Abstract
The transcriptional profiles of cardiac cells derived from murine embryos and from mouse embryonic stem cells (mESCs) have primarily been studied within a cell population. However, the characterization of gene expression in these cells at a single-cell level might demonstrate unique variations that cannot be appreciated within a cell pool. In this study, we aimed to establish a single-cell quantitative PCR platform and perform side-by-side comparison between cardiac progenitor cells (CPCs) and cardiomyocytes (CMs) derived from mESCs and mouse embryos. We first generated a reference map for cardiovascular single cells through quantifying lineage-defining genes for CPCs, CMs, smooth muscle cells (SMCs), endothelial cells (EDCs), fibroblasts and mESCs. This panel was then applied against single embryonic day 10.5 heart cells to demonstrate its ability to identify each endocardial cell and chamber-specific CM. In addition, we compared the gene expression profile of embryo- and mESC-derived CPCs and CMs at different developmental stages and showed that mESC-derived CMs are phenotypically similar to embryo-derived CMs up to the neonatal stage. Furthermore, we showed that single-cell expression assays coupled with time-lapse microscopy can resolve the identity and the lineage relationships between progenies of single cultured CPCs. With this approach, we found that mESC-derived Nkx2-5+ CPCs preferentially become SMCs or CMs, whereas single embryo-derived Nkx2-5+ CPCs represent two phenotypically distinct subpopulations that can become either EDCs or CMs. These results demonstrate that multiplex gene expression analysis in single cells is a powerful tool for examining the unique behaviors of individual embryo- or mESC-derived cardiac cells.
Keywords: Cardiac progenitor cells, Cardiogenesis, Embryonic stem cell, Nkx2-5, Differentiation
Highlighted article: Multiplex gene expression analysis on single cardiac lineage cells provides a high-resolution view of cardiac fate choice in the mouse embryo and during stem cell differentiation.
INTRODUCTION
The heart is the first organ to form during mouse embryonic development (DeRuiter et al., 1992; Icardo, 1996), a process involving multiple cell fate decisions. One such important decision is the differentiation of cardiac progenitor cells (CPCs) into multiple types of functional cells, such as cardiomyocytes (CMs), endothelial cells (EDCs) and smooth muscle cells (SMCs) (Harvey, 2002; Laugwitz et al., 2008; Martin-Puig et al., 2008; Sturzu and Wu, 2011). Our group and others have described the isolation and characterization of multipotent CPCs derived from mouse embryonic stem cells (mESCs) and embryos (Cai et al., 2003; Kattman et al., 2006; Moretti et al., 2006; Wu et al., 2006; Yang et al., 2008). We have previously generated a transgenic mouse line that expresses eGFP reporter driven by a 2.1 kb cardiac-specific enhancer of Nkx2-5, an important transcription factor in embryonic heart development (Lien et al., 1999). The endogenous level of Nkx2-5 expression is low in CPCs and increases in CMs; however, the eGFP expression driven by Nkx2-5 cardiac enhancer (Nkx2-5–eGFP) appears only in CPCs and early immature CMs (Lien et al., 1999; Wu et al., 2006). Consequently, the Nkx2-5–eGFP+ cells represent CPCs only in the early embryonic stage, and those progenitor cells were shown to differentiate into multiple types of cells when cultured in vitro (Moretti et al., 2006; Wu et al., 2006). However, no previous study has addressed heterogeneity and lineage relationships among progenitor cells during differentiation at a single-cell level.
Single-cell gene expression analysis offers a way to address these variations within a cell population (Guo et al., 2010; Tay et al., 2010). Microfluidic-enabled multiplex PCR arrays (Fluidigm) can perform quantitative real-time PCR (qPCR) reactions for up to 96 genes in 96 cells simultaneously (Citri et al., 2011; Narsinh et al., 2011; Sanchez-Freire et al., 2012). These arrays have been used successfully to dissect cellular composition and transcriptional heterogeneity in human colon tumors (Dalerba et al., 2011). Additionally, Buganim et al. profiled single cells at various stages during cellular reprogramming and found that this process consisted of an early stochastic and a late hierarchic phase (Buganim et al., 2012). These findings support the use of single-cell PCR arrays as a powerful system to explore mechanisms regulating cellular heterogeneity and cell lineage determination. Interestingly, Guo et al. employed this technology to generate an expression signature of hematopoietic stem cell differentiation in mice and found significant variation within the apparently uniform myeloid and lymphoid progenitor cell population (Guo et al., 2013). However, no study thus far has employed this approach to closely examine the heterogeneity of CPCs at a single-cell level.
In this study, we used the Fluidigm expression system to profile cardiac lineage-associated genes at single-cell level in six standard cell types: CPCs, CMs, fibroblasts (FBs), SMCs, EDCs and undifferentiated mESCs (Boyer et al., 2005; Souders et al., 2009; Sturzu and Wu, 2011). Using this as a reference map, we characterized single mESC-, mouse embryo- and adult heart-derived cardiac cells. Furthermore, we evaluated the lineage choice made by a single CPC during in vitro differentiation by assessing the final identity of each of its progenies under identical culturing condition. Our results demonstrate the utility of single-cell gene expression profiling to study cell fate, maturity and lineage choices in the developing heart.
RESULTS
Transcriptional analysis of single cardiovascular cells
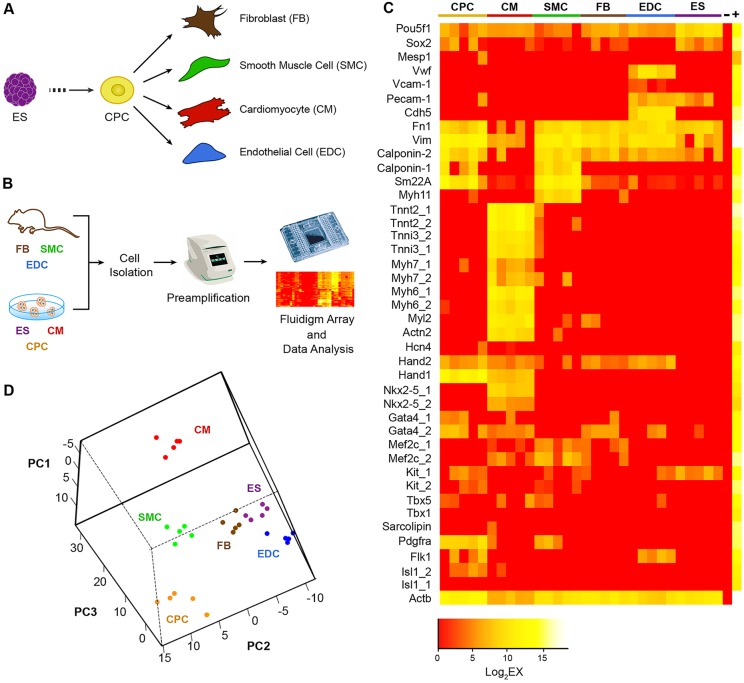
The cardiovascular lineage consists of multiple cell types during embryonic development. During the early stage, CPCs predominate and give rise to CMs, SMCs and EDCs upon differentiation (Fig. 1A). In an adult heart, cardiac FBs are present and account for a significant proportion of cell nuclei (Ieda et al., 2009). These cardiovascular cell types can also be generated using in vitro differentiation of mESC. Employing a previously described Nkx2-5 cardiac enhancer-driven eGFP (Nkx2-5–eGFP) mESC (Wu et al., 2006), we differentiated these cells towards the cardiovascular lineages and purified the eGFP+ CPCs or CMs by fluorescence-activated cell sorting (FACS). We first confirmed that the differences in expression of pluripotent and cardiomyocyte genes can be reliably detected in single ESCs and CMs by single-cell real-time qPCR analysis (supplementary material Fig. S1; Table S1). Next, we established a reference set of genes for the determination of the main cardiovascular cell types as well as undifferentiated mESC (supplementary material Table S2). This gene set was then applied to single cells from the five main cardiovascular cell types (CPCs, CMs, SMCs, EDCs and FBs) and undifferentiated mESCs, using a microfluidic-enabled multiplex PCR platform from Fluidigm (Biomark HD) (Fig. 1B). We show that a high degree of plate-to-plate consistency in gene expression can be achieved based on the expression of the housekeeping gene β-actin (supplementary material Fig. S2). Furthermore, we found a distinct expression signature from this reference gene panel for each of our cell types of interest (Fig. 1C). As expected, the expression levels of Pou5f1 (also known as Oct4) and Sox2 were high in undifferentiated mESCs, whereas the expression of sarcomeric or endothelial cell genes was absent. In designing mESC-specific probes, the lack of introns in Pou5f1 and Sox2 prevents the creation of intron-spanning primers that would exclude genomic DNA during cDNA amplification. We therefore detected a low background signal in non-ESCs. To further validate the expression of Pou5f1 and Sox2 in mESCs but not in CMs, we performed additional amplification reactions using different primer sets against these genes (supplementary material Fig. S3; Table S3). Similarly, the expression of EDC lineage genes, such as Vwf, Vcam1, Pecam1 and Cdh5, were expectedly high in EDCs, as was that of Calponin 1 (Cnn1) and Myh11 in SMCs. Interestingly, fibroblast and SMCs shared a number of expressed genes, supporting the phenotypic similarity between these cell types. This is not entirely surprising, as it is well known that FBs and coronary SMCs in the heart are both descendants from the developing epicardium (Cai et al., 2008; Zhou et al., 2008).
