Abstract
The Hippo pathway is an evolutionarily conserved regulator of tissue growth and cell fate during development and regeneration. Conversely, deregulation of the Hippo pathway has been reported in several malignancies. Here, we used integrative functional genomics approaches to identify TAZ, a transcription co-activator and key downstream effector of the Hippo pathway, as an essential driver for the propagation of TNBC malignant phenotype. We further showed in non-transformed human mammary basal epithelial cells that expression of constitutively active TAZ confers cancer stem cell (CSC) traits that are dependent on the TAZ and TEAD interacting domains. In addition, to gain a better understanding of how TAZ functions, we performed genetic-function analysis of TAZ. Significantly, we identified that both the WW and transcriptional activation domains of TAZ are critical for the induced CSC properties as well as tumorigenic potential as manifested in vitro and in human breast cancer xenograft in vivo. Collectively, our data suggest that pharmacological inhibition of TAZ activity may provide a novel means of targeting and eliminating breast CSCs.
Keywords: Hippo pathway, TAZ, breast cancer, cancer stem cells, mammary tumorigenesis
Introduction
Until recently, cancer research efforts have primarily focused on characterizing the cellular and molecular events that enable the malignant transformation of cells harboring oncogenes.1,2 Unfortunately, the identity of cells that acquire the first genetic lesions contributing to the initiation of carcinogenesis has remained elusive. At least 2 non-mutually exclusive models have been proposed to account for their complex intratumoral heterogeneity and inherent differences in tumor-initiating/regenerating capacity: the cancer stem cell (CSC) and stochastic clonal evolution models, respectively.3,4 The CSC hypothesis is based on the observation that many cancers are driven by a subpopulation of cells that display stem cell properties. In addition, previous studies have suggested those CSCs that mediate metastasis are resistant to conventional anticancer therapies and contribute to disease relapse. Consequently, the development of novel technologies for both CSC detection and therapeutic targeting is gaining considerable interest.
The Hippo pathway is an evolutionarily conserved regulator of tissue growth and cell fate during development and regeneration.5,6 On the other hand, deregulation of the Hippo pathway caused by gene mutation(s) or anomalous protein expression and/or activity has been linked to many human diseases including cancer.7,8 Central to the Hippo pathway are 2 key downstream effector proteins, YAP (Yes-associated protein) and TAZ (transcriptional co-activator with PDZ-binding motif). YAP/TAZ do not contain intrinsic DNA-binding domains but instead are recruited to target genes by interacting with multiple transcription factors and mediating diverse sets of transcriptional programs.9 YAP/TAZ are tightly regulated by a number of evolutionarily conserved upstream molecules, such as Mst1/2, Lats1/2 and RASSF family proteins. Inactivation of YAP/TAZ by the Hippo pathway via cytoplasmic sequestration from 14–3–3 binding is well known.10,11 In addition, a novel Hippo pathway-independent restriction of YAP/TAZ by angiomotin and their ubiquitin-mediated degradation has been recently reported,12-14 adding another layer to the complexity of YAP/TAZ regulation.
TAZ, also known as WWTR1 for WW domain containing transcription regulator 1, was first identified in a cDNA screen for 14–3–3-binding proteins.15 The TAZ gene was mapped to chromosome 3q24.16 Human TAZ shares 45% amino acid identity with YAP and also contains the WW domain, coiled-coil region and PDZ-binding motif. Therefore, TAZ is regarded as a paralog of YAP; and both can function as transcriptional co-activators.15 In addition, a recent study reported that YAP/TAZ are nuclear relays of mechanical signals exerted by extracellular matrix (ECM) rigidity and cell shape, thus serving as sensors and mediators of mechanical cues instructed by the cellular microenvironment.17 TAZ has also been indicated responsible for resistance to Taxol in human breast cancer cells.18
Recently, 2 independent research groups identified TAZ as an essential regulator of the CSC properties.19,20 Still, detailed biochemical analysis of the TAZ protein domains and key amino acid residues that contribute to its breast cancer transcriptional activity and oncogenic function are unclear. Here, we show that the Hippo pathway components are highly deregulated in basal-like breast cancer patient samples. Importantly, expression of constitutively active TAZ confers experimental cancer stem-like traits through interaction with the TEAD/TEF transcription factor family. Furthermore, we demonstrate that both the WW domain and transcription activation domain (TAD) of TAZ are critical for the induction of breast CSC properties and mammary tumorigenesis.
Results
Generation of a human Hippo functional interaction network
This study was initiated to investigate the role of the Hippo pathway on breast cancer (BC) tumorigenesis and disease subtype specificity. Since the functional interactions (FI) within the human Hippo pathway and its cross-talk with other signaling pathways are context-dependent and remain largely unknown, we generated a computational network model that represents an ensemble of potentially significant interactions and genetic linkage/association events. Specifically, we mined all evolutionarily conserved Hippo pathway orthologs from several high-quality pathway databases.21-23 In addition, we used Lats1 and Lats2 protein kinases and the transcriptional co-activators YAP and TAZ as seed nodes to select primary, secondary and tertiary-order binary protein-protein interactions (PPI). Finally, we extended our FI network to include non-PPI and protein-DNA interactions relevant to the Hippo pathway, allowing us to detect crosstalk among pathways based on protein interactions. To reduce the complexity of our network model while maximizing relevant protein information, Dijkstra's algorithm was used to calculate the optimal set of interactions by computing the shortest paths between all candidate pathway members. Collectively, a novel human Hippo FI network involving 327 genes (herein denoted Hippo-FIN) was generated (Table S1).
Quantitative prediction of Hippo pathway activity from multi-dimensional cancer genomics data
To assess Hippo-FIN activity among different BC subtypes, we used multi-dimensional genomics data sets from a TCGA patient panel (n = 515 patients) that consisted of basal-like, Her2-enriched, Luminal A and Luminal B BC subtypes with the PAM50-defined subtype predictor as a classification metric.24 There were only 8 claudin-low tumors, thus we did not perform focused analyses on this subtype. First, we applied a rank-based probabilistic pathway activity algorithm to RNA-Seq expression data.25 We proposed a null model generated by randomly permuting the TCGA transcriptome data set while preserving the observed alterations across both genes and samples. Significantly, our analysis identified the Hippo pathway components as being highly deregulated in basal-like patient samples (>89 % of patients, P < 0.003) compared to other BC subtypes (Fig. 1A).
