Abstract
The rhizobacterium Pseudomonas fluorescens SS101 inhibits growth of oomycete and fungal pathogens, and induces resistance in plants against pathogens and insects. To unravel regulatory pathways of secondary metabolite production in SS101, we conducted a genome-wide search for sRNAs and performed transcriptomic analyses to identify genes associated with the Rsm (repressor of secondary metabolites) regulon. In silico analysis led to the identification of 16 putative sRNAs in the SS101 genome. In frame deletion of the sRNAs rsmY and rsmZ showed that the Rsm system regulates the biosynthesis of the lipopeptide massetolide A and involves the two repressor proteins RsmA and RsmE, with the LuxR-type transcriptional regulator MassAR as their most likely target. Transcriptome analyses of the rsmYZ mutant further revealed that genes associated with iron acquisition, motility and chemotaxis were significantly upregulated, whereas genes of the type VI secretion system were downregulated. Comparative transcriptomic analyses showed that most, but not all, of the genes controlled by RsmY/RsmZ are also controlled by the GacS/GacA two-component system. We conclude that the Rsm regulon of P. fluorescens SS101 plays a critical role in the regulation of lipopeptide biosynthesis and controls the expression of other genes involved in motility, competition and survival in the plant rhizosphere.
Introduction
Computational searches of intergenic regions, promoters and rho-independent transcription terminators (Livny et al., 2005; 2006; Sridhar and Gunasekaran, 2013; Wright et al., 2013) combined with experimental approaches (Sharma and Vogel, 2009) have revealed the presence of several small RNAs (sRNAs) in bacterial genomes. In general, two types of regulatory sRNAs have been described (Majdalani et al., 2005; Gottesman et al., 2006; Pichon and Felden, 2007; Gottesman and Storz, 2011). The first targets specific messenger RNAs (mRNAs) by base pairing. An example is RyhB in Escherichia coli which interacts with the mRNA encoding SodB, an iron-containing superoxide dismutase (Salvail et al., 2010). The second type interacts with RNA-binding proteins of the RsmA/CsrA family. RsmA (regulator of secondary metabolism) and CsrA (carbon storage regulator) act as translational repressors and their sequestration by activated sRNAs can relieve repression of the target mRNAs.
In Pseudomonas, relatively few sRNAs have been studied in detail for their functions. In Pseudomonas protegens strain CHA0, the sRNAs RsmX, RsmY and RsmZ are under the control of the GacS/GacA two-component system and regulate the production of a range of secondary metabolites (Heeb et al., 2002a; Valverde et al., 2003; Kay et al., 2005; Lapouge et al., 2007; 2008). In P. protegens CHA0, Gac/Rsm-mediated regulation of secondary metabolites involves sequestration of the repressor proteins RsmA and RsmE that act post-transcriptionally by binding to the target mRNA (Blumer et al., 1999; Reimmann et al., 2005; Lapouge et al., 2008). In Pseudomonas aeruginosa, the two sRNAs, RsmY and RsmZ, regulate quorum sensing and the biosynthesis of several exoproducts (Brencic et al., 2009; Frangipani et al., 2014). Other sRNAs described for P. aeruginosa are PhrS, PrrF1 and PrrF2: PhrS is involved in the regulation of quinolone biosynthesis (Sonnleitner and Haas, 2011; Sonnleitner et al., 2011), and PrrF1 and PrrF2 contribute to iron acquisition (Wilderman et al., 2004; Sonnleitner and Haas, 2011).
Most of the known sRNAs in Pseudomonas and other Gram-negative bacterial genera are under the control of the Gac/Rsm signal transduction pathway. Based on the proposed model, the phosphorylated regulator GacA binds to a conserved element upstream of the sRNA promoter, referred to as the GacA box, to activate their expression (Lapouge et al., 2008). In many cases, mutations or deletions of the sRNAs result in phenotypes similar to that of GacS/GacA mutants. For example, ΔrsmYZ and ΔgacA mutants of P. aeruginosa are both deficient in the synthesis of the quorum sensing signal N-butanoyl-homoserine lactone, hydrogen cyanide (HCN), pyocyanin, elastase and chitinase as well as in biofilm formation (Kay et al., 2006; Brencic et al., 2009). In Pseudomonas entomophila, ΔrsmYZ and ΔgacA mutants were both deficient in the production of entolysin (Vallet-Gely et al., 2010). Similarities in phenotypes of rsm and gac mutants have also been described for Pectobacterium carotovorum (Liu et al., 1998), E. coli (Weilbacher et al., 2003), Salmonella enterica (Fortune et al., 2006) and Legionella pneumophila (Sahr et al., 2009).