Fig. 1.
Transcriptional analysis of cardiac lineage cells at a single-cell level. (A) Mouse ESCs in vitro differentiate to become CPCs that are capable of giving rise to CMs, SMCs, EDCs and FBs. (B) Process diagram of cardiovascular lineage single-cell profiling using the microfluidic-enabled multiplex PCR platform (Fluidigm). (C) Reference gene expression panel for standard cardiac cells. Each column represents one cell, and each row represents one gene assay. Note that for some genes there are two probes used (e.g. Tnnt2_1 and Tnnt2_2) against the same target cDNA to safeguard against rare cases of amplification failure. Yellow to red color scale indicates high to low/no expression for a particular marker. ‘−’ represents a negative control containing no cDNA, whereas ‘+’ represents mouse universal cDNA. (D) PCA on the reference single-cell gene expression data. Notably, the same type of cell colorized with the same color clustered together. Data represent the results from two independent experiments.
The expression of transcripts for sarcomeric proteins and cardiac transcription factors was expectedly high in CMs, whereas genes that represent CPC (e.g. Pdgfra, Flk1/Kdr – Mouse Genome Informatics, cKit, and cardiac transcription factors, such as Isl1, Nkx2-5, Hand1, Hand2 and Gata4) were generally present at modest to low levels. To determine the consistency in gene expression for each single cell within the same cell type, we performed a principal component analysis (PCA) (Fig. 1D) and a Pearson correlation coefficient analysis (supplementary material Table S4) on the single-cell expression data and found that cells within each cell type show highly similar expression profiles across all genes examined. These results indicate that our cardiovascular gene expression panel can uniquely identify each single cardiovascular cell, especially CMs and EDCs, which exhibit a more distinct gene signature.
Identification of single embryonic day 10.5 heart cells
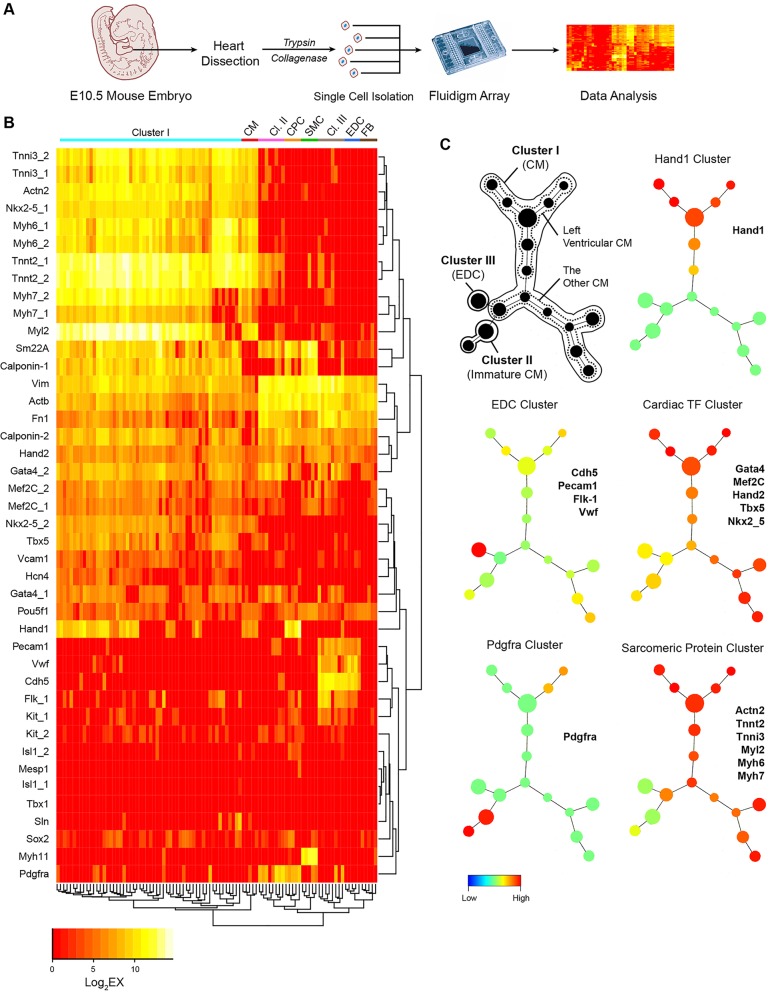
To demonstrate the utility of the single-cell expression panel, we employed it to determine the identity of unselected single embryonic day 10.5 (E10.5) heart cells (Fig. 2A). When these single E10.5 heart cells were profiled along with standard cell types, we found by unsupervised hierarchical clustering analysis that they organized into three distinct clusters (I, II, III) (Fig. 2B). Cluster I cells resemble standard CMs, which highly express sarcomeric protein genes (e.g. Myh6, Tnnt2) and many cardiac transcription factors (e.g. Mef2c, Gata4). Cluster II cells appear to represent immature CMs, which grouped between CMs and CPCs and expressed both Pdgfra, a marker for CPCs, as well as Tnnt2, a sarcomeric protein marker. Cluster III cells expressed EDCs genes (e.g. Cdh5, PECAM/CD31) at a high level and are most likely EDCs (Fig. 2B). The identity of these E10.5 embryonic mouse heart cells as CMs, immature CMs and EDCs is further supported by PCA of these cells against standard cell types (supplementary material Fig. S4A) and the Pearson correlation coefficient of the cluster I, II and III cells against their standard cells (supplementary material Fig. S4B).
Fig. 2.
Characterization of the identities of unselected single E10.5 heart cells. (A) Schematic showing isolation and identification of unselected single embryonic heart cells. (B) Unsupervised hierarchical clustering analysis of the unselected single embryonic heart cells against the profiles of reference cardiac cells. Note that the unselected E10.5 heart cells self-organized into three distinct clusters (Cl. I, II, III). (C) SPADE analysis of E10.5 heart cells. Single-cell data from unsupervised hierarchical clustering were plotted as cell-lineage tree based on the similarity of their gene expression. Cells with nearly identical gene expression are grouped within one node and the number of cells within the node is represented by the size of each node. Nodes that are most similar in gene expression are displayed closest to each other in the tree diagram. The SPADE tree in black displays the cell identity for each node in the tree. The colored SPADE trees display the expression levels of the representative genes listed next to each tree. The color legend below indicates the level of gene expression, in which red represents the highest expression level, whereas blue represents little to no expression. Data represent the results from two independent experiments.