Figure 1.
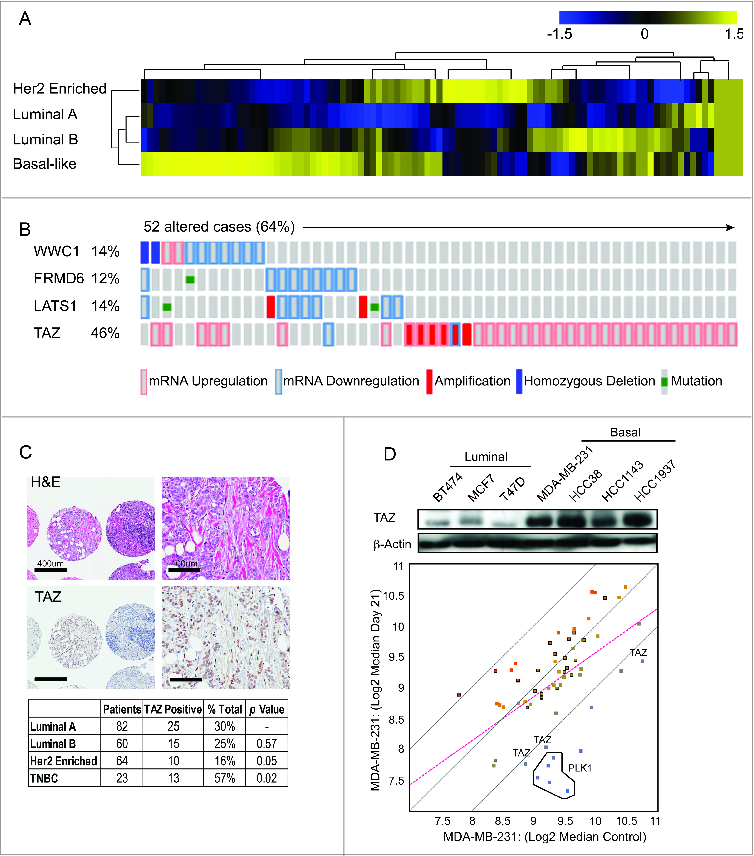
TAZ is highly expressed in basal/triple-negative breast cancer (TNBC) patients. (A) Heat map and heirarchical agglomerative clustering showing Hippo-FIN activity among different breast cancer subtypes using RNA-Seq data sets from a TCGA patient panel (n = 515 patients) that consists of basal-like, Her2 enriched, Luminal A and Luminal B BC subtypes using the PAM50-defined subtype predictor as a classification metric. (B) Alterations in the Hippo-FIN are mutually exclusive. Integrated analysis of mRNA, mutation and copy number events identify TAZ, FRMD6, LATS1 and WWC1 genes as deregulated in basal-like breast cancer tumors to a maximum p-value of 0.05 by Fisher's exact test. Tumor samples are shown in columns and genes in rows. Only samples with >4 % alterations are shown. Shown are genes with statistically significant levels of: (i) mutation (MutSig, false discovery rate <0.1) and mutation types, (ii) deletions and amplifications for genomic regions with statistically significant focal copy number changes (GISTIC2.0) and (iii) RNA expression level for selected genes, expressed as fold change from the median value for all patient tumor samples. (C) TAZ protein is highly expressed in triple-negative breast cancer (TNBC) TMAs. Representative examples of TNBC TMA are shown. Upper, H&E staining; bottom, IHC staining exhibiting high TAZ nuclear expression. (D) TAZ expression in different types of breast cancer cells was revealed by immunoblot. β-Actin was used as loading control. (Upper panel) A population of MDA-MB-231 cells was infected with a pooled shRNA library of a subset of Hippo pathway genes. Log2 median fold change in shRNA abundance of experimental or control (neutral) shRNAs at day 0 vs day 21 tumors (n = 3). The frequency of shRNA-encoding constructs was determined by deep sequencing. An enrichment score was calculated for each shRNA using the probability distribution of the rank product statistics for replicated experiments.
In addition to the alterations in gene expression, genomic perturbations encompassing several distinct classes of DNA sequence changes may also lead to deregulation of the Hippo pathway. Consistent with previous reports, our analyses suggest that the rewiring of Hippo signaling results from molecular events other than the somatic mutation and structural genetic variations of Hippo-FIN genes.7 A plausible explanation for the low frequency of genetic mutations observed in our analyses highlights the essential developmental role of the Hippo pathway, in which haplo-insufficient developmental phenotypes preclude transmission of loss-of-function alleles. Nonetheless, integrative computational analysis successfully identified several genes that were recurrently altered across multiple BC patients. As summarized in Figure 1B, WWC1, FRMD6, LATS1 and TAZ genes were collectively altered in >64 % of basal-like BC patients. FRMD6 and LATS1 co-occurred (P < 0 .0001) but were mutually exclusive with WWC1 or TAZ, with the latter being ranked as the key Hippo-FIN deregulated gene. Notably, TAZ was over-expressed in >44 % of basal-like BC patients and approximately 18% of these patients harbored a corresponding copy number amplification (Fig. 1B). Correspondingly, basal-like BC patients with elevated TAZ mRNA expression levels were more likely to develop metastasis and had a reduced survival compared to those having tumors characterized by normal (physiological) TAZ mRNA expression levels (Log rank Test P-Value: 0.0067 and data not shown).
TAZ is overexpressed in triple-negative breast cancers (TNBCs) and confers cancer stem-like properties on non-transformed mammary epithelial cells
Basal-like tumors are frequently assimilated to TNBC and used interchangeably to identify breast cancers that lack expression of ER, PR, and HER2 expression.26 To independently evaluate the clinical impact of TAZ protein expression and localization in TNBC, we tested 229 unique BC tumor tissue samples for the expression of TAZ by immunohistochemistry (IHC). TAZ, but not YAP, was highly over-expressed and readily detected in both the cytoplasmic and nuclear compartments in a large subset of TNBC patients but not in other BC subtypes (Fig. 1C; Figure S1).
