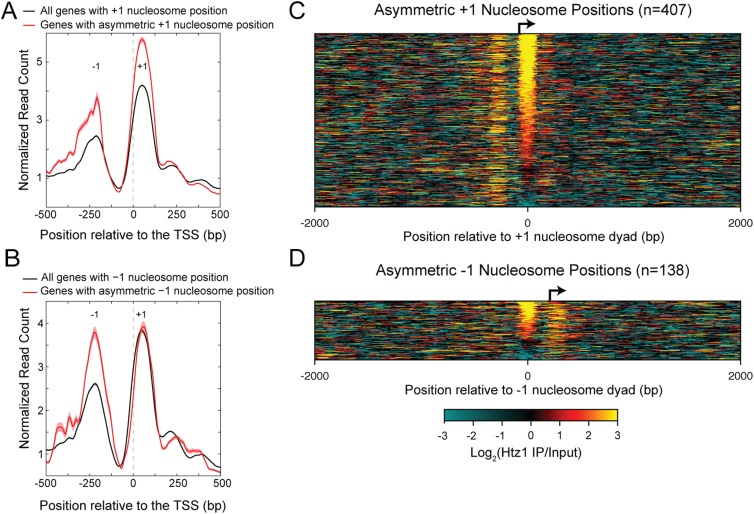
Previous studies have indicated that RSC interacts with H2A.Z-enriched nucleosomes (Hartley and Madhani 2009; Floer et al. 2010). To determine if asymmetric nucleosome positions are enriched for H2A.Z (encoded by HTZ1), we performed H2A.Z ChIP-seq and confirmed previous observations of H2A.Z enrichment at ±1 positions (Fig. 5A,B; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005). Most of the asymmetric ±1 nucleosome positions are enriched for H2A.Z containing nucleosomes (Fig. 5C,D). Furthermore, enrichment of H2A.Z at asymmetric ±1 nucleosome positions is even higher than at all ±1 positions, as expected for sites of RSC action (Hartley and Madhani 2009; Floer et al. 2010).
Figure 5.
Genes with asymmetric nucleosome positions are enriched for nucleosomes containing H2A.Z. (A) Genes with asymmetric +1 nucleosome positions are enriched for H2A.Z at the +1 position to a greater extent on average than are all genes. (B) Same as A for genes with asymmetric −1 nucleosome positions. Data are averaged over a 20-bp sliding window. (C) Heatmap of the log2 enrichment of the H2A.Z IP over input, plotted relative to the dyad axis of asymmetric +1 nucleosome positions. (D) Same as C for asymmetric −1 nucleosome positions.
Discussion
Using H4S47C-anchored cleavage mapping, we have identified a subset of nucleosome positions flanking promoters that are characterized by asymmetric H4-DNA contacts. These asymmetric nucleosomes are inaccessible to MNase on the side of the dyad axis that is cleaved less frequently by H4S47C. The loss of H4-DNA contacts with a gain of MNase protection suggests the binding of a protein that is capable of distorting the nucleosome structure at a specific location, while at the same time restricting access of MNase to nucleosomal DNA (Fig. 6). In support of this model, we found that asymmetric positions flanking promoters are bound by the RSC chromatin remodeler. High H2A.Z levels further support RSC enrichment at asymmetric positions, as RSC is known to bind nucleosomes that contain H2A.Z (Hartley and Madhani 2009; Floer et al. 2010). The functional significance of the RSC-asymmetric nucleosome complex is demonstrated by the requirement of RSC for the robust expression of genes containing an asymmetric +1 nucleosome position. This requirement of RSC reflects a role in both relieving the nucleosomal barrier to RNAPII and in the maintenance of nucleosome depleted regions in the promoters of the genes containing an asymmetric +1 nucleosome position.
Figure 6.
