Abstract
Auxin elicits diverse cell behaviors through a simple nuclear signaling pathway initiated by degradation of Aux/IAA co-repressors. Our previous work revealed that members of the large Arabidopsis Aux/IAA family exhibit a range of degradation rates in synthetic contexts. However, it remained an unresolved issue whether differences in Aux/IAA turnover rates played a significant role in plant responses to auxin. Here, we use the well-established model of lateral root development to directly test the hypothesis that the rate of auxin-induced Aux/IAA turnover sets the pace for auxin-regulated developmental events. We did this by generating transgenic plants expressing degradation rate variants of IAA14, a crucial determinant of lateral root initiation. Progression through the well-established stages of lateral root development was strongly correlated with the engineered rates of IAA14 turnover, leading to the conclusion that Aux/IAAs are auxin-initiated timers that synchronize developmental transitions.
Keywords: Arabidopsis, Phytohormone, Saccharomyces, Ubiquitin
Highlighted article: Degradation dynamics of the auxin-responsive inhibitor IAA14 act as a tunable timer, controlling both the density and timing of lateral root emergence in Arabidopsis.
INTRODUCTION
The plant hormone auxin directs many developmental responses, including the elaboration of branching patterns in the root. Among the first steps in auxin signal transduction is the ubiquitylation and degradation of transcriptional co-repressor proteins called Aux/IAAs (Chapman and Estelle, 2009), which belong to a large gene family. Auxin induces a high-affinity interaction between a 13 amino acid degron sequence in the Aux/IAA (DII) and an F-box protein belonging to the TIR1/AFB family (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007). This allows de-repression of the AUXIN RESPONSE FACTORS (ARFs), which in turn activate auxin-responsive genes. Stabilizing degron mutations in different Aux/IAAs provoke distinct phenotypes, as do loss-of-function mutations in activator ARFs, suggesting functional specificity within these families (Lokerse and Weijers, 2009). Additionally, stabilized versions of one Aux/IAA cannot fully recapitulate the phenotype of stabilized versions of a different family member, even when expressed from the same promoter (Muto et al., 2007; Weijers et al., 2005). Divergence in Aux/IAA function is likely multifaceted, including potential differences in affinity for the receptor-hormone complex, turnover rates and interaction strength with ARFs (Pierre-Jerome et al., 2013).
The dynamics of cellular signaling, in addition to absolute changes in signal abundance, can play a crucial role in determining cellular outcomes (Purvis and Lahav, 2013). For example, calcium dynamics have recently been shown to play an essential role in communication among gametophytic cells during fertilization (Ngo et al., 2014). There is significant variation among Aux/IAAs in both auxin-induced affinity for TIR1/AFBs and degradation rates (Calderón Villalobos et al., 2012; Havens et al., 2012). This variation can be translated into quantitative differences in auxin-induced transcription in a synthetic context (Pierre-Jerome et al., 2014).
Lateral root development has a well-defined sequence of developmental stages (Fig. 1A), many of which are controlled by auxin, and is non-essential for plant survival in laboratory conditions (Malamy and Benfey, 1997). Thus, it is an ideal testing ground for interrogating the possibility that nuclear auxin signaling dynamics act as a checkpoint to set the timing of auxin-regulated cell behaviors. To test this hypothesis, we used a synthetic yeast system to guide the engineering of plants expressing degradation rate variants of IAA14, an Aux/IAA that is crucial for the first asymmetric divisions that mark the initiation of new root primordia (Fukaki et al., 2002; Vanneste et al., 2005). By expressing these variants in plants under the native IAA14 promoter, we found that the dynamics of lateral root initiation and emergence were plastic and could be tuned by altering the pace of auxin-induced IAA14 turnover.