Next, to determine the functional relevance of increased TAZ expression in BC patients and its importance in promoting tumorigenicity, we performed an in vivo RNAi screen on a focused subset of core Hippo pathway genes (Table S2). A pooled screening format was chosen because testing the effects of individual shRNAs on in vivo tumor growth would require a prohibitively large number of mice. MDA-MB-231 was chosen, among several high TAZ-expressing TNBC cell lines examined, because these cells were capable of forming tumors upon injection of the fewest number of cells into the mammary fat pad of severe combined immunodeficiency (SCID) mice (Fig. 1D). Nine hippo pathway genes targeted by a redundant set of 5 validated shRNAs per gene, as well as 5 shRNAs targeting Polo-Like Kinase (PLK1) as positive control and 20 shRNAs targeting luciferase as negative controls (previously optimized for target specificity and minimal off-target effects) were investigated. Twenty-one days later, mammary tumors were harvested and massively parallel sequencing was used to determine the abundance of each shRNA in genomic DNA from tumors and initially injected cells. As shown in Figure 1D, multiple independent shRNAs targeting TAZ were identified.
Central to the Hippo pathway is a kinase cascade, wherein Mst1/2 kinases and Sav1 form a complex to phosphorylate and activate LATS1/2, which in turn phosphorylates TAZ on several serine amino acid residues (S66, S89, S117 and S311) resulting in the inactivation of TAZ10 (Fig. 2A). We next sought to generate a constitutively active form of TAZ, insensitive to both cytoplasmic sequestration and protein degradation mediated by the upstream Hippo kinase cascade. We used previously characterized TAZ mutants (S66, S89, S117 and S311 substituted to alanine) to mimic permanent dephosphorylation (herein termed TAZ-4SA) and an additional point mutation in the TEAD-binding domain (S51 to alanine) that results in an activated form of TAZ lacking TEAD binding (4SA-S51A)27. Retroviral expression of constitutively active Flag-tagged TAZ-4SA was accomplished using pools of infected non-transformed mammary epithelial MCF10A cells to avoid clonal selection bias (Fig. 2B). Compared with control vector-transduced cells, TAZ-4SA-expressing cells demonstrated a dramatic change of cell morphology, i.e., epithelial cells were transformed to an elongated fibroblast-like morphology with pronounced cellular scattering, indicating that TAZ-transduced epithelial cells have undergone epithelial-to-mesenchymal transition (EMT)10,28,29 (Fig. S2A).
Figure 2.
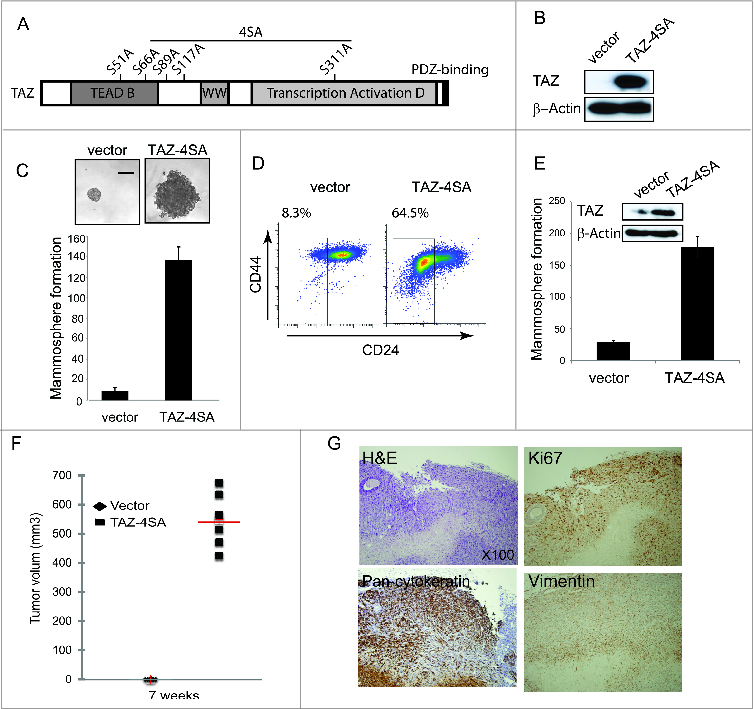
TAZ induces the breast cancer stem cell (CSC) properties and breast tumor formation. (A) Schematic of human TAZ protein showing the TEAD-interaction domain (TEAD-B), WW domain (WW), transcription activation domain (TAD) and PDZ-binding motif. Four serine–to-alanine point mutations (S66A, S89A, S117A and S311A) are introduced into wild-type TAZ construct (TAZ-4SA); additional serine-to-alanine mutation (S51A) is introduced into TAZ-4SA construct (TAZ-4SA-S51A), which leads to the loss of interaction of TAZ with TEAD. (B) Ectopic expression of constitutively active TAZ-4SA in human non-transformed breast epithelial MCF10A cells as revealed by immunoblot. β-Actin was used as the loading control. (C) Images and quantifications of mammosphere formation of vector or TAZ-4SA transduced MCF10A cells. Bars denote standard errors (n = 6). Representative images are shown. (Scale bar, 100μm). (D) Flow cytometry analysis of CD44high/CD24low population in vector or TAZ-4SA transduced MCF10A cells. Percentage of CD44high/CD24low subpopulation is indicated. (E) Quantification of mammosphere formation in vector or TAZ-4SA transduced HMEC cells. Bars denote standard errors (n = 6). Insert: Ectopic expression of constitutively activate TAZ-4SA in human breast epithelial HMEC cells was revealed by immunoblot. β-Actin was used as the loading control. (F) TAZ-4SA-transduced MCF10A cells induce mammary tumor formation when injected into the mammary fat pad of NOD/SCID mice (n = 6). (G) Histological analysis of tumors from the TAZ-4SA injected mice. Shown are H&E and IHC staining of Ki67, pan-cytokeratin AE1/3 and human-specific vimentin.