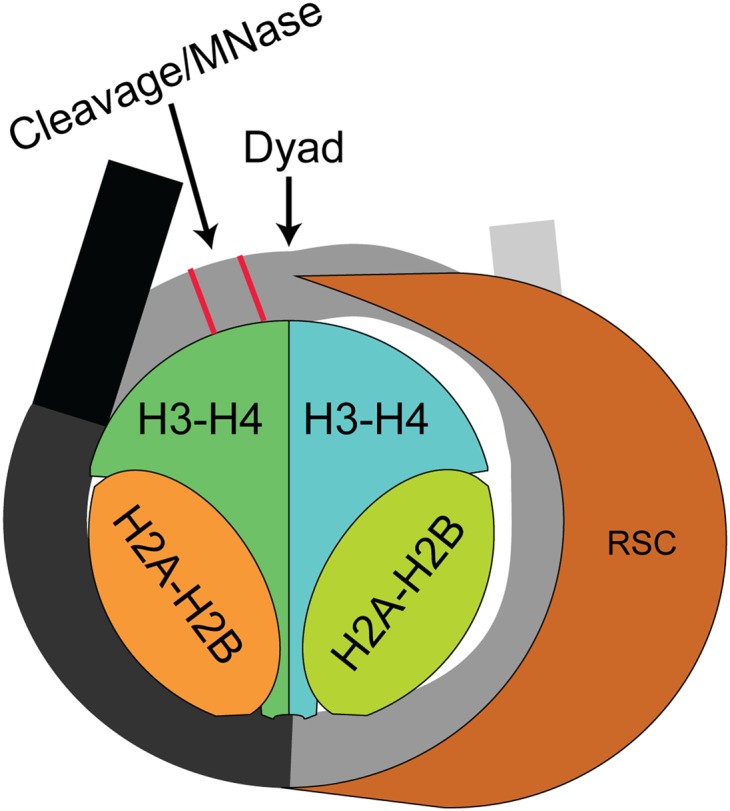
Model for interaction of RSC with nucleosomes to generate asymmetric histone-DNA interactions. RSC binds to the nucleosome and distorts histone-DNA contacts on one side.
Cryo-electron microscopic structures of RSC bound to a nucleosome have shown that RSC can envelop the whole nucleosome (Leschziner et al. 2007; Chaban et al. 2008), suggesting that RSC translocates nucleosomal DNA while remaining bound to the histones (Lorch et al. 2011). Based on the RSC-nucleosome interactions, the RSC-asymmetric nucleosome complex we have identified could reflect either of two different processes. The first is a stalled byproduct in the course of DNA translocation, similar to a backtracked RNA polymerase. In this scenario, the asymmetric nucleosome positions could reflect a barrier to RSC translocation. The second possibility is that the RSC-asymmetric nucleosome complex represents an intermediate step in the RSC remodeling pathway. In vitro studies have shown that in the absence of ATP, RSC can unwrap nucleosomal DNA right up to the dyad while remaining bound to the histones and also to the nucleosomal DNA, protecting the DNA from nuclease attack (Lorch et al. 2010). Upon addition of ATP, the nucleosomal DNA is highly susceptible to nuclease attack, indicating that ATP-dependent translocation of the RSC-histone complex exposes DNA that was originally bound to the histones (Lorch et al. 2010). Asymmetric nucleosome positions observed in vivo closely resemble the RSC-nucleosome complex in the absence of ATP, because at asymmetric nucleosome positions, histone-DNA interactions on one side of the dyad are lost but the nucleosomal DNA is still protected from nuclease attack. Thus, based on in vitro results, we can infer that the RSC-bound asymmetric nucleosome observed in vivo is a metastable intermediate in the remodeling process formed prior to ATP binding and DNA translocation. However, it remains unknown how the transition from the ATP-free form to the ATP bound form is regulated in vivo.
Our observation of half-nucleosome footprints at asymmetric +1 positions is similar to an earlier observation by Lee and Garrard at a transcriptionally active locus, which they attributed to nucleosome splitting (Lee and Garrard 1991). Tracking “old” and “new” histones had also demonstrated splitting of nucleosomes in yeast (Katan-Khaykovich and Struhl 2011) and in human cells (Huang et al. 2013). Our results suggest that the half-nucleosome footprints at active loci in yeast are due to RSC action at these nucleosome positions, suggesting a possible mechanism for nucleosome splitting.
The core ATPase subunits of SWI/SNF family remodelers are conserved between yeast and metazoans. However, the addition of a large number of metazoan-specific subunits results in the formation of several distinct remodeling complexes, complicating the study of chromatin remodeling in metazoans. In contrast, the substrate of the remodelers, the nucleosome, is universal and highly conserved. Thus, identifying alternative nucleosome structural intermediates provides a feasible way to understand chromatin remodeling in all eukaryotes.