In this study, we conducted a genome-wide search for sRNAs in Pseudomonas fluorescens strain SS101 and performed transcriptomic analyses to identify genes associated with the Rsm regulon and with the Gac regulon. We addressed the function of the Rsm regulon, involving the two sRNAs RsmY (PflSS101_4962) and RsmZ (PflSS101_1168), and the two repressor proteins RsmA (PflSS101_4138) and RsmE (PflSS101_3491), in lipopeptide biosynthesis and predicted the potential target genes of the Rsm repressor proteins. Strain SS101 was originally isolated from the rhizosphere of wheat (de Souza et al., 2003), has activity against various oomycete and fungal pathogens (de Souza et al., 2003; Tran et al., 2013; van de Mortel et al., 2009) and induces systemic resistance in tomato and Arabidopsis against several pathogens and insect pests (Tran et al., 2013; van de Mortel et al., 2012). Comparative genome analyses of multiple Pseudomonas species and strains (Loper et al., 2012) revealed that strain SS101 harbours 350 unique genes, which include prophage and genomic islands. Unlike many other P. fluorescens and P. protegens biocontrol strains, SS101 does not produce the typical secondary metabolites such as 2,4-diacetylphloroglucinol (DAPG), phenazines, pyrrolnitrin, pyoluteorin and HCN (Loper et al., 2012). The main secondary metabolite produced by SS101 is the cyclic lipopeptide massetolide A, whose biosynthesis is governed by the non-ribosomal peptide synthetase (NRPS) genes massABC and regulated by the GacS/GacA system (de Bruijn and Raaijmakers, 2009a). Massetolide A contributes to biofilm formation, swarming motility, antimicrobial activity and defense against protozoan predators (Mazzola et al., 2009; Raaijmakers et al., 2010). Here, genome-wide transcriptional analysis of mutants with deletions in rsmY and rsmZ revealed that the NRPS genes massA, massB, massC as well as the LuxR-type transcriptional regulator massAR were significantly downregulated. Via mutational and phenotypic analyses, we show that the Rsm system regulates massetolide biosynthesis as well as several other genes and traits in the rhizobacterium P. fluorescens SS101.
Results and discussion
Small RNAs in P. fluorescens SS101
A total of 68 tRNAs and 19 rRNAs were found in the SS101 genome (Table S1). Genome-wide analyses revealed 16 predicted sRNAs including homologues of the two signal recognition particle RNAs SrpB_1 (PflSS101_3911) and SrpB_2 (PflSS101_3926) (Table 1). Signal recognition particle (Srp) is a ribonucleoprotein complex that participates in multiple protein targeting pathways in bacteria (Koch et al., 1999) and is primarily involved in the incorporation of proteins in the inner membrane (Rosenblad et al., 2009). Furthermore, we also found a 6S SsrS RNA (PflSS101_5226) in the SS101 genome. In E. coli, 6S RNA is encoded by the ssrS gene which regulates transcription during late exponential and stationary growth (Wassarman, 2007). Bacterial Ribonuclease P (PflSS101_0956) was found in the SS101 genome and represents a ribonucleoprotein complex comprised of a single RNA (∼ 400 nt) and a single small protein subunit (∼ 14 kDa) with the RNA as the catalytic subunit of the enzyme involved in the maturation of tRNA transcripts (Ellis and Brown, 2009). We also found homologues of PhrS (PflSS101_4081), PrrF1 (PflSS101_4589) and PrrF2 (PflSS101_3274), which are known to repress or activate the translation of target mRNAs by a base pairing mechanism. In P. aeruginosa, the two prrF sRNA genes are found in tandem. Homologous genes in other Pseudomonas species are located considerably distant from each other on the chromosome (Wilderman et al., 2004). Also in SS101, PrrF1 (PflSS101_4589) and PrrF2 (PflSS101_3274) are found at different locations in the genome. We also found RgsA (PflSS101_1357) in the SS101 genome, which is an sRNA probably regulated indirectly by GacA and directly by the stress sigma factor RpoS (Gonzalez et al., 2008).