Acquisition of an EMT phenotype is a critical process for switching early-stage carcinomas into invasive malignancies and is associated with tumor aggressiveness and metastasis. Furthermore, cells undergoing EMT and CSCs share many similar molecular characteristics, suggesting that there could be common regulatory programs.30,31 We next sought to determine whether TAZ confers cancer stem-like traits by an EMT-mediated transcriptional program. We assessed the capacity of TAZ-4SA to form and propagate mammospheres of MCF10A cells in vitro. It was found that TAZ-4SA-expressing MCF10A cells gave rise to mammospheres of increased size and number (Fig. 2C). Notably, FACS analysis revealed that TAZ-4SA expressing cells demonstrated an 8 fold increase in the CD44high/CD24low population compared with the vector-transduced control cells (Fig. 2D). Consistently, we also detected a significant increase in the mammosphere formation and CD44high/CD24low population in the TAZ-4SA-expressing primary human mammary epithelial cells (HMEC) (Fig. 2E; Figures 2B).
To further examine the contribution of endogenous TAZ to the BC stem-like traits, we performed stable knockdown of TAZ in MDA-MB-231 cells using RNAi. Western analysis of 2 independent shRNA hairpins targeting non-overlapping regions of the TAZ mRNA transcript showed >90 % reduction of TAZ expression compared with a non-targeting control (Figure S2C). Consistent with previous report, self-renewal as measured by mammosphere formation was reduced in both TAZ-knockdown cell lines19 (Figure S2D).
To evaluate the effects of constitutively activated TAZ on in vivo mammary tumor-forming potential, we injected TAZ-4SA-expressing MCF10A cells into the mammary fat pad of SCID mice. Expression of TAZ-4SA resulted in high-grade tumors and enhanced in vivo growth (Fig. 2F). As shown in Figure 2G, representative hematoxylin and eosin staining for TAZ-4SA expressing tumor sections showed prominent spindle and epithelioid cell morphology, whereas immunohistochemistry identified tumor cells expressing cytokeratin and vimentin, indicating poorly differentiated carcinomas. In addition, high Ki67 expression was observed, which is typically associated with high histologic tumor grade.
Induction of cancer stem-like properties is mediated through TEAD/TEF transcription factors
TAZ interacts with numerous transcription factors to regulate its complete spectrum of target genes. We and others have previously demonstrated that TAZ interacts with members of the TEAD/TEF family and this plays a leading role in the TAZ-induced EMT.29,32 We then established MCF10A cell lines that stably express Flag-tagged TAZ-4SA-S51A, which is constitutively active but lacks interactions with the TEAD family of transcription factors (Fig. 3A). Significantly, mammosphere formation was completely inhibited in TAZ-4SA-S51A clones when compared with the TAZ-4SA expressing cells (Fig. 3B). Furthermore, FACS analysis of TAZ-4SA-S51A expressing cells had no detectable increase in the CD44high/CD24low subpopulation, which was independently confirmed by stable inhibition of TEAD1, TEAD3 and 4 by RNAi (Fig. 3D-E). Together, these findings clearly demonstrate that disruption of the TAZ-TEAD interaction inhibits the ability of TAZ to confer cancer stem-like traits.
Figure 3.
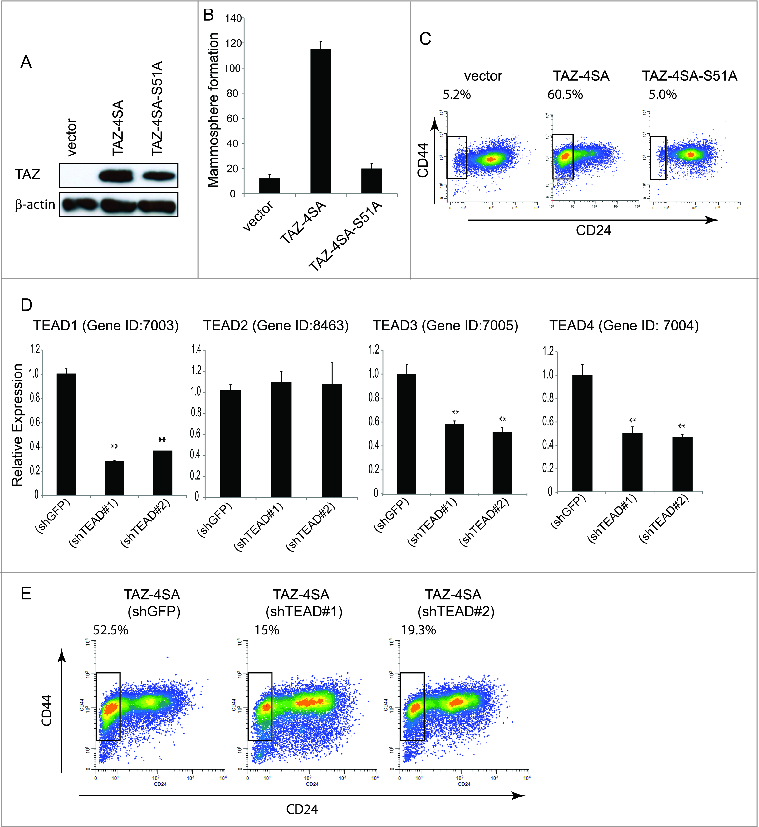
TAZ-induced breast CSC properties are dependent on the TAZ-TEAD interaction. (A) Ectopic expression of TAZ-4SA or TAZ-4SA-S51A in human breast epithelial MCF10A cells as revealed by immunoblot. β-Actin was used as the loading control. (B) Quantification of mammosphere formation in TAZ-4SA or TAZ-4SA-S51A transduced MCF10A cells. Bars denote standard errors (n = 6). (C) Flow cytometry analysis of the CD44high/CD24low population in vector, TAZ-4SA or TAZ-4SA-S51A transduced MCF10A cells. Percentage of CD44high/CD24low subpopulation is indicated. In contrast to TAZ-4SA, TAZ-4SA-S51A fails to increase the CD44high/CD24low cell subpopulation. (D) Real-time RT-PCR examination of TEAD1, TEAD2, TEAD3 and TEAD4 mRNA followed by treatment with control or 2 independent shRNAs (shTEAD#1, 2) in the TAZ-4SA-transduced MCF10A cells. GAPDH was used as an internal control (**, p < 0 .001). (E) Flow cytometry analysis of the CD44high/CD24low population in control, shTEAD#1 or shTEAD#2 treated cells.