Methods
ChIP
All strains used in this study are listed in Supplemental Table S5. The Sth1-3FLAG strain (GZY9) and the Swr1-3FLAG strain (GZY33) were generated using p3FLAG-KanMX as described (Gelbart et al. 2001). The Ino80-3FLAG strain (YTT1728) and the Htz1-3FLAG strain (YTT3249) were a gift from Toshi Tsukiyama. Yeast were grown in YPD to an OD600 of 0.6–0.8. Nuclear isolation, MNase digestion, and chromatin preparation were performed as described (Zentner et al. 2013). Native ChIP was performed using FLAG M2 magnetic beads (Sigma) in all cases except for H2A.Z, for which anti-FLAG magnetic beads (Lake Pharma) were used. Native ChIP-seq was performed as described (Zentner et al. 2013), and sequencing libraries were constructed as described (Henikoff et al. 2011). Libraries were sequenced for 25 cycles in paired-end mode on the Illumina HiSeq platform at the FHCRC Genomics Shared Resource.
Analysis of H4S47C-anchored cleavage mapping data
H4S47C cleavage data obtained from three independent experiments (GEO accession numbers GSM1255647, GSM1255648, GSM1255649) (Henikoff et al. 2014) were combined for the analyses, except for Supplemental Figure S3, where each data set was analyzed separately. To minimize size-related biases, we used a modified Illumina paired-end DNA sequence library preparation that does not include a size-selection step (Henikoff et al. 2011). The fraction of ends of fragments mapped at each genomic position was multiplied by the total number of nucleotides mapped genome-wide to obtain the normalized cleavage frequency. Nucleosome dyad axis positions were obtained from the published “unique map” that consists of 67,543 nucleosome positions and not the 351,264 overlapping positions (Brogaard et al. 2012a). There is an inevitable background of non-H4S47C-dependent cleavages that we previously showed are enriched in linker regions (Henikoff et al. 2014). Therefore, we did not use the 351,264 overlapping positions (Brogaard et al. 2012a) because these would be more enriched for background cleavages relative to the 67,543 nonoverlapping positions that we used.
Left-right distance probabilities were calculated for fragments mapping within each dyad axis position ±30 bp (Henikoff et al. 2014). The left-right probability distribution features two peaks: at −2 and +5. Based on the left-right probability distribution plot, we calculated the adjusted probabilities at −2 and +5 by averaging over adjacent positions with high probability and subtracting out the probabilities of background positions. After calculating the adjusted probabilities at −2 and +5, we ordered the −2 and +5 probabilities independently using Z-scores. We identified asymmetric nucleosomes as those that had a +5 Z-score greater than −1 (to pick positions with a good signal to noise ratio) and where the +5 Z-score was greater than the −2 Z-score by at least 2.5 (to pick positions with asymmetry). The nucleosome positions in Figure 1E (middle and bottom) were ordered based on their degree of asymmetry, which was calculated as:
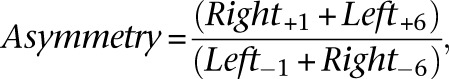 |
where “Left” and “Right” each refer to the normalized frequency of left and right fragment ends at a given position (using the + strand as reference), and the subscript refers to the position relative to the nucleosome dyad axis. The specific positions were chosen based on the directional nature of H4S47C cleavages (Supplemental Fig. S1B). This degree of asymmetry is plotted for different groups of nucleosome positions in Supplemental Figure S3.
Determination of positional enrichment
Each nucleosome position we considered was assigned a genic position (−1, 0, +1, +2, and so on) based on published positioning nomenclature for budding yeast (Jiang and Pugh 2009). Those nucleosomes not bearing a genic position were assigned as intergenic. We calculated the fraction of nucleosome positions in each of the genic positions and in the intergenic region that were asymmetric and normalized the fractions so that the intergenic region had an enrichment value of one.
MNase-seq analysis
To map subnucleosomal footprints, we used our published MNase-seq data (GEO accession number GSM754390). We computationally selected fragments of length 73 ± 20 and calculated the average distribution of fragment centers around the nucleosome dyad axis of asymmetric nucleosomes.
For plotting the frequencies of MNase cuts, we combined published and new MNase-seq data sets, which are detailed in Supplemental Table S6.
Remodeler ChIP analysis
ChIP enrichment at a given base-pair position in the yeast genome was calculated as the logarithm to the base 2 of the ratio of fraction of reads mapped at the position in the IP to the fraction of reads mapped at that position in the input. Enrichment over a given nucleosome position was calculated as the logarithm to the base 2 enrichment of ChIP over input in the interval (dyad ± 70 bp).