Given the ability of our single-cell expression profile to identify each cell, we wished to determine whether cells from the E10.5 heart exhibit additional variation (e.g. higher order dimensions) that could be revealed when these cells are plotted in a lineage tree diagram based on the similarity of their gene expression. To accomplish this, we used the spanning-tree progression of density-normalized events (SPADE) program (Linderman et al., 2012; Qiu et al., 2011) to convert the multi-dimensional expression data into a lineage tree of interconnected cell populations (Fig. 2C). Cells with nearly identical gene expression are grouped within one node and the number of cells within the node is represented by the size of each node. The black-colored SPADE tree indicates the most likely cell identity of each node or collection of nodes. The multi-colored SPADE trees show the expression levels of cell type-specific genes listed next to each tree. For example, all EDCs are found within one node that exhibits the highest level of endothelial cell gene expression. Furthermore, differentiated CMs (e.g. cluster I) exhibit a high level of sarcomeric protein gene expression. When we further analyzed all CMs based on the expression of Hand1, which is known to be expressed preferentially in first heart field and in the left ventricle (LV) (McFadden et al., 2005; Thomas et al., 1998), we found a distinct cluster of nodes that are Hand1+, most likely representing CMs in the LV, as well as Hand1– CMs that are most likely CMs in the right ventricle (RV) and atria (Fig. 2C). We performed additional single-cell Fluidigm studies to assess the specificity of Hand1 for LV CMs compared with other genes, such as Cited1 and connexin 40 (Gja5 – Mouse Genome Informatics), and confirmed that Hand1 expression is the most specific marker for LV CMs (supplementary material Fig. S5; Table S5). Together, these results demonstrate the power of single-cell multiplexed gene expression profiling to uncover previously unsuspected subpopulations of cells at a defined stage of cardiac development.
Transcriptional similarity between immature cardiac cells derived from mESCs and mouse embryos
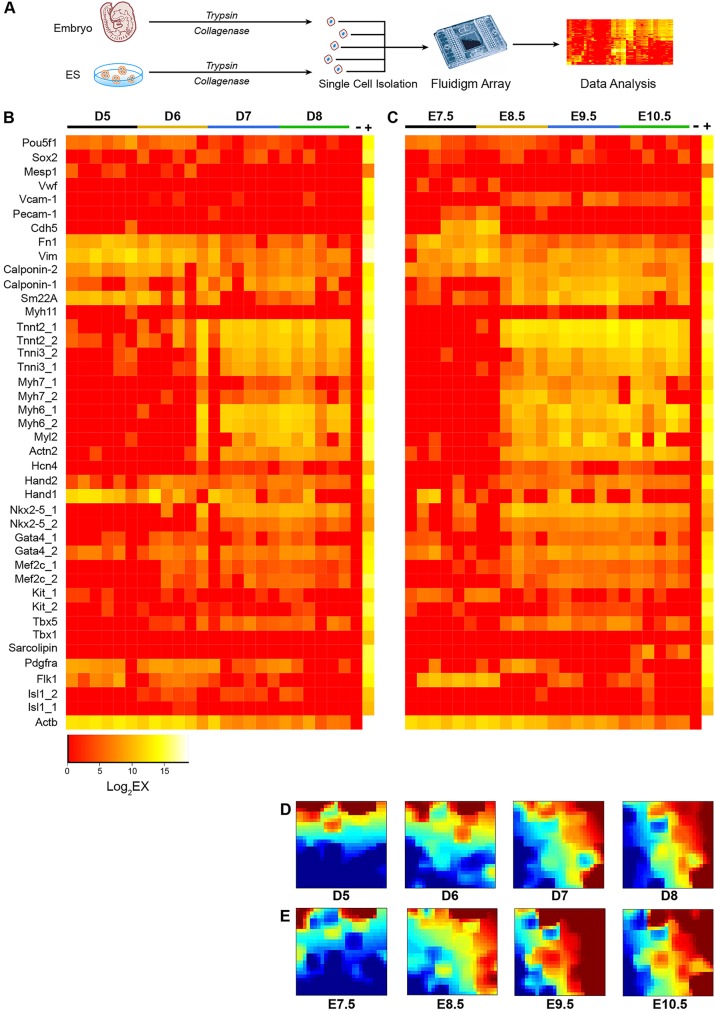
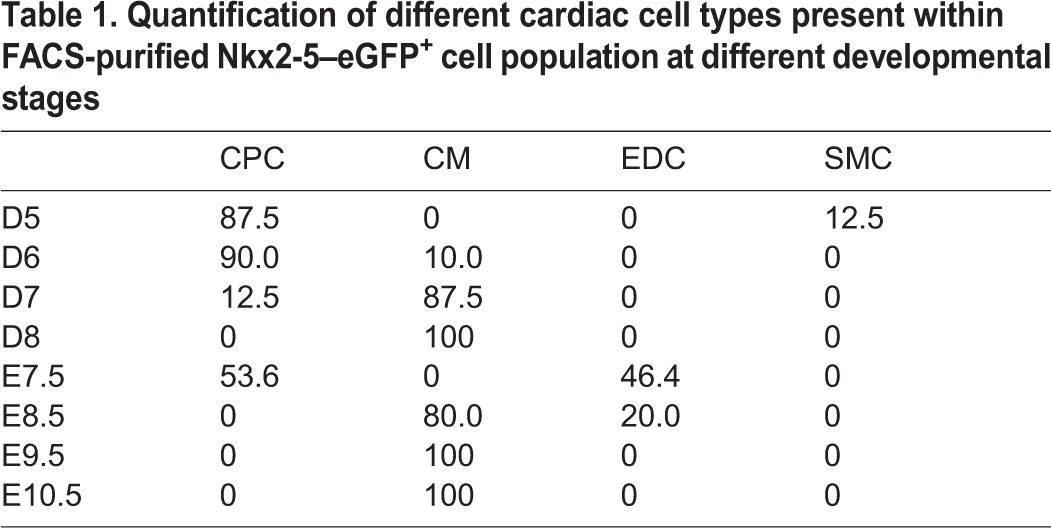
Whereas in vitro differentiation of mESCs provides a useful platform to generate cardiac lineage cells for in vitro studies, these cardiac cells have not been directly compared with mouse embryo-derived cardiac cells in a stage-by-stage fashion at the transcriptional level. Here, we isolated single eGFP+ cells from in vitro differentiation of Nkx2-5–eGFP mESCs by FACS at days 5, 6, 7 and 8 of differentiation, along with eGFP+ cells from Nkx2-5–eGFP transgenic mouse embryos at embryonic days 7.5, 8.5, 9.5 and 10.5 (supplementary material Fig. S6), and performed expression profiling on the Fluidigm arrays (Fig. 3A). We found highly similar expression profiles between mESC-derived and embryo-derived cardiac cells at these early stages of development (Fig. 3B,C). Interestingly, whereas single eGFP+ cell derived from mESC at day 5 of differentiation consisted mainly of CPCs, these cells quickly transitioned into CMs (87.6%) by day 7 of differentiation (Table 1). This pattern of gene expression change appears to characterize embryo-derived eGFP+ cells at days 7.5 and 8.5 of development as well, with the exception that a fraction of the early embryo-derived eGFP+ cell population expressed EDC genes, whereas a few of the early mESC-derived eGFP+ cells expressed SMC genes (Table 1). To determine whether eGFP+ cells from embryonic day 7.5 heart contain EDCs, we performed additional anti-CD31 flow cytometry analysis and confirmed that a proportion of these eGFP+ cells are indeed EDCs (supplementary material Fig. S7).
Fig. 3.
The transcriptional profiles of cardiac cells derived from mESCs and mouse embryos are highly similar. (A) Schematic diagram of mESC- and embryo-derived single cardiac cell profiling. (B) Expression profiles for single eGFP+ cells from in vitro differentiation of Nkx2-5–eGFP mESCs at days 5, 6, 7 and 8 of differentiation. (C) Expression profiles for single eGFP+ cells from Nkx2-5–eGFP transgenic mouse embryos at embryonic days 7.5, 8.5, 9.5 and 10.5. (D,E) GEDI plots depicting the gene expression signatures of single cells at different developmental stages. Each tile of the mosaic represents an SOM cluster of expressed genes. The SOM organizes genes by grouping together genes with the most similar expression levels. The color of each tile indicates gene expression levels, in which brown and red represent high levels of expression, and green and blue represent low to no expression. Data represent the results from two independent experiments.