Analysis of important TAZ motifs that contribute to the induced cancer stem-like traits
It has been previously reported that several serine amino acid residues (S66, S89, S117 and S311) of TAZ can be phosphorylated by the Hippo pathway upstream Lats1/2 kinase.10 We previously showed that individual TAZ serine mutants induced various degree of cell migration and cellular transformation.29 To test the stem-like properties conferred by each TAZ serine mutant, we transduced the MCF10A cells with vector control, TAZ-S66A, TAZ-S89A, TAZ-S89A, TAZ-117A or TAZ-S311A and performed the FACS analysis (Fig. 4A; Figure S3A). Of note, TAZ-S66A and TAZ-117A had no effect on the CD44high/CD24low population whereas TAZ-S89A and TAZ-S311A increased the CD44high/CD24low population by 2 fold (Fig. 4C; Figure S3B). It is known that phosphorylation of S89 leads to sequestration of TAZ in the cytoplasm and phosphorylation of S311 primes TAZ to degradation.10,33 We thus focused on the single mutation of TAZ S89 or S311 as well as their double mutations (S89A, S311A). To test their effect on TAZ-induced cancer stem-like traits, we transduced MCF10A cells with vector control, TAZ-S89A, TAZ-S311A, TAZ-2SA (S89A, S311A) or TAZ-4SA (Fig. 4A). As a result, TAZ-S89A induced the mammosphere formation to a similar extent as TAZ-4SA, whereas TAZ-S311A failed to do so (Fig. 4B). Of particular note, TAZ-2SA promoted mammopsphere formation and increased the CD44high/CD24low population to an extent comparable to TAZ-4SA (Fig. 4B and C). These results indicate that both serine 89 and 311 are important to the TAZ-induced breast stem-like properties.
Figure 4.
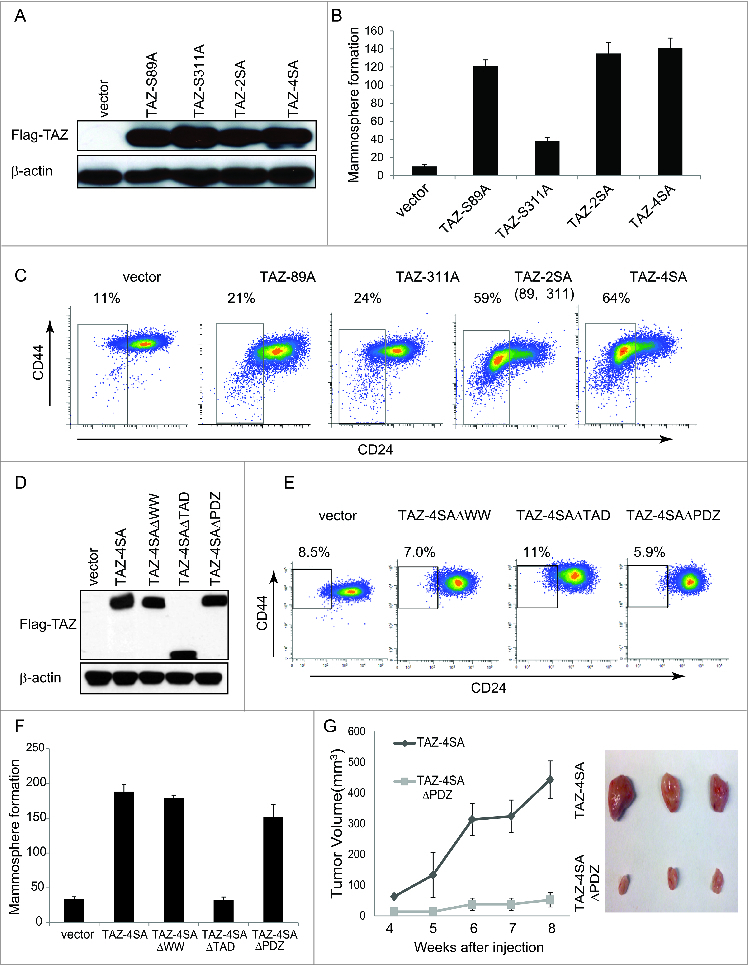
For figure legend, see page 166.Figure 4 (See previous page). The WW and TAD domains are important for TAZ induced mammary tumor formation. (A) Ectopic expression of TAZ-S89A, TAZ-S311A, TAZ-2SA (S89A, S311A), TAZ-4SA in human non-transformed breast epithelial MCF10A cells as revealed by immunoblot. β-Actin was used as the loading control. (B) Quantifications of mammosphere formation of vector or TAZ-S89A, TAZ-S311A, TAZ-2SA (S89A, S311A) and TAZ-4SA transduced MCF10A cells. Bars denote standard errors (n = 6). (C) Flow cytometry analysis of CD44high/CD24low population in vector or TAZ-S89A, TAZ-S311A, TAZ-2SA (S89A, S311A) and TAZ-4SA transduced MCF10A cells. Percentage of CD44high/CD24low subpopulation is indicated. (D) Ectopic expression of TAZ-4SA, TAZ-4SAΔWW, TAZ-4SAΔTAD or TAZ-4SAΔPDZ in human non-transformed breast epithelial MCF10A cells as revealed by immunoblot. β-Actin was used as the loading control. (E) Flow cytometry analysis of CD44high/CD24low population in vector, TAZ-4SAΔWW, TAZ-4SAΔTAD and TAZ-4SAΔPDZ transduced MCF10A cells. Percentage of CD44high/CD24low subpopulation is indicated. (F) Quantifications of mammosphere formation of vector, TAZ-4SAΔWW, TAZ-4SAΔTAD, TAZ-4SAΔPDZ and TAZ-4SA transduced MCF10A cells. Bars denote standard errors (n = 6). (G) TAZ-4SA and TAZ-4SAΔPDZ-transduced MCF10A cells induce mammary tumor formation when injected into the mammary fat pad of NOD/SCID mice (n = 6). Representative images of induced mammary tumors are shown (Right panel).