RSC ChIP analysis
We generated fragment midpoint vs. length maps of RSC (Sth1 subunit) ChIP and input sequencing data sets and then calculated the enrichment map as the logarithm to the base 2 of the ratio of ChIP map to the input map. In the RSC enrichment map, we observed high enrichment at fragment lengths of 200 ± 20 bp over the nucleosome dyad axis. These fragment lengths correspond to remodeler-nucleosome complexes. Hence, we averaged the enrichment at each position relative to the nucleosome dyad axis for fragment lengths of 200 ± 20 bp from the enrichment maps.
RSC depletion data
For analysis of nucleosome positions following RSC depletion, we used published single-end nucleosome positioning data, in which the STH1 promoter was replaced by the MET3 promoter, allowing for twofold repression by a 2-h methionine treatment (Van de Vosse et al. 2013; SRA accession number SRP011944). Reads from control and Sth1 depletion experiments were extended to 150 bp and smoothed with a kernel density function using a Gaussian kernel and a bandwidth of 20.
For analysis of gene expression following RSC depletion, we used published expression data from a degron-containing strain, in which no Sth1 was detectable after 5 h under degron-inducing conditions (Hartley and Madhani 2009). The log2 ratios comparing the Sth1 degron strain to the control strain for each probe was obtained directly from GEO (GSM358298-301). The expression change of a given gene was calculated as described (Hartley and Madhani 2009).
NET-seq data
A previously published NET-seq data set (Churchman and Weissman 2011) was used for analysis of RNAPII stalling in wild-type yeast (GEO accession number GSM617027). Well-expressed genes were picked as those with an average of four reads/bp in the first 500 bp of an mRNA transcript. The stall density plotted here is identical to the “Mean Pause Density” defined in the original study.
Data access
The ChIP-seq and MNase-seq data sets generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE59523.
Supplementary Material
Acknowledgments
We thank Christine Codomo for sequencing library preparation, Jorja Henikoff for help with data analysis, the FHCRC Genomics Shared Resource for Illumina sequencing, and Christopher Weber, Pete Skene, Florian Steiner, and Paul Talbert for critical comments on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.182618.114.
Freely available online through the Genome Research Open Access option.
References
- Annunziato AT. 2005. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 280: 12065–12068. [DOI] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. 2012a. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard KR, Xi L, Wang JP, Widom J. 2012b. A chemical approach to mapping nucleosomes at base pair resolution in yeast. Methods Enzymol 513: 315–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87: 1249–1260. [DOI] [PubMed] [Google Scholar]
- Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. 2008. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol 15: 1272–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408. [DOI] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol 21: 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ramachandran S, Krassovsky K, Bryson TD, Codomo CA, Brogaard K, Widom J, Wang JP, Henikoff JG. 2014. The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. eLife 3: e01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhang Z, Xu M, Li Y, Li Z, Ma Y, Cai T, Zhu B. 2013. H3.3-H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet 9: e1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Rando OJ. 2014. Mechanisms underlying nucleosome positioning in vivo. Annu Rev Biophys 43: 41–63. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K. 2011. Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc Natl Acad Sci 108: 1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Garrard WT. 1991. Transcription-induced nucleosome ‘splitting’: an underlying structure for DNase I sensitive chromatin. EMBO J 10: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. 2007. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci 104: 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci 102: 18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. 2010. Mechanism of chromatin remodeling. Proc Natl Acad Sci 107: 3458–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Griesenbeck J, Boeger H, Maier-Davis B, Kornberg RD. 2011. Selective removal of promoter nucleosomes by the RSC chromatin-remodeling complex. Nat Struct Mol Biol 18: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle-Heyrman G, Zaichuk T, Xi L, Zhang Q, Uhlenbeck OC, Holmgren R, Widom J, Wang JP. 2013. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc Natl Acad Sci 110: 20158–20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. 2013. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R, Jones A. 1976. Genomic transcriptional activity and the structure of chromatin. Nature 260: 495–500. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 27: 6987–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol 19: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, et al. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26: 4920–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152: 969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. 2013. Mot1 redistributes TBP from TATA-containing to TATA-less promoters. Mol Cell Biol 33: 4996–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tsukiyama T, Henikoff S. 2013. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet 9: e1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.