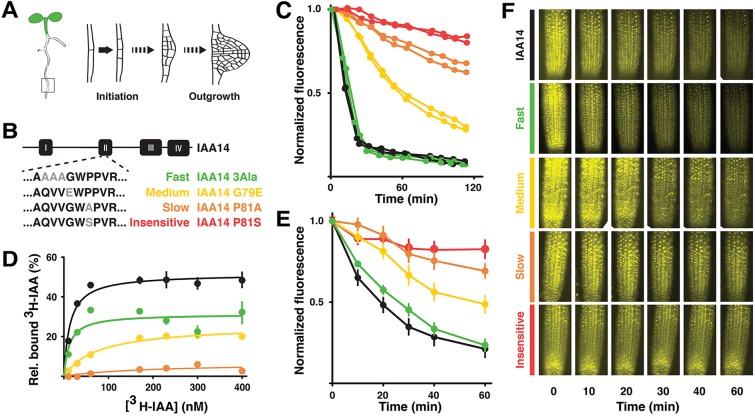
Fig. 1.
Engineered IAA14 variants exhibit a range of degradation rates and auxin sensitivities. (A) Lateral root development. (B) Amino acid substitutions engineered in domain II of IAA14. (C) IAA14 rate variants exhibited a range of auxin-induced degradation rates in yeast when co-expressed with the AFB2 auxin receptor. Time-lapse flow cytometry was used to monitor YFP-IAA14 signal after auxin treatment (Time 0) and then normalized to initial fluorescence. Two biological replicates are shown with 10,000 measurements taken per sample per time-point. (D) When combined with TIR1, IAA14 variants had different auxin-binding affinities. In vitro saturation binding experiments showed high auxin binding affinity by TIR1:14-control (Kd=12.2±5.7) and TIR1:14-Fast (Kd=13.6±2.7) co-receptor complexes. Auxin binding affinity was substantially reduced in TIR1:14-Medium complex (Kd=77.3±11.4) and TIR1:14-Slow complexes (Kd=187±206). A representative saturation binding experiment is shown. Error bars indicate s.e.m. Some error bars are within the boundaries of the markers. (E) Degradation rates in plants were similar to those observed in yeast. Venus-tagged IAA14 variants were induced by heat-shock, then sprayed with auxin (Time 0). Each data point represents the average of two biological replicates (four or five plants per replicate) from two independent lines. Error bars indicate s.e.m. (F) Images of representative heat-shock-inducible Venus-IAA14 variant plants. Treatments are as in E.
RESULTS AND DISCUSSION
IAA14 degradation rate and auxin sensitivity can be tuned by point mutations
We engineered a library of full-length IAA14 degradation rate variants by mutating the DII domain, using either point mutations or domain swaps (Fig. 1B; supplementary material Fig. S1A). Point mutations were guided by previously identified variants that slowed or abolished turnover of DII-luciferase fusions in plants (Ramos et al., 2001). Chimeras were designed by replacing a DII-containing region of IAA14 with that of a slower-degrading Aux/IAA. In a synthetic yeast degradation assay (Havens et al., 2012), the engineered IAA14 variants exhibited a range of auxin-induced degradation dynamics (Fig. 1B,C; supplementary material Fig. S1A-C). For simplicity of interpretation, we focused our analysis on the point mutants that spanned the range of observed degradation rates. We designated the variants as wild-type (14), fast (F), medium (M), slow (S) or insensitive (I). The I-variant protein harbors the identical mutation found in the solitary root (slr-1) mutant (Fukaki et al., 2002), and was insensitive to auxin when expressed in yeast (Fig. 1B,C). Consistent with their degron location, variants were weaker co-receptors in in vitro auxin-binding assays (Fig. 1D). Auxin dose response assays provided additional evidence that auxin sensitivity of the IAA14 variants correlated well with observed degradation rates (supplementary material Fig. S1D).
To test whether observed degradation rate differences among the IAA14 variants in yeast were maintained in their native context, we developed a fluorescent Aux/IAA degradation assay in plants (Fig. 1E,F). Expression levels of Aux/IAAs are generally low, so we employed a previously characterized heat-shock-inducible promoter to provide detectable levels of initial fluorescence (Gray et al., 2001). Heat-shock treatment led to largely equal induction of all VENUS-IAA fusion proteins across plant lines (Fig. 1F). Time-lapse quantification of fluorescence following auxin treatment led to a relative rate of degradation that was strikingly similar to that observed in yeast (Fig. 1E).