TAZ contains several typical functional protein domains, such as the WW protein-protein interaction domain, transcriptional activation domain (TAD) and PDZ-binding domain, which are important for TAZ functions (Fig. 2A). For example, it has been shown that Zonula occludens-1 (Zo-1) and -2 (ZO2) interact with the TAZ PDZ-binding motif and negatively regulate TAZ function.34 To further explore whether these domains play a role in TAZ-induced breast CSC traits, we generated retroviral expression constructs with deleted WW, TAD or PDZ-binding domains in TAZ-4SA. As shown in Figure 4D, MCF10A cells were transduced with vector control or TAZ-4SAΔWW, TAZ-4SAΔTAD, TAZ-4SAΔPDZ or TAZ-4SA expression constructs. Surprisingly, all TAZ mutants tested failed to induce CD44high/CD24low population changes (Fig. 4E). Moreover, although TAZ-4SAΔWW and TAZ-4SAΔPDZ promoted mammosphere formation in vitro, TAZ-4SAΔTAD completely abolished such capability (Fig. 4F). To independently test the effects of TAZ mutants on mammary tumorigenesis in vivo, we injected vector, TAZ-4SAΔWW, TAZ-4SAΔTAD, TAZ-4SAΔPDZ or TAZ-4SA transduced cells into the mammary fat pad of SCID mice. Interestingly, TAZ-4SAΔWW and TAZ-4SAΔTAD expressing cells completely lost their ability to form mammary tumors, whereas TAZ-4SAΔPDZ transduced cells formed much smaller tumors as compared to TAZ-4SA (Fig. 4G). Taken together, these results indicated that the WW, TAD and PDZ-binding domains of TAZ play important roles in the TAZ-induced breast CSC properties and primary mammary tumor formation.
Discussion
In the current study, we identified the Hippo pathway as being highly transcriptionally deregulated in basal-like but not the other breast cancer subtypes. Furthermore, mutual exclusivity analysis revealed several genes that were recurrently altered across multiple BC patients using copy number, somatic exonic mutations and concordant mRNA expression from large-scale TCGA data sets. For example, consistent with previous published findings, TAZ was highly overexpressed in basal-like/triple-negative breast cancers (TNBCs).19,28,35 Since altered levels of gene expression do not corroborate a causal role in carcinogenesis, we used an RNAi-mediated gene essentiality profiling to unambiguously determine the functional requirements of core Hippo pathway genes in tumor growth in vivo. Significantly, we demonstrate that TAZ is the key Hippo pathway effector in basal-like BC and the post-transcriptional silencing of TAZ using RNAi abrogated the malignant phenotype of TNBC cells.
Overlapping functions have been defined for TAZ and YAP.10,28,29,36 Although the net effect of deregulated YAP/TAZ activity appears in many tissues to be similar, our analyses indicate that their activity in basal-like and triple-negative breast cancer (TNBC) cells are controlled by disparate regulatory mechanisms. For example, it was previously reported that expression of constitutively expressed TAZ stimulates cell proliferation and dramatically increases the S-phase cell population.10 Here, we demonstrated that expression of constitutively active TAZ conferred cancer stem-like traits, including increased self-renewal and tumor formation potential in vivo. We previously showed that YAP/TAZ increases the expression of amphiregulin (AREG), an epidermal growth factor receptor (EGFR) ligand, which contributes to the YAP/TAZ-induced growth factor independent growth.29,37 Interestingly, we found here that increased AREG expression has no effect on the TAZ-conferred stem-like traits (Figure S4A and B), indicating that the stem-like traits may not be coupled with cell proliferation. Two major mechanisms contribute to the pro-tumorigenic transcriptional program mediated by TAZ: 1) the transcriptional output of Hippo signaling is context/cell-type dependent; and 2) TAZ has multiple transcription factor binding partners. Disruption of TAZ-TEAD binding or silencing of TEAD1, TEAD3 and TEAD4 by RNAi completely abolished the ability of TAZ to confer CSC-like traits, as we demonstrated directly in the present study.
We have previously reported that different TAZ serine mutants display various degree of malignant behavior.29 Here we expand our studies on the TAZ-induced stem-like traits and found that TAZ-S66A and TAZ-S117A mutants failed to change the CD44high/CD24low population. TAZ-S89A or TAZ-S311A elicited partial stem-like traits, but neither induced tumor formation (data not shown). Interestingly, TAZ S89A and TAZ S311A double mutants conferred strong breast CSC-like traits to the non-transformed mammary epithelial cells. In addition, we provide genetic evidence of both TAZ WW and PDZ-binding domains playing an important role in self-renewal and mammary tumorigenesis. It has been reported that the WW domain of TAZ is important for its oncogenic capability as assessed by anchorage-independent cell growth in soft agar in that MCF10A cells transduced with the WW domain mutation formed significantly fewer colonies compared to the wild-type TAZ.38 It was further demonstrated that the reduced transforming ability of TAZ WW mutant was due to its lost interaction with the WW domain binding protein 2 (Wbp2). Interestingly, Wbp2 has also been shown to interact with YAP.39 Nevertheless, the mechanisms by which Wbp2 regulates TAZ-mediated transformation and gene expression are currently unknown. For example, it will be of interest to investigate whether Wbp2 has any effect on the loss of tumorigenic potential of TAZ-4SAΔWW.
Finally, the C-terminus of TAZ contains a highly conserved PDZ-binding motif that localizes TAZ into discrete nuclear foci and is essential for TAZ-stimulated gene transcription.15 In the current study, we demonstrate that loss of the PDZ-binding motif of TAZ abrogated mammary tumor formation potential in vivo. The specific PDZ protein responsible for the regulation of TAZ localization and BC oncogenic function thus will be of considerable interest to pursue.