Table 1.
Quantification of different cardiac cell types present within FACS-purified Nkx2-5–eGFP+ cell population at different developmental stages
To provide a global unbiased assessment of the similarity or difference in gene expression profile between mESC- and embryo-derived eGFP+ cells, we analyzed our data from the expression heat maps using the gene expression dynamics inspector (GEDI) program that plots signature genes from an expression dataset that provides the best discrimination between different biological samples (Eichler et al., 2003). This analysis confirmed that, whereas mESC- and embryo-derived eGFP+ cells show some degree of variability early on, they become highly similar beyond D7 of mESC in vitro differentiation or day 8.5 of embryonic development (Fig. 3D,E).
Comparison of gene expression profile from single mESC-derived CMs after extended culturing with those from an adult mouse heart
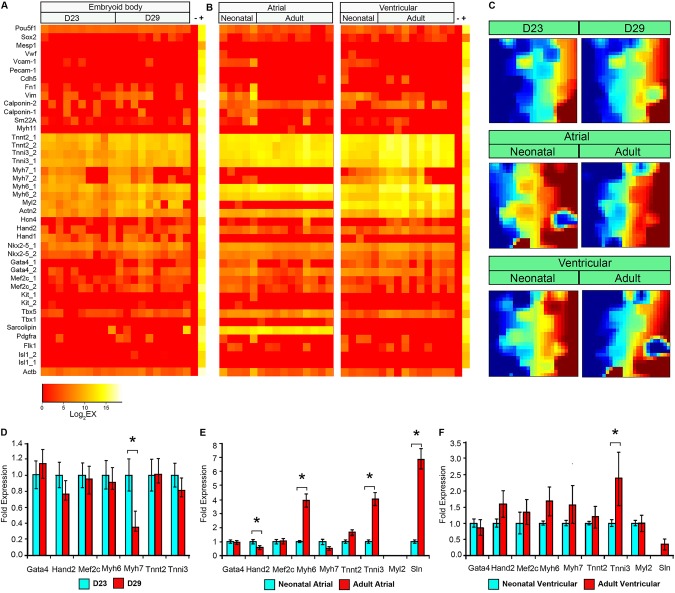
To address whether a single mESC-derived CM can achieve a transcriptional profile similar to a single CM derived from a neonatal or an adult mouse heart, we profiled GFP+ CMs from day 23 and 29 of α-MHC-CRE/ROSA26-mTmG mESC in vitro differentiation, as well as dispersed single atrial and ventricular CMs from the hearts of neonatal and adult mice of the same genotype (Fig. 4; supplementary material Fig. S8; supplementary material Movies 1 and 2). The gene-expression heat maps showed that the mESC-derived CMs from both day 23 and 29 are highly similar to each other (Fig. 4A). Interestingly, the expression of genes in single CMs derived from neonatal and adult mouse hearts differs according to their chamber of origin. For example, atrial CMs expressed a high level of sarcolipin (Sln) and no MLC2v (Myl2), whereas ventricular CMs expressed a high level of Myl2 and no Sln. Furthermore, atrial CMs downregulated their expression of cardiac β-myosin heavy chain (Myh7) from neonatal to adult stages, whereas ventricular CMs maintained some expression of Myh7 even in adult CMs. Between adult mouse-derived CMs and mESC-derived CMs, it appears that the expression of sarcomeric protein genes is generally higher in adult mouse-derived CMs (Fig. 4A,B). These findings were further validated bioinformatically using GEDI analysis (Fig. 4C). To confirm these single-cell expression data, we isolated neonatal and adult mouse CMs as cell populations and quantified the expression of sarcomeric protein genes and cardiac transcription factors using real-time qPCR analysis (Fig. 4D-F; supplementary material Table S6). These results show general consistency with our single-cell data, in that the expression of most sarcomeric protein genes is modestly to significantly increased in both atrial and ventricular CMs from neonatal to adult heart, whereas the expression of cardiac transcription factors are relatively unchanged. These results are also distinct from the mESC-derived single-cell and pooled qPCR data, in which the sarcomeric protein gene expression is relatively unchanged even with extended culturing (Fig. 4A,D).
Fig. 4.
Comparison of gene expression profiles of single mESC-derived CMs after extended culture and those from adult mouse heart. (A) Expression profiles of single eGFP+ cells from in vitro differentiation of α-MHC-CRE/ROSA26-mTmG mESCs at days 23 and 29 of differentiation. (B) Expression profiles of single CMs from neonatal and adult mouse atrial and ventricular chambers. (C) GEDI plots for gene expression signatures of single cells at various stages. Note the difference between adult atrial and ventricular CMs and CMs generated from mESC differentiation. Some differences can also be observed between neonatal and adult CMs in the atria and ventricles. (D) qPCR analysis of sarcomeric protein genes and cardiac transcription factors from pooled eGFP+ cells at days 23 and 29 of in vitro α-MHC-CRE/ROSA26-mTmG mESC differentiation. (E) Same as in D, except that pooled neonatal and adult CMs were studied. The expression of Myl2 in these atrial CMs is relative to neonatal ventricular CMs. (F) Same as in D, except that pooled neonatal and adult ventricular CMs were studied. The expression of Sln in these ventricular CMs is relative to neonatal atrial CMs. Error bars represent s.e.m. *P<0.01. Data represent two independent experiments performed in triplicates.
Characterization of lineage decision by single CPCs during in vitro differentiation
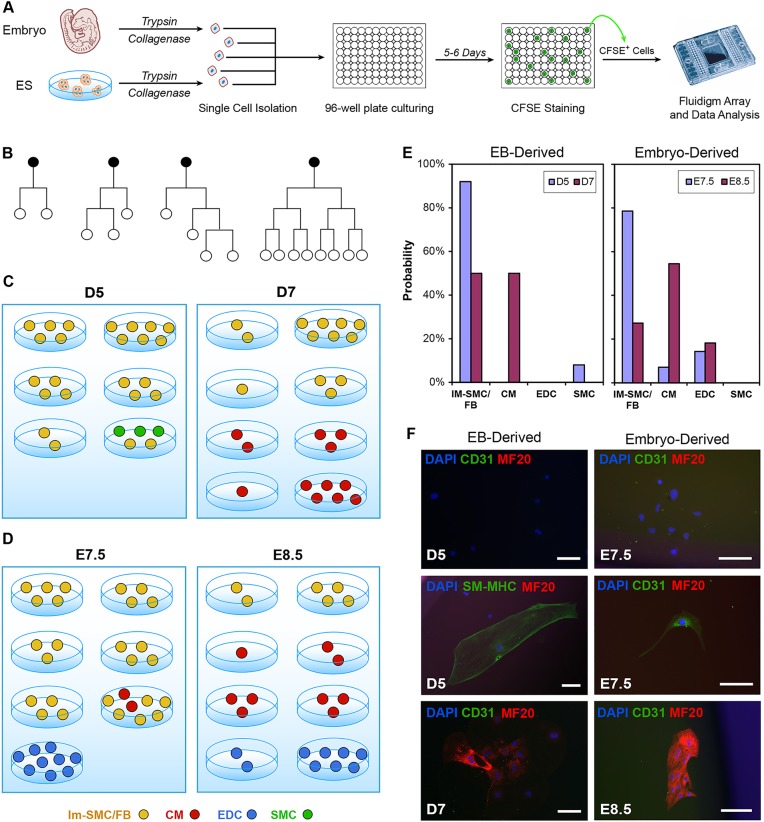
We have previously described the ability of single eGFP+ CPCs derived from Nkx2-5–eGFP mESCs to undergo bi-potential differentiation into CMs and SMCs (Wu et al., 2006). However, we were unable to determine quantitatively the frequency of CPC differentiation into either CMs or SMCs, as there was no tool available to assess exact identity of each individual progeny. By combining time-lapse video microscopy with single-cell multiplex PCR array, we are now able to observe each cell division event that takes place during the 5 days of CPC in vitro differentiation. Furthermore, we can assess the identity of each progeny as well using the Fluidigm array (Fig. 5A). Our representative video microscopy data showed that single CPCs undergo cell division with varying rates (Fig. 5B; supplementary material Movie 3). By carboxyfluorescein succinimidyl ester (CFSE) staining to re-capture live progenies (supplementary material Fig. S9), we found that, under the basal differentiation media condition, mESC-derived day 5 eGFP+ CPCs become mostly immature SMCs/FBs (Im-SMCs/FBs), with a rare number of cells becoming mature SMCs after 5 days of culturing (Fig. 5C). Only by isolating day 7 eGFP+ cells did we observe the ability of a single cell to display a firm CM phenotype after culturing, a finding that suggests that some of these eGFP+ cells have already committed to immature CM fate. Interestingly, when we performed a similar assay using Nkx2-5–eGFP mouse embryo-derived cells at E7.5 and E8.5 stages of development (equivalent to mESC days 5 and 7), we observed that these cells become either differentiated CMs or endothelial/endocardial cells (EDCs) (Fig. 5D), a cell type that was not observed in the mESC study.