Lateral root density is quantitatively decreased by slowing IAA14 degradation
Next, we analyzed root architecture of transgenic plants expressing 14-, F-, M-, S- or I-variants from the native IAA14 promoter. Relative stability of the IAA14 variants was inversely proportional to lateral root density (Fig. 2A). Expressing additional copies of wild-type IAA14 had no effect on root development (supplementary material Fig. S2A), and expression of IAA14 was similar across variant lines (supplementary material Fig. S2B). Complete stabilization of IAA14 in the I-variant inhibited lateral root development, as in slr-1 mutants. With intermediate degradation rate constructs, slower IAA14 degradation rate correlated well with the development of fewer lateral roots (Fig. 2A). This trend became clearer 14 days post-germination (dpg), when the densities of emerged roots of M- and S-variant plants were similar to the densities of 14- and M-variant plants at 7 dpg, respectively. These findings suggest that slowing the dynamics of IAA14 degradation translates into a quantitative reduction in lateral root emergence.
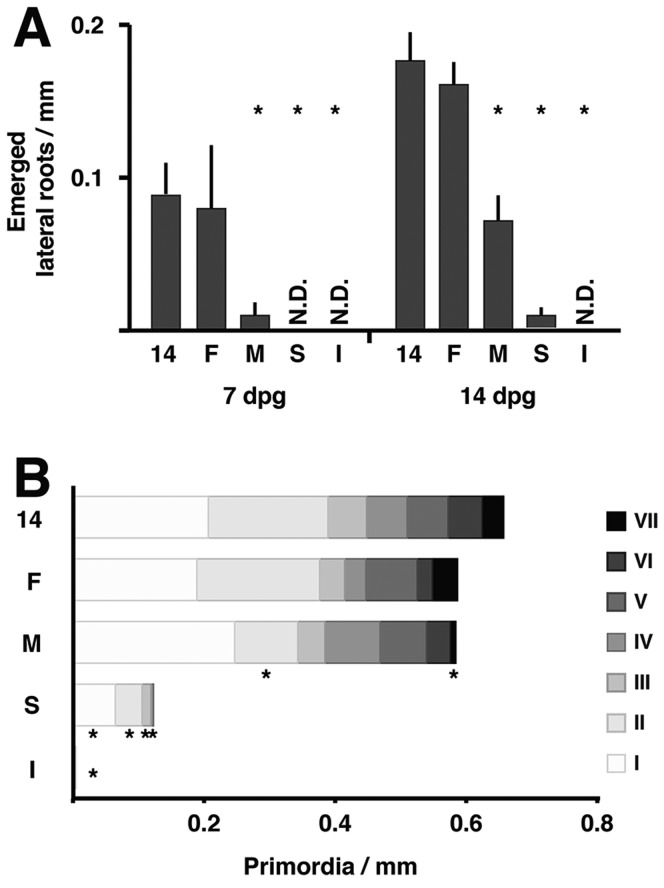
Fig. 2.
Slowing IAA14 degradation rate quantitatively decreases lateral root density. (A) 14-control (14) and 14-Fast (F) lines had similar densities of lateral roots, whereas plants expressing 14-Medium (M) and 14-Slow (S) variants had fewer lateral roots. No emerged lateral roots were detected in 14-Insensitive (I) lines. Averages calculated from three independent experiments with 6-10 seedlings per experiment. N.D., not detectable. (B) The density of lateral root primordia per developmental stage I-VII showed a similar trend to emerged roots. Medium variants had significantly fewer stage II and stage VII primordia than 14-control plants. All stages were significantly reduced in Slow and Insensitive lines. Averages calculated from 7 dpg seedlings from three independent experiments with six seedlings per experiment. Asterisks indicate significant differences (P<0.05) between variants and the 14-control line using a Student's t-test with a Hommel P-value correction for multiple testing. In B, no asterisks appear where variants had zero primordia at a given stage, but primordia absence at these stages was significant.