Materials and Methods
Cell culture, transfection and transduction
MCF10A cells were cultured as described.40 Human Mammary Epithelial Cells (HMEC) cells were cultivated in MEGM™ Mammary Epithelial Cell Growth Medium (Lonza, Walkersville, MD). MDA-MB-231 cells were cultivated in RPMI-1640 medium with 10% fetal bovine serum (FBS). All media were supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin and 2mM glutamine. All cells were cultured in a humidified atmosphere of 95% air and 5% CO2 at 37ºC. MDA-MB-231cells were purchased from the American Type Culture Collection (ATCC, VA). Transfection was performed using X-tremeGENE 9 DNA Transfection Reagent following the manufacturer's protocol (Roche). Packaging of retrovirus and lentivirus, cell transduction and drug selection were performed following standard protocols. For knockdown experiments, shRNA hairpins targeting human TAZ were obtained from the RNAi Consortium (The Broad Institute). The target sequences are listed (in the 5′-3′ direction): shNon-target-Ctrl: CAACAAGATGAAGAGCACCAA; shTAZ-1: CCTGCCGGAGTCTTTCTTTAA and shTAZ-2: GAAACTGCGGCTTCAGAGAAT. shAREG, shTEAD hairpins were generated as described.29,41
Plasmid construction
The human TAZ-S66A, TAZ-S89A, TAZ-S117A, TAZ-S311A and TAZ-4SA expression constructs are described previously.29 TAZ-2SA and TAZ-4SA-S51A was generated by PCR-based mutagenesis as described previously. Briefly, PCR was performed with complementary primers containing the desired mutation(s) (synthesized by IDT) using Expand High Fidelity PCR System (Roche) and cloned into the pBABE-puro vector as a Flag-tagged BamHI-EcoRI fragment.
Total RNA extraction and real-time RT-PCR
For RNA preparation and qRT-PCR, RNA was extracted using the Trizol reagent (Invitrogen). cDNA synthesis was performed using the First-Strand cDNA Synthesis Kit (GE Healthcare) and quantitative real-time RT-PCR was performed using Power SYBR Green PCR Master Mix (Invitrogen). Sequences of the qPCR primer pairs (in the 5′-3′ direction) are as follows: GAPDH-F: GGTGAAGGTCGGAGTCAACGG; GAPDH-R: GAGGTCAATGAAGGGGTCATTG; TAZ-F: AGTACCCTGAGCCAGCAGAA; TAZ-R: GATTCTCTGAAGCCGCAG-TT; TEAD1-F: CCCATTCCAGGGTTTGAGC; TEAD1-R: TGCACGAAGAGGTGTTTGTTG; TEAD2-F: GCCCGCT-ACATCAAGCTGA; TEAD2-R: TGGTTGCCATTGTCTG-GAAAG; TEAD3-F: GCCAGTGTCCTGCAGAACAA; TEAD3-R: CAAAGGGCTTGATGTCCTGAG; TEAD4-F: TCTCTGCCTTCCTGGAGCA and TEAD4-R: TCATAGATTTGGCGGATGTCC. All measurements were performed in triplicate and standardized to the levels of GAPDH.
Antibodies and immunoblot analysis
TAZ antibody was purchased from Cell Signaling Technology; YAP antibody from Santa Cruz; β-actin antibody from Upstate, fibronectin and Flag (M2) antibodies from Sigma-Aldrich, CD24-PE and CD44-APC antibodies from Invitrogen, amphiregulin antibody from R&D Systems. For protein extraction, cells were washed with phosphate-buffered saline (PBS) and collected with IP buffer: 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20% glycerol, 0.5% NP-40, plus 1x CompleteTM EDTA-free Protease Inhibitor Cocktail (Roche) or 1x HaltTM EDTA-free Protease and Phosphotase Inhibitor (Thermo Scientific). Cell lysate was cleared by centrifugation at 14,000 rpm for 20 min at 4 °C. Lysate was loaded onto 4–15% MINI-PROTEAN TGX gel (Bio-Rad) with 4X SDS sample buffer. For immunoblot, proteins were transferred onto Immobilon-P membrane (Millipore), detected by various antibodies and visualized with ECL Plus Western Blotting Detection Reagents (GE Healthcare).
Immunohistochemistry
Formalin-fixed paraffin embedded (FFPE) tissue blocks were sectioned in 5 micron thickness and subjected to immunohistochemistry (IHC) studies. Quality of histomorphology of tumor samples was assessed on hematoxylin and eosin (H & E) stained sections before immunostaining. Antibodies against pan-cytokeratin (AE1/3), TAZ and YAP were obtained from Cell Signaling Technology, and vimentin and Ki67 from Dako. Paraffin sections were placed on charged slides and IHC staining was carried out in a Dako AutostainerPlus (Carpinteria, CA) as previously described.42 Histomorphology and immunostaining results were interpreted by a board-certified pathologist (WB; BX).
Flow cytometry analysis
Cells were passed through a 35-μm filter, pelleted, washed in 1X phosphate buffered saline (PBS) + 0.5% fetal calf serum (FBS) and counted. One million cells were suspended in 1X PBS + 0.5% FBS and stained with anti-CD44-APC conjugate and anti-CD24-PE conjugate (BD Biosciences) for 30 min at 4°C. Cells were washed 3 times and then analyzed by flow cytometry.
Mammosphere formation assay
Mammosphere formation assay was performed by plating 5 × 104 cells in serum-free DMEM/F12 1:1 media (Gibco) supplemented with EGF (20 ng/mL) and B27 (2%) into ultra-low attachment 6-well plates (Corning). Mammospheres were allowed to grow for 7 d Total mammospheres greater than 100 μm in diameter were counted. Each experimental group was done in triplicate and the same experiments were repeated at least 3 times.