Table 1.
Small non-coding RNAs in P. fluorescens SS101
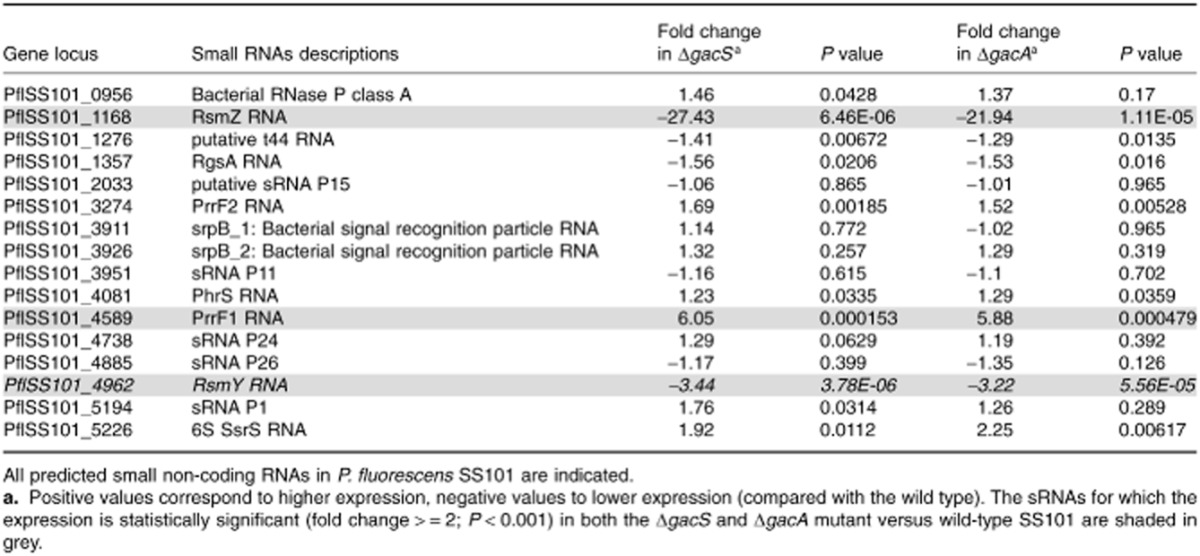
Two other sRNAs found in the SS101 genome were RsmY (PflSS101_4962) and RsmZ (PflSS101_1168) (Table 1). In P. protegens and P. aeruginosa, RsmY and RsmZ regulate secondary metabolite production by sequestering RNA-binding proteins (e.g. CsrA, RsmA) that act as translational repressors (Kay et al., 2005; Gottesman and Storz, 2011). In P. aeruginosa, the expression of all Gac-regulated genes was shown to be RsmY/Z dependent (Brencic et al., 2009). For the other sRNAs detected in the SS101 genome (Table 1), the functions are poorly understood or not known from other Pseudomonas species. Here, we will specifically focus on the sRNAs in strain SS101 that are regulated by the GacS/GacA two-component system.
Small RNAs in P. fluorescens SS101 regulated by the GacS/A system
Transcriptomic analyses of both gacS and gacA mutants of P. fluorescens SS101 (Tables S2, S3) revealed that the expression of three sRNAs (rsmY, rsmZ and prrF1) was significantly (> 2-fold, P < 0.001) altered (Table 1). Expression of rsmY and rsmZ was significantly downregulated in both gacS and gacA mutants, whereas expression of prrF1 was approximately six-fold upregulated in both gac mutants. The predicted sizes of the rsmY, rsmZ and prrF1 transcripts were 118 bp, 133 bp and 112 bp respectively. Subsequent prediction of their secondary structures revealed eight GGA motifs in both RsmY and RsmZ, with three in predicted loop regions respectively (Fig. 1A). In contrast, only one GGA motif was found in PrrF1, which is localized to a predicted stem (Fig. 1A). Repeated GGA motifs in loop regions of the secondary structure, as predicted for RsmY and RsmZ, are an essential characteristic of sRNAs for sequestration of RsmA and homologous repressor proteins (Lapouge et al., 2008). Previous work also showed that the regions upstream of these sRNAs contain a conserved 18 bp sequence which corresponds to the GacA-binding site for activation of these sRNAs (Heeb et al., 2002b; Kay et al., 2005). For SS101, we indeed found this typical GacA-binding box upstream of rsmY and rsmZ (Fig. 1B and C), but not for prrF1. Therefore, our subsequent functional analyses focused on rsmY and rsmZ.