Fig. 5.
Characterization of lineage decisions by single CPCs during in vitro differentiation. (A) The lineage decision for each single CPC during in vitro differentiation was determined through profiling the gene expression in each individual progeny of that CPC. (B) Representative lineage trees of single CPCs (black dots) based on time-lapse video microscopy data. Note the varying behaviors of each CPC. (C) Identity of lineage progenies from single eGFP+ CPCs that have been isolated from day 5 (D5) and 7 (D7) of in vitro mESC differentiation. (D) Same as in C, except that Nkx2-5–eGFP mouse embryo-derived eGFP+ cells at E7.5 and E8.5 of development were studied. (E) Summary of the frequency that each single Nkx2-5–eGFP+ cell became a CM, SMC/FB or EDC during in vitro differentiation. (F) Immunofluorescence staining of CPC progenies for specific cell-lineage markers. Scale bars: 100 µm. Data represent the results from two independent experiments.
Statistically, we found that almost all (92%) of the day 5 eGFP+ cells from mESC committed to Im-SMC/FB cell fate; however, in day 7 eGFP+ cells, this percentage drops to 50%, as the rest of the cells adopt a CM fate (Fig. 5E). We did not observe any EDC differentiation from mESC-derived Nkx2-5-eGFP+ cells under our assay conditions. For mouse embryo-derived eGFP+ cells, we found a significant number of immature SMCs/FBs from differentiation of E7.5 eGFP+ cells; however, this quickly resolved into three populations – CMs, immature SMCs and EDCs – when E8.5 eGFP+ cells were isolated and cultured. To test whether the cell culture medium harbors an instructive role in cell lineage decision, we cultured both mESC- and embryo-derived progenitor cells in endothelial cell culture medium (EDC medium). We found that, for embryo-derived progenitor cells, a significant number of the surviving progeny represent EDCs, whereas none of the surviving progeny from mESC-derived progenitor cells represent EDCs (supplementary material Fig. S10). Although it might seem that the EDC medium played an instructive role in directing embryo-derived progenitor cells to differentiate into EDCs, we found that CMs exhibit differential survival in EDC and SMC medium (data not shown). Hence, it is likely that the increased frequency of EDC in the progeny is due to selective survival of committed EDCs in the eGFP+ cell population. To confirm the specificity of the Fluidigm results for different cell lineages, we assessed the expression of CM, SMC and EDC genes at the protein level by immunofluorescence staining against lineage-specific markers, such as MF20, SM-MHC and CD31, respectively. We show that CM, SMC and EDC markers are expressed robustly in the differentiated single-cell progenies (Fig. 5F).
DISCUSSION
The comparison of cardiac cells derived from mESCs in vitro and murine heart in vivo on a population basis might overlook important behavioral differences at the single-cell level (Reiser et al., 1994; van den Heuvel et al., 2014; Xu et al., 2009; Yang et al., 2014). To address this issue, we employed a microfluid-enabled multiplex quantitative single-cell PCR assay to generate a reference gene expression panel to uniquely identify five main types of cardiac lineage cells (e.g. cardiac progenitor cells, cardiomyocytes, smooth muscle cells, endothelial cells and fibroblasts) based on their specific gene expression pattern. This reference panel was applied to single cells from unselected E10.5 embryonic mouse hearts to demonstrate its ability to identify each cell as CM, EDC or SMC/FB. In addition, we compared the transcriptional profiles for single cells derived from in vitro differentiated mESCs and murine hearts at different developmental stages. We found that mESC-derived CPCs and CMs are transcriptionally similar to embryo-derived CPCs and CMs up to the neonatal stage of murine development. Furthermore, we compared the lineage choices for single CPCs derived from mESCs and mouse embryos, and found a differential propensity of mESC-derived CPCs for becoming SMCs or CMs, and of single embryo-derived CPCs for becoming EDCs or CMs. Taken together, our single-cell transcriptional profiling revealed similarities between mESCs and in vivo-derived CMs up to the neonatal stage of development and showed distinct phenotypes in single isolated CPCs from mESCs and embryonic heart. These findings highlight the utility of a single-cell gene expression profiling approach combined with bioinformatics analysis to observe CPC behavior at greater resolution, and might enable us to identify new markers and pathways that regulate CPC differentiation.
Although our single-cell analysis has revealed unique properties between mESC- and embryo-derived cardiac cells, an important caveat regarding this assay should be considered. The sensitivity of this assay for identifying the cell type of interest is highly dependent on the choice of probes (i.e. lineage-specific primer pairs) and the starting cell population. For example, we set out to develop a single-cell gene panel that can identify the five main cell types from different stages of the developing heart (e.g. CPCs, CMs, SMCs, EDCs and FBs). Therefore, we were unable to detect any epicardial cells or neural crest cells because we did not choose to incorporate markers that are specific for these cell types. Furthermore, as epicardial and neural crest cells represent a relatively rare cell population in the E10.5 heart, we would not have been able to discover a distinct subpopulation of these rare cells (<1-2% of total heart cells), as we only profiled ∼72 E10.5 cells. Similarly, other cell types in the heart (e.g. blood cells) or contaminants (e.g. hepatocytes or neurons) will most likely appear indistinguishable from one another, due to their generally absent expression of cardiovascular lineage genes.
Use of single-cell transcriptional profiling to explore phenotypically similar subgroups within a cell population
The panel of lineage-specific markers that we established for this study has enabled the determination of the identity of individual cardiovascular cells within a cell population. We applied this marker panel against five known cardiovascular cell types and found in a PCA that each standard cell type clustered tightly together and away from other standard cell types (Fig. 1). These data support the robustness of our single-cell profiling strategy to reliably determine the identity of each single cardiovascular cell. When we applied this reference gene panel to an unselected population of single cells derived from E10.5 heart, we found three distinct clusters of cell types that represent CMs, immature CMs and ECs (Fig. 2B). Interestingly, we show that, within the CM population (cluster I and II), there are subpopulations that can be identified in SPADE analysis based on their expression of Hand1 (Fig. 2C). Given the relatively restricted expression of Hand1 in the developing LV (McFadden et al., 2005; Thomas et al., 1998), we believe this Hand1-expressing subpopulation most likely represents single CMs derived from the LV or the interventricular septum of the E10.5 heart. Consistent with this, we found that Hand1 expression best correlates with an LV CM identity compared with other reported LV markers, such as Cited1 or Cx40 (supplementary material Fig. S5). The presence of an immature and a more mature CM subpopulation within the E10.5 cells also raises interesting questions regarding the differences in their developmental origin [e.g. first heart field (FHF) versus second heart field (SHF)] and growth regulation. Although we are unable to determine whether some of these CMs are descendants from FHF versus SHF progenitor cells, as the expression of SHF markers such as Isl1 is largely absent in CMs (Cai et al., 2003), we found that immature CMs retain their expression of progenitor markers such as Pdgfra (Fig. 2B) and downstream targets of Wnt signaling, such as Lef1 and Axin2 (data not shown). Indeed, a recent paper by Buikema et al. has highlighted the importance of Wnt signaling activity in fetal cardiomyocyte proliferation and myocardial development (Buikema et al., 2013). Taken together, our results support the use of single-cell Fluidigm assays to identify and characterize phenotypically similar cardiomyocyte subgroups within the developing heart.
