Abstract
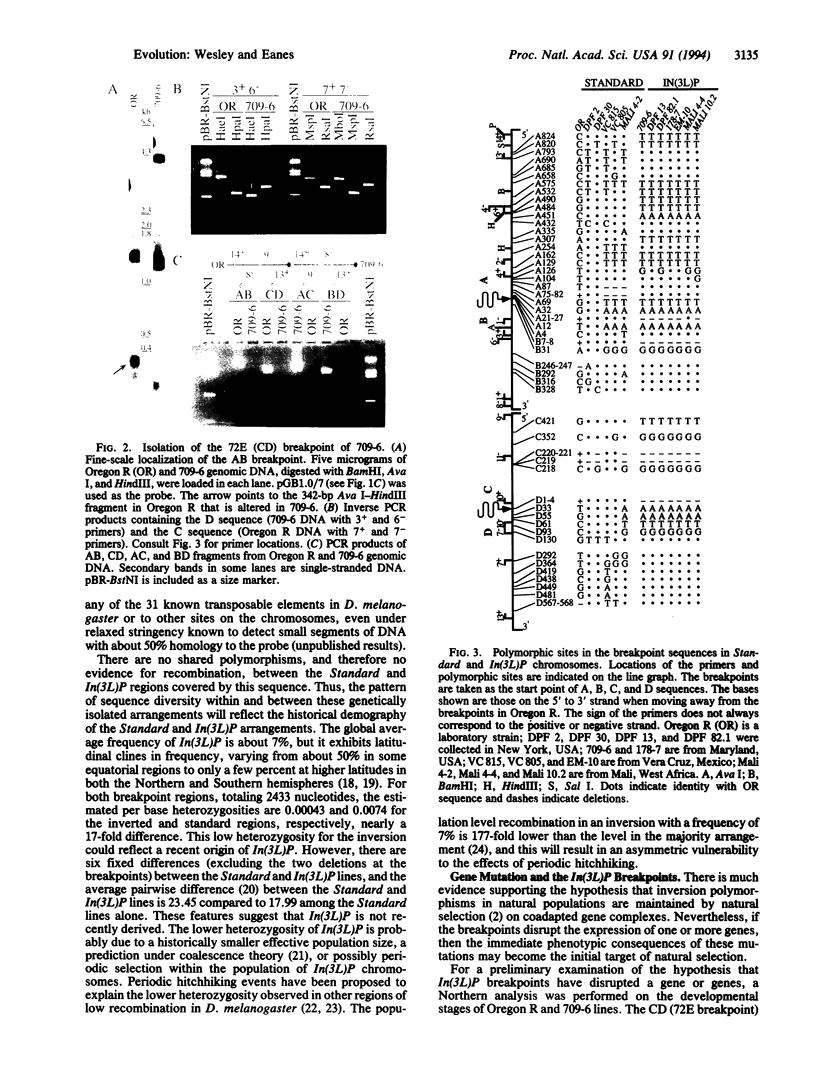
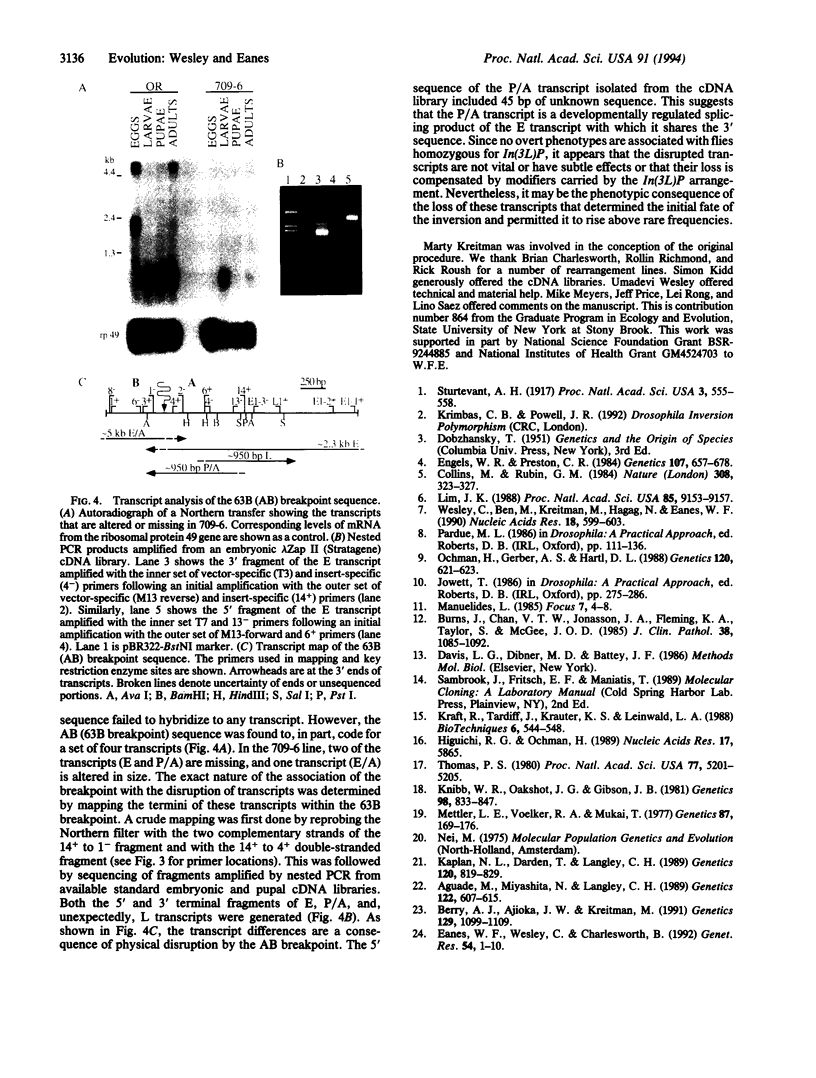
Chromosomal rearrangements constitute a significant feature of genome evolution, and inversion polymorphisms in Drosophila have been studied intensely for decades. Population geneticists have long recognized that the sequence features associated with inversion breakpoints would reveal much about the mutational origin, uniqueness, and genealogical history of individual inversion polymorphisms, but the cloning of breakpoint sequences is not trivial. With the aid of a method for rapid recovery of DNA clones spanning rearrangement breakpoints, we recover and examine the DNA sequences spanning the breakpoints of the cosmopolitan inversion In(3L)Payne in Drosophila melanogaster. By examining the sequence diversity associated with six standard and seven inverted chromosomes from natural populations, we find that the inversion is monophyletic in origin, the sequences are genetically isolated from recombination at the breakpoints, and there is no association with features such as transposable elements. The inverted sequences show 17-fold less nucleotide polymorphism, but there are eight fixed differences in the region spanning both breakpoints. This suggests that this inversion is not recently derived. Finally, Northern analysis and transcript mapping find that the distal breakpoint has disrupted three transcripts that are normally expressed in the standard arrangement. Incidentally, the method introduced here can be used to isolate breakpoint sequences of arrangements associated with many human diseases.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguade M., Miyashita N., Langley C. H. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics. 1989 Jul;122(3):607–615. doi: 10.1093/genetics/122.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J., Chan V. T., Jonasson J. A., Fleming K. A., Taylor S., McGee J. O. Sensitive system for visualising biotinylated DNA probes hybridised in situ: rapid sex determination of intact cells. J Clin Pathol. 1985 Oct;38(10):1085–1092. doi: 10.1136/jcp.38.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M., Rubin G. M. Structure of chromosomal rearrangements induced by the FB transposable element in Drosophila. Nature. 1984 Mar 22;308(5957):323–327. doi: 10.1038/308323a0. [DOI] [PubMed] [Google Scholar]