In addition to their lateral root defect, slr mutants lack root hairs (Fukaki et al., 2002). Root hair density was correlated with degradation rates in plants expressing IAA14 variants (supplementary material Fig. S2C). Also consistent with the slr mutant phenotype, hypocotyl elongation did not correlate with IAA14 stability (supplementary material Fig. S2D).
To further assess the effect of slowing IAA14 degradation rate, we analyzed the distribution of lateral root developmental stages (I-VII) among IAA14 variants (Fig. 2B). Across all I-variant plants analyzed, only one stage I primordia was observed, and none at any other stage. The density of primordia in S-variant lines was approximately one quarter of that observed in 14-variant plants, and all were in the earliest stages (I-IV). The distribution of primordia at stages III-VI was similar in plants expressing 14-, F- and M-variants. However, M-variant plants had significantly fewer stage II and stage VII primordia, consistent with a developmental delay leading to fewer emerged roots.
As treatment with higher auxin concentrations increases Aux/IAA degradation rates (supplementary material Fig. S1D) (Dreher et al., 2006; Havens et al., 2012), we assessed whether increased auxin levels could restore wild-type phenotypes in plants expressing the slower degrading variants. Although each variant exhibited more emerged lateral roots in response to auxin, the relative differences between variants remained similar (supplementary material Fig. S2E). Therefore, auxin availability alone cannot account for the observed differences in phenotype among the variants.
IAA14 degradation dynamics determine the dynamics of lateral root development
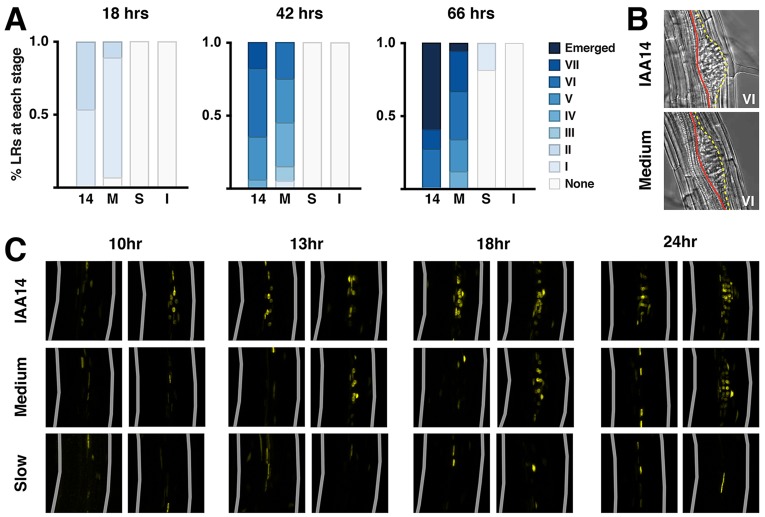
To assess developmental dynamics directly, we used gravistimulation to synchronize the location and timing of primordia initiation (De Smet et al., 2007; Lucas et al., 2008; Péret et al., 2012). When roots reorient after turning the plate 90°, auxin concentration increases and a lateral root primordium is induced on the outer side of the bend (Laskowski et al., 2008). By 18 hours post-induction (hpi), all 14-variant plants had initiated a new primordium at the expected position (Fig. 3A). In M-variant plants, initiation was delayed with several roots lacking primordia altogether at 18 hpi. This delay became even more apparent over time. In addition, many late stage primordia in M-variant plants exhibited an elongated and flattened shape, appearing to displace both the surrounding vasculature and endodermal layers (Fig. 3B). These phenotypes are reminiscent of those seen in mutants where primordia fail to emerge through the endodermis (Kumpf et al., 2013; Lucas et al., 2013; Vermeer et al., 2014). These defects suggest that early developmental delays might disrupt coordination between primordia and overlaying cell layers, and support a role for IAA14 in lateral root emergence (Kumpf et al., 2013; Swarup et al., 2008).