Characterization of lineage decisions from single CPCs during in vitro differentiation
Previous efforts to identify the lineage descendants of CPCs have assessed mRNA expression of multiple lineage-specific genes by qPCR or protein expression by immunostaining in a population of cells (Kattman et al., 2006; Moretti et al., 2006; Wu et al., 2006). Our single-cell multiplex PCR assay has enabled us to quantify 33 unique genes for each progeny of CPCs and can unequivocally identify the type of cell that is represented in the differentiated progeny pool. Our finding that Nkx2-5+ progenitor cells derived from mESCs differentiated preferentially into SMCs and CMs, and that similar cells derived from mouse embryos generated either EDCs or CMs, is quite intriguing. It has been reported recently by Lescroart et al. and Devine et al., using single-cell lineage tracing in vivo, that FHF cells give rise to only EDCs or CMs but not to both, and that SHF cells give rise to both CMs+SMCs or CMs+ECs (Devine et al., 2014; Lescroart et al., 2014). Our results are consistent with these findings, as we did not observe any bi-potent progenitor cell that gives rise to both CMs and ECs from a single cell (Fig. 5). However, we did find cells that can give rise to both immature SMCs and CMs from the same single cell, which might represent descendants from a SHF progenitor cell. Our finding suggests that the combination of time-lapse video microscopy, single-cell gene expression array and high-resolution immunofluorescence imaging might allow the characterization of progenies from single progenitor cell differentiation to assess cell fate commitment in vitro, provided that appropriate fluorescent reporters are available for the selective isolation of FHF and SHF progenitor cells.
It is worth pointing out that the propensity for mESC- and embryo-derived CPCs to differentiate and survive in vitro into a specific cell type can be highly media dependent. When we cultured embryo-derived CPCs in EDC medium, we found a greater number of EDCs within the progeny population. This might be due to either preferential differentiation or survival advantage. Indeed, we noted that the survival of mESC-derived CMs in EDC medium is reduced compared with those cultured in our standard differentiation medium, and is completely abolished in SMC medium (data not shown). Furthermore, Nkx2-5 expression is fairly broad in early mouse embryos and might encompass many true endocardial cells at the time of isolation (Stanley et al., 2002). Indeed, our single-cell gene expression data from mouse embryo-derived Nkx2-5+ CPCs showed that some of these CPCs already express a number of EDC genes (e.g. VE-Cadherin/Cdh5, Pecam1, Vwf, Flk-1/Kdr) upon isolation at E7.5 (Fig. 3). Taken together, these data support the early commitment of multipotent FHF CPCs to an EDC or CM fate (prior to E7.5), and suggest that our EDC media study most likely represents the differential survival of eGFP+ cells that have already committed to an EDC fate.
Transcriptional profiling of mESCs and adult heart-derived CMs reveals differences that might reflect the degree of cardiomyocyte maturation
Although ESC/iPSC-derived CMs may hold significant promise for cell-based therapy and as in vitro platforms for disease pathway and therapeutic drug discovery, their phenotype has been recognized as largely immature (Feinberg et al., 2013; Lieu et al., 2009; Liu et al., 2007; Martinez-Fernandez et al., 2013; Pillekamp et al., 2012; Robertson et al., 2013; Satin et al., 2008; Snir et al., 2003; Yang et al., 2014). Using our single-cell multiplex PCR expression analysis, we compared stage-by-stage the expression of main cardiac sarcomeric genes and transcription factors at distinct stages of development. We found that CMs derived from mESCs largely resemble CMs derived from the embryonic mouse heart up to the neonatal stage of development (Fig. 4A,B). However, CMs from adult mouse hearts exhibit modest variations compared with CMs from neonatal heart, particularly in the expression of sarcomeric protein genes, in which some genes show significant increases. We confirmed this finding by performing qPCR analysis on cell populations of purified adult atrial and ventricular CMs, and compared them with neonatal atrial and ventricular CM populations. These results suggest that CM maturation that normally takes place during neonatal development is absent during mESC in vitro differentiation. Further studies examining the difference in gene expression at the protein level between mESC-derived and neonatal/adult mouse-derived CMs should help to clarify these phenotypic differences.
Conclusion
We demonstrate in this study that application of single-cell multiplex PCR array might reveal the identity of each single cardiovascular cell derived from mESC differentiation in vitro as well as from embryonic heart in vivo. Our results support the transcriptional similarity between mESC and embryo-derived cardiac cells up to the neonatal stage of development. We believe that single-cell expression profiling is a powerful tool to address transcriptionally distinct subpopulations and might be useful to elucidate novel cell surface or intracellular markers when combined with genome-wide expression analysis.
MATERIALS AND METHODS
Cells preparation from embryoid bodies
Embryoid body-based (EB) differentiation of Nkx2-5–eGFP mESCs or α-MHC-CRE/ROSA26-mTmG mESCs derived from mice generated through mating of α-MHC-CRE mice with ROSA26-mTmG mice, was carried out according to Huang and Wu (2010). In brief, after specific days of differentiation, EBs were dissociated with trypsin and collagenase sequentially. Single cells were isolated by FACS, based on eGFP expression and lack of SSEA-1 expression, which is used to eliminate contaminating undifferentiated mESCs. For CPC isolation, eGFP+ cells were derived from 5 days differentiation of Nkx2-5–eGFP ESCs. For CM isolation, GFP+ cells were derived from α-MHC-CRE/ROSA26-mTmG ESCs after 29 days of differentiation. The numbers of cells that have been profiled at each developmental stage are listed in supplementary material Table S4.
Preparation of standard cells from embryonic or postnatal mice
Mouse embryos were harvested from pregnant Nkx2-5–eGFP female mice at 7.5, 8.5, 9.5 and 10.5 days post coitum and their hearts were treated with trypsin to dissociate into single cells. The eGFP+ cells were then isolated by FACS (supplementary material Fig. S6). To isolate postnatal CMs, neonatal [i.e. postnatal day 1 (P1)] and adult (three-months old) mice were sacrificed and their atrial and ventricular chambers dissociated into single cells using collagenase. The resulting single cells with intact shape and rectangular CM morphology were used for single-cell gene expression profiling. It was important to choose only the highest quality 2-5% of the adult CMs (e.g. ones that exhibit rod/rectangular shape and are able to beat in vitro) (see supplementary material Movies 1 and 2) for single-cell profiling, as adult CMs degrade easily after dissociation. To determine the expression of sarcomeric protein genes in pooled CMs, six high quality CMs from each cardiac chamber were pre-amplified with gene-specific primer pairs and the resultant cDNA was pooled for qPCR analysis. Mouse SMCs were derived from CD31– cells within the aorta of E14.5 embryos, and cardiac FBs and EDCs were derived from Thy1+ and CD31+ cells from E16.5 hearts, respectively. The numbers of cells that were profiled at each developmental stage are listed in supplementary material Table S7. All procedures involving animals were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee at Stanford University.
Pre-amplification and qPCR analysis with 48×48 Fluidigm dynamic arrays
Single cells derived from mESCs, mouse embryonic or adult hearts were added into one well of a 96-well plate at one cell per well in lysis buffer and kept in −20°C or immediately used. For pre-amplification, each plate underwent reverse transcription and PCR amplification with the following conditions: 50°C for 15 min, 70°C for 2 min, 20 cycles of 95°C for 15 s and 60°C for 4 min. After pre-amplification, the cells were either frozen at −20°C or used immediately. Amplified products were diluted fivefold with Tris-EDTA and used as templates for microfluidic PCR reactions using lineage-specific gene primers (Sanchez-Freire et al., 2012).