- Eanes W. F., Wesley C., Charlesworth B. Accumulation of P elements in minority inversions in natural populations of Drosophila melanogaster. Genet Res. 1992 Feb;59(1):1–9. doi: 10.1017/s0016672300030111. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Preston C. R. Formation of chromosome rearrangements by P factors in Drosophila. Genetics. 1984 Aug;107(4):657–678. doi: 10.1093/genetics/107.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. G., Ochman H. Production of single-stranded DNA templates by exonuclease digestion following the polymerase chain reaction. Nucleic Acids Res. 1989 Jul 25;17(14):5865–5865. doi: 10.1093/nar/17.14.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Darden T., Hudson R. R. The coalescent process in models with selection. Genetics. 1988 Nov;120(3):819–829. doi: 10.1093/genetics/120.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb W. R., Oakeshott J. G., Gibson J. B. Chromosome Inversion Polymorphisms in DROSOPHILA MELANOGASTER. I. Latitudinal Clines and Associations between Inversions in Australasian Populations. Genetics. 1981 Aug;98(4):833–847. doi: 10.1093/genetics/98.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Lim J. K. Intrachromosomal rearrangements mediated by hobo transposons in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9153–9157. doi: 10.1073/pnas.85.23.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler L. E., Voelker R. A., Mukai T. Inversion Clines in Populations of DROSOPHILA MELANOGASTER. Genetics. 1977 Sep;87(1):169–176. doi: 10.1093/genetics/87.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988 Nov;120(3):621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume A. G., Knecht D. A., Chovnick A. The rosy locus in Drosophila melanogaster: xanthine dehydrogenase and eye pigments. Genetics. 1991 Dec;129(4):1099–1109. doi: 10.1093/genetics/129.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H. Genetic Factors Affecting the Strength of Linkage in Drosophila. Proc Natl Acad Sci U S A. 1917 Sep;3(9):555–558. doi: 10.1073/pnas.3.9.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley C. S., Ben M., Kreitman M., Hagag N., Eanes W. F. Cloning regions of the Drosophila genome by microdissection of polytene chromosome DNA and PCR with nonspecific primer. Nucleic Acids Res. 1990 Feb 11;18(3):599–603. doi: 10.1093/nar/18.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]