Fig. 3.
IAA14 degradation dynamics determine the rate of lateral root development. (A) Progression of lateral root development was delayed in plants containing slower IAA14 variants. Root development was synchronized and developmental stages were analyzed at 18, 42 and 66 hpi for 14-control (14), 14-Medium (M), 14-Slow (S) and 14-Insensitive (I) lines. Percentages were calculated from 18-20 seedlings per independent line per time-point. (B) Late stage primordia in 14-Medium plants frequently exhibited aberrant morphology. Yellow line, outer layer of lateral root primordium; red line, vascular tissue. (C) Expression of the auxin-responsive reporter DR5-Venus is rapidly induced by root bending in IAA14-control lines, but delayed in 14-Medium and 14-Slow lines. The outer bend, where the primordia initiate, is towards the right. Two individuals are shown for each time-point post-induction to illustrate the observed range of DR5 expression levels within a line. Gray lines indicate outer root boundaries.
Developmental progression was further compromised in S- and I-variant plants. No primordia were detected in S-variant plants at 18 hpi, and by 66 hpi, only a subset of roots had stage I primordia. This is consistent with delayed induction of the auxin-responsive DR5-VENUS reporter (Brunoud et al., 2012) in these lines (Fig. 3C). Although a peak of reporter expression was detected in the outer bend of 14-variant roots as early as 10 hpi, induction was delayed until 13 hpi in M-variant plants. Little to no induction of reporter expression was observed in S-variant plants as late as 24 hpi. As expected, no primordia were detected in any I-variant roots at any time (Fig. 3A). In addition, four LATERAL ORGAN BOUNDARIES DOMAIN (LBD) genes that act downstream of IAA14 signaling show decreased auxin-induced expression in slower variant lines (supplementary material Fig. S3).
An auxin-induced re-programmable timer
In this work, we used a synthetic biology approach to examine auxin signaling in order to answer a fundamental question in plant development: what controls and coordinates the pace of organogenesis? By engineering Aux/IAA variants with reduced auxin sensitivity, we discovered that timing of organ initiation was plastic and could be tuned. Our engineering of degradation rates by single point mutations may be analogous to natural evolutionary processes of selecting Aux/IAA properties to tune emergent auxin signaling modules in new contexts. Optimization of auxin sensitivity among co-expressed Aux/IAAs could produce a hierarchical order of action through ordered turnover of different Aux/IAAs (Pierre-Jerome et al., 2014). In this way, sequential degradation of Aux/IAAs could lead to a wave of gene expression, similar to the temporal gradation of gene expression observed for Dpp signaling during Drosophila wing development (O'Keefe et al., 2014). Multiple Aux/IAA proteins are co-expressed during lateral root formation (De Smet et al., 2010; Lavenus et al., 2013). By slowing the degradation rate of IAA14, the persistent protein might gain the ability to interfere with other Aux/IAA proteins (i.e. compete for binding with the TIR1-auxin complex or ARFs) or to interact with additional partners. Higher order transcriptional complexes between Aux/IAAs and ARFs (Korasick et al., 2014; Nanao et al., 2014) are likely to influence Aux/IAA degradation rates, further modulating the dynamics of downstream transcription of target genes.
MATERIALS AND METHODS
Yeast transformation and strain construction
Yeast cells (Saccharomyces cerevisiae) were grown on Yeast Peptone Dextrose (YPD) and Synthetic Complete (SC) medium supplemented with 80 mg/ml adenine and made according to standard protocols. Transformations were performed using a standard lithium acetate protocol (Gietz and Woods, 2002) into MATa W303-1A or MATα W814-29B, a gift from D. Gottschling (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). Amino acid substitutions were introduced into the IAA14-coding region in pDONR entry vectors via Gibson cloning (Gibson et al., 2009). These coding regions were then cloned into pGP4GY-ccdB destination vectors (Havens et al., 2012) using LR clonase. Resulting constructs were transformed into MATa cells, and mated with MATα cells containing pGP5G-TIR1 or AFB2 constructs using standard procedures.