Single CPC culturing and analysis of gene expression in progenies by qPCR
Each single CPC derived from mESCs or mouse embryos was introduced into one well of a 96-well plate pre-loaded with CM differentiation media [82% (v/v) IMDM, 2 mM L-glutamin, 15% (v/v) serum, 0.01‰ (v/v) monothioglycerol and 50 µg/ml ascorbic acid] or EDC differentiation media [EGM-2 Bulletkit (Lonza, Basel, Switzerland), 50 ng/ml VEGF, 10 ng/ml bFGF]. After culturing for 6 days, CFSE (Invitrogen), a green fluorescent dye, was added to visualize live cells in each well and the wells with progenies were then dissociated with trypsin. Each dissociated progeny was then pipetted into one PCR tube pre-loaded with CellsDirect reaction mix for subsequent reverse transcription and cDNA synthesis (Invitrogen).
Analysis of gene expression in cell populations by qPCR
FACS-purified α-MHC-CRE/ROSA26-mTmG mESC-derived CMs after 23 days (D23) or 29 days (D29) of differentiation were lysed with TRIzol (Invitrogen) and underwent RNA extraction according to the manufacturer's specifications. Equal quantities of total RNA from D23 and D29 mESC-CMs were used for cDNA synthesis followed by qPCR. All qPCR gene expression data were normalized against the housekeeping gene gapdh.
Bioinformatics analysis of Fluidigm single-cell data
The heat maps of single-cell gene expression in this study reflect the Log2EX value, which is derived from the Ct value in each reaction subtracted from the limit of detection (LoD). The LoD for all single-cell data was set as 25, following a pre-amplification step of 20 cycles. Cells that failed to amplify β-actin or GADPH above our pre-specified threshold (Ct=21) were eliminated. As all of the Fluidigm chips were well controlled, because all samples were prepared with automatic platforms, the technical variations were minimized and allowed for parallel comparison of different samples without normalization (Guo et al., 2013). A detailed discussion on single-cell data normalization is also provided in the Fluidigm single-cell application guidance online (www.fluidigm.com/single-cell-guidance-request.html). The quantile-quantile plots and Pearson correlation coefficients of the housekeeping gene β-actin among different Fluidigm arrays were analyzed and supported the premise that gene expression among different plates can be directly compared due to low plate-to-plate variation (supplementary material Fig. S2).
The hierarchical clustering and PCA were both performed with the manufacturer-supplied program SINGuLAR 1.3.1 analysis toolset in R (Fluidigm). The signature GEDI plot for each cell type was generated using GEDI 2.1 (Eichler et al., 2003), which translated high-dimensional gene expression data into a two-dimensional (2D) mosaic image through unsupervised learning neural networks, known as self-organizing map (SOM). SOM organizes genes by grouping together those with similar expression levels. In the mosaic image, each tile represents an individual SOM cluster, and their color correlates with the level of gene expression (Eichler et al., 2003). The expression signature for each of the cell types was generated based on the average of all signatures for that cell type at a particular stage of development.
A SPADE analysis was performed with CytoSPADE software (Bendall et al., 2011; Linderman et al., 2012; Qiu et al., 2011). Specifically, cardiac lineage gene assays were divided into five clusters (e.g. sarcomeric protein cluster, EDC cluster, cardiac transcription factor cluster, Pdgfra cluster and Hand1 cluster) according to their expression patterns. For those clusters with more than one assay, the average value (Ct) of all gene assays in each single cell was calculated and used as the cluster expression value in that single cell. The cluster expression values of all single cells were then analyzed with the SPADE program, which examined the data geometry with a topological method, extracted a hierarchy in an unsupervised manner and visualized the hierarchy in a branched tree structure.
The Fluidigm single cell profiling data have been deposited in the GEO database with the accession number GSE64938.
Immunofluorescent staining of progenies of single CPCs after culturing
Single Nkx2-5–eGFP+ CPCs were isolated by FACS from EBs at day 5 or day 7 and from embryos at E7.5 or E8.5, and were cultured for 5 additional days in one well of a 96-well plate. Cells in 96-well plates were then directly fixed with 4% PFA and permeabilized with 0.025% Triton X-100 (Sigma). These cells were then blocked with blocking buffer (TBS containing 1% BSA, 0.1% Tween20 and 10% normal goat serum) and incubated overnight with primary antibodies against CD31 (BectonDickinson, 558737; 1:200), sarcomeric myosin heavy chain (MF20; 1:500) (Developmental Studies Hybridoma Bank, University of Iowa, USA) and smooth muscle myosin heavy chain (SM-MHC) (Biomedical Tech, BT-562; 1:200). The following day, secondary antibodies were used to detect for the presence of protein expression.
Supplementary Material
Acknowledgements
We thank Sopheak Sim at the Stanford Institute for Stem Cell Biology and Regenerative Medicine for assistance with the Fluidigm Biomark HD array, Drs Ronglih Liao and Weiting Zhang at the Harvard Brigham and Women's Hospital for providing purified adult CMs, and members of the Wu laboratory for manuscript critique.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Financial support for this work was provided in part by grants from the National Institutes of Health/National Heart, Lung and Blood Institute (NIH/NHBI) [U01 HL099776-5], the NIH Director's New Innovator Award [DP2 OD004411-2], the California Institute of Regenerative Medicine [RB3-05129], the American Heart Association [14GRNT18630016] and the Endowed Faculty Scholar Award from the David and Lucile Packard Foundation for Children and the Child Health Research Institute at Stanford (to S.M.W.). Deposited in PMC for release after 12 months.