TMA score and statistical analysis
The TMAs were built at the Pathology Resource Network (PRN) at Roswell Park Cancer Institute (RPCI) using the pathology paraffin archives. All patients in these TMAs had surgeries to remove the primary breast cancer and metastasis, when applicable, between 1996 and 2009. The staining intensity (0, 1, 2, 3) was multiplied by the percentage of cells (0, 1, 2, 3) to generate a final staining score. Staining score greater than 0 was defined as positive. Fisher's exact test was used to test the association between TAZ or YAP1 staining in the nucleus (nucleus only or both nucleus and cytoplasm) and in each cancer subtype. Cancer subtype was defined as following: Luminal A if estrogen (ER) and progesterone (PR) are both positive but Her 2 is negative; Luminal B if ER, PR and Her2 are all positive; Her2 enriched if ER and PR are both negative but Her2 is positive; and triple-negative breast cancer (TNBC) if ER, PR and Her2 are all negative. Statistical analysis of data was performed using the SPSS statistics software package (SPSS, IL). All results are expressed as mean ± SD. *, P < .05; **, P < .001; ***, P < .0001.
In vivo tumor growth assays
For TAZ tumor formation, vector control, TAZ-4SA, TAZ-4SAΔWW, TAZ-4SAΔTAD or TAZ-4SAΔPDZ transduced MCF10A cells in exponential growth phase were harvested and suspended in PBS (50% matrigel); and 1 × 106 (in 0.1 mL) cells were injected into the mammary fat pad of female NOD/SCID mice at 6–8 weeks old. The mice were generated at the Roswell Park Cancer Institute. Tumor sizes were measured twice a week using calipers. Mammary tumor formations were also detected by the In Vivo Luminescence Imaging System. The care and use of animals was approved by the Institutional Animal Care and Use Committee of the Roswell Park Cancer Institute (Buffalo, NY).
Supplementary Material
Acknowledgments
We thank Ellen Karasik for her expertise in immuno- histochemistry.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Roswell Park Cancer Institute and National Cancer Institute (NCI) grant #P30 CA016056; and in part by National Cancer Institute (NCI) R21 CA179693 and the American Cancer Society Institutional Research Grant 02–197–04 (to J.Z).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100(1):57-70. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144(5):646-674. [DOI] [PubMed] [Google Scholar]
- 3. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 2012; 21(3):283-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10(6):717-728; http://dx.doi.org/ 10.1016/j.stem.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 5. Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010; 19(4):491-505; PMID:20951342; http://dx.doi.org/ 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development 2011; 138(1):9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013; 13(4):246-257; http://dx.doi.org/ 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- 8. Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev 2013; 27(4):355-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 2011; 13(8):877-883; http://dx.doi.org/ 10.1038/ncb2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mole Cell Biol 2008; 28(7):2426-2436; http://dx.doi.org/ 10.1128/MCB.01874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 2008; 283(9):5496-5509. [DOI] [PubMed] [Google Scholar]
- 12. Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev 2010; 24(1):72-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem 2011; 286(9):7018-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 2011; 25(1):51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 2000; 19(24):6778-6791; PMID:11118213; http://dx.doi.org/ 10.1093/emboj/19.24.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN. A method for constructing radiation hybrid maps of whole genomes. Nat Genet 1994; 7(1):22-28; http://dx.doi.org/ 10.1038/ng0594-22 [DOI] [PubMed] [Google Scholar]
- 17. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature 2011; 474(7350):179-183. [DOI] [PubMed] [Google Scholar]
- 18. Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res 2011; 71(7):2728-2738. [DOI] [PubMed] [Google Scholar]
- 19. Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011; 147(4):759-772. [DOI] [PubMed] [Google Scholar]
- 20. Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev 2011; 25(24):2594-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Science Signal 2013; 6(302):rs15. [DOI] [PubMed] [Google Scholar]
- 22. Kwon Y, Vinayagam A, Sun X, Dephoure N, Gygi SP, Hong P, Perrimon N. The Hippo signaling pathway interactome. Science 2013; 342(6159):737-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Defining the protein-protein interaction network of the human hippo pathway. Mol Cell Proteomics : MCP 2014; 13(1):119-131; http://dx.doi.org/ 10.1074/mcp.M113.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490(7418):61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su J, Yoon BJ, Dougherty ER. Accurate and reliable cancer classification based on probabilistic inference of pathway activity. PloS One 2009; 4(12):e8161; http://dx.doi.org/ 10.1371/journal.pone.0008161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. New Eng J Med 2010; 363(20):1938-1948; http://dx.doi.org/ 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 27. Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol 2012; 23(7):785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 2008; 68(8):2592-2598. [DOI] [PubMed] [Google Scholar]
- 29. Yang N, Morrison CD, Liu P, Miecznikowski J, Bshara W, Han S, Zhu Q, Omilian AR, Li X, Zhang J. TAZ induces growth factor-independent proliferation through activation of EGFR ligand amphiregulin. Cell Cycle 2012; 11(15):2922-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133(4):704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011; 331(6024):1559-1564. [DOI] [PubMed] [Google Scholar]
- 32. Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem 2009; 284(20):13355-13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{β}-TrCP E3 ligase. J Biol Chem 2010; 285(48):37159-37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Remue E, Meerschaert K, Oka T, Boucherie C, Vandekerckhove J, Sudol M, Gettemans J. TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett 2010; 584(19):4175-4180. [DOI] [PubMed] [Google Scholar]
- 35. Skibinski A, Breindel JL, Prat A, Galvan P, Smith E, Rolfs A, Gupta PB, Labaer J, Kuperwasser C. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep 2014; 6(6):1059-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A 2006; 103(33):12405-12410; http://dx.doi.org/ 10.1073/pnas.0605579103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol 2009; 11(12):1444-1450; http://dx.doi.org/ 10.1038/ncb1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF, Hong W. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene 2011; 30(5):600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A 1995; 92(17):7819-7823; http://dx.doi.org/ 10.1073/pnas.92.17.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003; 30(3):256-268. [DOI] [PubMed] [Google Scholar]
- 41. Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008; 22(14):1962-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu B, Thong N, Tan D, Khoury T. Expression of thyroid transcription factor-1 in colorectal carcinoma. Appl Immunohistochem Mol Morphol 2010; 18(3):244-249; http://dx.doi.org/ 10.1097/PAI.0b013e3181c29407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