Yeast degradation assays
Cell cultures were prepared for degradation assays as described previously (Havens et al., 2012). For each strain, one replicate was mock treated (95% [v/v] ethanol) and one replicate was treated with 10 µM indole-3-acetic acid. Time 0 readings were taken immediately after auxin addition. Subsequent readings were taken at 10 min intervals over 2 h. Control experiments were measured every hour.
Auxin-binding assays
Radioligand binding assays were performed as previously described (Calderón Villalobos et al., 2012). In brief, recombinant TIR1-ASK1 protein complex and N-terminal GST-tagged IAA14 variant proteins were purified to high purity. Duplicate samples containing both proteins, radiolabeled indole-3-acetic acid (IAA) and cold competitor (unlabeled IAA) were incubated for 1 h on ice, subsequently filter immobilized, and washed with binding buffer. Filters were incubated overnight and retained radiolabeled auxin was measured via scintillation counting. Nonspecific binding was determined using a 10,000-fold excess of cold IAA with respect to [3H]-IAA. Data analysis was performed using GraphPad Prism 5 software. Kd values were obtained applying One-site binding (hyperbola) model. Specific binding was calculated as the difference of average total binding and nonspecific binding. Samples for total and nonspecific binding were in duplicates. Experiments were repeated twice either as saturation or competition binding with consistent results.
Plant growth
Seeds were sown on 0.5× LS, 1.8% agar plates, stratified at 4°C for 2 days, and grown in continuous light conditions for 7 or 14 dpg. For lateral root induction assays, plants were rotated 90° after 4 dpg. Plants were cleared and imaged 18, 42 and 66 h after rotation. For auxin-treatment assays, 7 dpg seedlings were transplanted to plates containing 1 µM IAA and grown for 7 additional days.
Transgenic plant lines
DR5::VENUS-N7 seeds were provided by T. Vernoux. A 2 kb IAA14 promoter fragment (Fukaki et al., 2002) was cloned into pGREEN vectors, and IAA14 variant CDSs were cloned downstream. IAA14 variants were crossed with DR5::VENUS-N7 plants. HS::VENUS-IAA-NLS constructs were generated by fusing IAA14 CDSs to an N-terminal VENUS and C-terminal SV40 NLS repeat using Gly-Ala linkers via Gibson cloning. The resulting fragments were cloned into pGREEN vectors containing the soybean heat-shock promoter HS6871 (Gray et al., 2001). All constructs were transformed into Col-0 wild-type plants using the floral dip method (Clough and Bent, 1998).
Time-lapse fluorescence detection following heat-shock
Plates were placed on a slide warmer set at 37°C for 2 h to induce expression. Seedlings were arranged on agar blocks containing 5 μM IAA or mock treatment, and sprayed with liquid 0.5× LS containing 5 μM IAA or vehicle. A coverslip was placed over agar blocks. Plants were imaged at 0, 10, 20, 30, 40 and 60 m post-treatment.
Histology and microscopy
IAA14 variant plants were cleared and prepared for phenotyping as described previously (Malamy and Benfey, 1997). Plants were imaged using a Leica DMI 3000B microscope fitted with a Leica long working 40× HCX PL FLUORTAR objective. Images were captured using Leica LAS AF version 2.6.0 software and a Leica DFC 345FX camera. For fluorescent images, Fiji software was used to quantify fluorescence in a region of interest for each image. Non-fluorescent siblings were used to calculate and subtract background. Fluorescence was then normalized to initial values.
RNA isolation and qPCR
Roots from 7 dpg plants were excised and flash frozen at −80°C. RNA was extracted using an Illustra kit (GE Healthcare), and 1 µg of complementary DNA was prepared using iScript kit (Bio-Rad). qPCR was performed using SYBR Green Supermix (Bio-Rad) and the CFX96 Real-Time System (Bio-Rad). Expression for each gene was calculated using the formula (Etarget)−ΔCPtarget(control−sample)/(Eref)−ΔCPref(control−sample) and normalized to reference gene At1g13320 (Pfaffl, 2001).