Author contributions
G.L. and S.M.W. designed the experiments; G.L., K.P., R.K. and A.S. performed the experiments; G.L., K.P. and S.M.W. performed data analysis; G.L., K.P., R.K., A.S. and S.M.W. prepared the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.116897/-/DC1
References
- Bendall S. C., Simonds E. F., Qiu P., Amir E.-a. D., Krutzik P. O., Finck R., Bruggner R. V., Melamed R., Trejo A., Ornatsky O. I. et al. (2011). Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687-696 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G. et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947-956 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D. A., Cheng A. W., Itskovich E., Markoulaki S., Ganz K., Klemm S. L., van Oudenaarden A. and Jaenisch R. (2012). Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209-1222 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema J. W., Mady A. S., Mittal N. V., Atmanli A., Caron L., Doevendans P. A., Sluijter J. P. G. and Domian I. J. (2013). Wnt/beta-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development 140, 4165-4176 10.1242/dev.099325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S. L., Chen J. and Evans S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877-889 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.-L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X. et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104-108 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A., Pang Z. P., Südhof T. C., Wernig M. and Malenka R. C. (2011). Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat. Protocols 7, 118-127 10.1038/nprot.2011.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Kalisky T., Sahoo D., Rajendran P. S., Rothenberg M. E., Leyrat A. A., Sim S., Okamoto J., Johnston D. M., Qian D. et al. (2011). Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 29, 1120-1127 10.1038/nbt.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRuiter M. C., Poelmann R. E., VanderPlas-de Vries I., Mentink M. M. T. and Gittenberger-de Groot A. C. (1992). The development of the myocardium and endocardium in mouse embryos. Fusion of two heart tubes? Anat. Embryol. (Berl.) 185, 461-473 10.1007/BF00174084 [DOI] [PubMed] [Google Scholar]
- Devine W. P., Wythe J. D., George M., Koshiba-Takeuchi K. and Bruneau B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife 3, e03848 10.7554/eLife.03848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler G. S., Huang S. and Ingber D. E. (2003). Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics 19, 2321-2322 10.1093/bioinformatics/btg307 [DOI] [PubMed] [Google Scholar]
- Feinberg A. W., Ripplinger C. M., van der Meer P., Sheehy S. P., Domian I., Chien K. R. and Parker K. K. (2013). Functional differences in engineered myocardium from embryonic stem cell-derived versus neonatal cardiomyocytes. Stem Cell Rep. 1, 387-396 10.1016/j.stemcr.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D. and Robson P. (2010). Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675-685 10.1016/j.devcel.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Guo G., Luc S., Marco E., Lin T.-W., Peng C., Kerenyi M. A., Beyaz S., Kim W., Xu J., Das P. P. et al. (2013). Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 13, 492-505 10.1016/j.stem.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. P. (2002). Patterning the vertebrate heart. Nat. Rev. Genet. 3, 544-556 10.1038/nrg843 [DOI] [PubMed] [Google Scholar]
- Huang X. and Wu S. M. (2010). Isolation and functional characterization of pluripotent stem cell-derived cardiac progenitor cells. Curr. Protoc. Stem Cell Biol. Chapter 1, Unit 1F 10 10.1002/9780470151808.sc01f10s14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icardo J. M. (1996). Developmental biology of the vertebrate heart. J. Exp. Zool. 275, 144-161 [DOI] [PubMed] [Google Scholar]
- Ieda M., Tsuchihashi T., Ivey K. N., Ross R. S., Hong T.-T., Shaw R. M. and Srivastava D. (2009). Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell 16, 233-244 10.1016/j.devcel.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S. J., Huber T. L. and Keller G. M. (2006). Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell 11, 723-732 10.1016/j.devcel.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Laugwitz K.-L., Moretti A., Caron L., Nakano A. and Chien K. R. (2008). Islet1 cardiovascular progenitors: a single source for heart lineages? Development 135, 193-205 10.1242/dev.001883 [DOI] [PubMed] [Google Scholar]
- Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A. et al. (2014). Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16, 829-840 10.1038/ncb3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C. L., Wu C., Mercer B., Webb R., Richardson J. A. and Olson E. N. (1999). Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development 126, 75-84. [DOI] [PubMed] [Google Scholar]
- Lieu D. K., Liu J., Siu C.-W., McNerney G. P., Tse H.-F., Abu-Khalil A., Huser T. and Li R. A. (2009). Absence of transverse tubules contributes to non-uniform Ca(2+) wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev. 18, 1493-1500 10.1089/scd.2009.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman M. D., Bjornson Z., Simonds E. F., Qiu P., Bruggner R. V., Sheode K., Meng T. H., Plevritis S. K. and Nolan G. P. (2012). CytoSPADE: high-performance analysis and visualization of high-dimensional cytometry data. Bioinformatics 28, 2400-2401 10.1093/bioinformatics/bts425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Fu J. D., Siu C. W. and Li R. A. (2007). Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem Cells 25, 3038-3044 10.1634/stemcells.2007-0549 [DOI] [PubMed] [Google Scholar]
- Martin-Puig S., Wang Z. and Chien K. R. (2008). Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2, 320-331 10.1016/j.stem.2008.03.010 [DOI] [PubMed] [Google Scholar]
- Martinez-Fernandez A., Li X., Hartjes K. A., Terzic A. and Nelson T. J. (2013). Natural cardiogenesis-based template predicts cardiogenic potential of induced pluripotent stem cell lines. Circ. Cardiovasc. Genet. 6, 462-471 10.1161/CIRCGENETICS.113.000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden D. G., Barbosa A. C., Richardson J. A., Schneider M. D., Srivastava D. and Olson E. N. (2005). The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189-201 10.1242/dev.01562 [DOI] [PubMed] [Google Scholar]
- Moretti A., Caron L., Nakano A., Lam J. T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S. et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151-1165 10.1016/j.cell.2006.10.029 [DOI] [PubMed] [Google Scholar]
- Narsinh K. H., Sun N., Sanchez-Freire V., Lee A. S., Almeida P., Hu S., Jan T., Wilson K. D., Leong D., Rosenberg J. et al. (2011). Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 121, 1217-1221 10.1172/JCI44635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillekamp F., Haustein M., Khalil M., Emmelheinz M., Nazzal R., Adelmann R., Nguemo F., Rubenchyk O., Pfannkuche K., Matzkies M. et al. (2012). Contractile properties of early human embryonic stem cell-derived cardiomyocytes: beta-adrenergic stimulation induces positive chronotropy and lusitropy but not inotropy. Stem Cells Dev. 21, 2111-2121 10.1089/scd.2011.0312 [DOI] [PubMed] [Google Scholar]
- Qiu P., Simonds E. F., Bendall S. C., Gibbs K. D. Jr, Bruggner R. V., Linderman M. D., Sachs K., Nolan G. P. and Plevritis S. K. (2011). Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886-891 10.1038/nbt.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser P. J., Westfall M. V., Schiaffino S. and Solaro R. J. (1994). Tension production and thin-filament protein isoforms in developing rat myocardium. Am. J. Physiol. 267, H1589-H1596. [DOI] [PubMed] [Google Scholar]
- Robertson C., Tran D. D. and George S. C. (2013). Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells 31, 829-837 10.1002/stem.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Freire V., Ebert A. D., Kalisky T., Quake S. R. and Wu J. C. (2012). Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat. Protoc. 7, 829-838 10.1038/nprot.2012.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satin J., Itzhaki I., Rapoport S., Schroder E. A., Izu L., Arbel G., Beyar R., Balke C. W., Schiller J. and Gepstein L. (2008). Calcium handling in human embryonic stem cell-derived cardiomyocytes. Stem Cells 26, 1961-1972 10.1634/stemcells.2007-0591 [DOI] [PubMed] [Google Scholar]
- Snir M., Kehat I., Gepstein A., Coleman R., Itskovitz-Eldor J., Livne E. and Gepstein L. (2003). Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 285, H2355-H2363. [DOI] [PubMed] [Google Scholar]
- Souders C. A., Bowers S. L. K. and Baudino T. A. (2009). Cardiac fibroblast: the renaissance cell. Circ. Res. 105, 1164-1176 10.1161/CIRCRESAHA.109.209809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E. G., Biben C., Elefanty A., Barnett L., Koentgen F., Robb L. and Harvey R. P. (2002). Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 46, 431-439. [PubMed] [Google Scholar]
- Sturzu A. C. and Wu S. M. (2011). Developmental and regenerative biology of multipotent cardiovascular progenitor cells. Circ. Res. 108, 353-364 10.1161/CIRCRESAHA.110.227066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S., Hughey J. J., Lee T. K., Lipniacki T., Quake S. R. and Covert M. W. (2010). Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature 466, 267-271 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Yamagishi H., Overbeek P. A., Olson E. N. and Srivastava D. (1998). The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev. Biol. 196, 228-236 10.1006/dbio.1998.8849 [DOI] [PubMed] [Google Scholar]
- van den Heuvel N. H. L., van Veen T. A. B., Lim B. and Jonsson M. K. B. (2014). Lessons from the heart: mirroring electrophysiological characteristics during cardiac development to in vitro differentiation of stem cell derived cardiomyocytes. J. Mol. Cell. Cardiol. 67, 12-25 10.1016/j.yjmcc.2013.12.011 [DOI] [PubMed] [Google Scholar]
- Wu S. M., Fujiwara Y., Cibulsky S. M., Clapham D. E., Lien C.-L., Schultheiss T. M. and Orkin S. H. (2006). Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 127, 1137-1150 10.1016/j.cell.2006.10.028 [DOI] [PubMed] [Google Scholar]
- Xu X. Q., Soo S. Y., Sun W. and Zweigerdt R. (2009). Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cells 27, 2163-2174 10.1002/stem.166 [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M. H., Adler E. D., Roepke T. K., Kattman S. J., Kennedy M., Henckaerts E., Bonham K., Abbott G. W., Linden R. M. et al. (2008). Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453, 524-528 10.1038/nature06894 [DOI] [PubMed] [Google Scholar]
- Yang X., Pabon L. and Murry C. E. (2014). Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 114, 511-523 10.1161/CIRCRESAHA.114.300558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R. et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109-113 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.