Root hair density and hypocotyl measurements
Vertically grown 7 dpg plants were imaged on a Leica dissecting microscope (S8APO, Leica Microsystems) and camera (DFC290, Leica Microsystems) for root hairs or scanned for hypocotyl measurement. Root hair number and hypocotyl length was determined using ImageJ. Two replicates were quantified, with four to six plants per replicate for root hairs and eight to ten plants for hypocotyls.
Supplementary Material
Acknowledgements
We thank Keiko Torii, Takato Imaizumi, Christine Queitsch, David Raible and members of the Nemhauser Lab, and the Seattle Developmental Biology Group for excellent discussions and critical evaluation of this manuscript; Autumn Walker, Danny Liang, Minki Kim, Kevin Ford and Wai Pang Chan for technical assistance; Teva Vernoux for providing seeds.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the Paul G. Allen Family Foundation, the National Institutes of Health (NIH) [R01 GM107084] and the National Science Foundation (NSF) [CISE-0832773 to E.K. and IOS-0919021 to J.L.N.]. J.M.G. was supported by the Developmental Biology Predoctoral Training Grant [T32HD007183] from the National Institute of Child Health and Human Development (NICHD). A.H. and L.I.A.C.V. were supported by funds from the IPB-Leibniz Institute of Plant Biochemistry. Deposited in PMC for release after 12 months.
Author contributions
J.M.G., B.L.M., A.H., L.I.A.C.V., E.K. and J.L.N. conceived and designed experiments. J.M.G., A.H., A.L. and T.P.F. performed experiments. J.M.G. and J.L.N. wrote the paper.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.117234/-/DC1
References
- Brunoud G., Wells D. M., Oliva M., Larrieu A., Mirabet V., Burrow A. H., Beeckman T., Kepinski S., Traas J., Bennett M. J. et al. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103-106 10.1038/nature10791 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L. I. A., Lee S., De Oliveira C., Ivetac A., Brandt W., Armitage L., Sheard L. B., Tan X., Parry G., Mao H. et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477-485 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E. J. and Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43, 265-285 10.1146/annurev-genet-102108-134148 [DOI] [PubMed] [Google Scholar]
- Clough S. J. and Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- De Smet I., Tetsumura T., De Rybel B., Frey N. F. d., Laplaze L., Casimiro I., Swarup R., Naudts M., Vanneste S., Audenaert D. et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681-690 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- De Smet I., Lau S., Voss U., Vanneste S., Benjamins R., Rademacher E. H., Schlereth A., De Rybel B., Vassileva V., Grunewald W. et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 107, 2705-2710 10.1073/pnas.0915001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S. and Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441-445 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dreher K. A., Brown J., Saw R. E. and Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699-714 10.1105/tpc.105.039172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H. and Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153-168 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. and Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343-345 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gietz R. D. and Woods R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87-96 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- Gray W. M., Kepinski S., Rouse D., Leyser O. and Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414, 271-276 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Havens K. A., Guseman J. M., Jang S. S., Pierre-Jerome E., Bolten N., Klavins E. and Nemhauser J. L. (2012). A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160, 135-142 10.1104/pp.112.202184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S. and Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446-451 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Korasick D. A., Westfall C. S., Lee S. G., Nanao M. H., Dumas R., Hagen G., Guilfoyle T. J., Jez J. M. and Strader L. C. (2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111, 5427-5432 10.1073/pnas.1400074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R. P., Shi C.-L., Larrieu A., Stø I. M., Butenko M. A., Péret B., Riiser E. S., Bennett M. J. and Aalen R. B. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 110, 5235-5240 10.1073/pnas.1210835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M., Grieneisen V. A., Hofhuis H., ten Hove C. A., Hogeweg P., Marée A. F. M. and Scheres B. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6, e307 10.1371/journal.pbio.0060307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J., Goh T., Roberts I., Guyomarc'h S., Lucas M., De Smet I., Fukaki H., Beeckman T., Bennett M. and Laplaze L. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450-458 10.1016/j.tplants.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Lokerse A. S. and Weijers D. (2009). Auxin enters the matrix--assembly of response machineries for specific outputs. Curr. Opin. Plant Biol. 12, 520-526 10.1016/j.pbi.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Lucas M., Godin C., Jay-Allemand C. and Laplaze L. (2008). Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J. Exp. Bot. 59, 55-66 10.1093/jxb/erm171 [DOI] [PubMed] [Google Scholar]
- Lucas M., Kenobi K., von Wangenheim D., Voß U., Swarup K., De Smet I., Van Damme D., Lawrence T., Péret B., Moscardi E. et al. (2013). Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc. Natl. Acad. Sci. USA 110, 5229-5234 10.1073/pnas.1210807110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J. E. and Benfey P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33-44. [DOI] [PubMed] [Google Scholar]
- Muto H., Watahiki M. K., Nakamoto D., Kinjo M. and Yamamoto K. T. (2007). Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol. 144, 187-196 10.1104/pp.107.096628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao M. H., Vinos-Poyo T., Brunoud G., Thévenon E., Mazzoleni M., Mast D., Lainé S., Wang S., Hagen G., Li H. et al. (2014). Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 5, 3617 10.1038/ncomms4617 [DOI] [PubMed] [Google Scholar]
- Ngo Q. A., Vogler H., Lituiev D. S., Nestorova A. and Grossniklaus U. (2014). A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev. Cell 29, 491-500 10.1016/j.devcel.2014.04.008 [DOI] [PubMed] [Google Scholar]
- O'Keefe D. D., Thomas S., Edgar B. A. and Buttitta L. (2014). Temporal regulation of Dpp signaling output in the Drosophila wing. Dev. Dyn. 243, 818-832 10.1002/dvdy.24122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., Li G., Zhao J., Band L. R., Voß U., Postaire O., Luu D.-T., Da Ines O., Casimiro I., Lucas M. et al. (2012). Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14, 991-998 10.1038/ncb2573 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E., Moss B. L. and Nemhauser J. L. (2013). Tuning the auxin transcriptional response. J. Exp. Bot. 64, 2557-2563 10.1093/jxb/ert100 [DOI] [PubMed] [Google Scholar]
- Pierre-Jerome E., Jang S. S., Havens K. A., Nemhauser J. L. and Klavins E. (2014). Recapitulation of the forward nuclear auxin response pathway in yeast. Proc. Natl. Acad. Sci. USA 111, 9407-9412 10.1073/pnas.1324147111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis J. E. and Lahav G. (2013). Encoding and decoding cellular information through signaling dynamics. Cell 152, 945-956 10.1016/j.cell.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. A., Zenser N., Leyser O. and Callis J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349-2360 10.1105/tpc.13.10.2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., Benková E., Swarup R., Casimiro I., Péret B., Yang Y., Parry G., Nielsen E., De Smet I., Vanneste S. et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946-954 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderón Villalobos L. I. A., Sharon M., Zheng C., Robinson C. V., Estelle M. and Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Vanneste S., De Rybel B., Beemster G. T. S., Ljung K., De Smet I., Van Isterdael G., Naudts M., Iida R., Gruissem W., Tasaka M. et al. (2005). Cell cycle progression in the Pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035-3050 10.1105/tpc.105.035493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer J. E. M., von Wangenheim D., Barberon M., Lee Y., Stelzer E. H. K., Maizel A. and Geldner N. (2014). A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343, 178-183 10.1126/science.1245871 [DOI] [PubMed] [Google Scholar]
- Weijers D., Benkova E., Jäger K. E., Schlereth A., Hamann T., Kientz M., Wilmoth J. C., Reed J. W. and Jürgens G. (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24, 1874-1885 10.1038/sj.emboj.7600659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.