Figure 1.
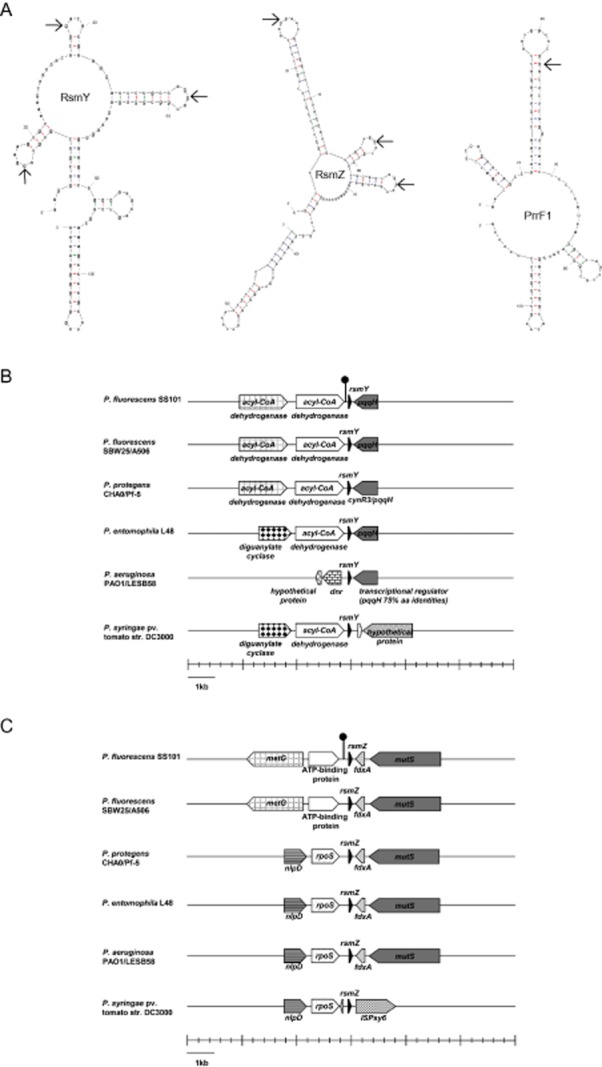
Secondary structures of small RNAs, RsmY, RsmZ, PrrF1 in P. fluorescens SS101 and the genetic organization of rsmY and rsmZ in strain SS101 and other Pseudomonas species and strains.A. Predicted secondary structures of RsmY, RsmZ and PrrF1 of P. fluorescens SS101 by MFOLD (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form). The typical GGA motifs located in the loop regions are indicated with arrows.B. Genetic organization of rsmY regions in different Pseudomonas species and strains. Block arrows indicate directionality of the open reading frame, and orthologous genes are represented by color and pattern. The loop symbol in front of rsmY indicates the position of the upstream activating sequence (UAS for rsmY: TGTAAGCATTCTCTTACA). Abbreviations: pqqH/cynR3/dnr: transcriptional regulator.C. Genetic organization of rsmZ regions in different Pseudomonas species and strains. Block arrows indicate directionality of the open reading frame, and orthologous genes are represented by colour and pattern. The loop symbol in front of rsmZ indicates the position of the UAS (UAS for rsmZ: TGTAAGCATTCGCTTACT). Abbreviations: metG: methionyl-tRNA synthetase; fdxA: ferredoxin; mutS: DNA mismatch repair protein; nlpD: lipoprotein; rpoS: RNA polymerase sigma factor; ISPsy6: transposase.
Role of RsmY and RsmZ in lipopeptide biosynthesis in P. fluorescens SS101
The location of rsmY and rsmZ in the genomes appears to be conserved, at least to some extent, for the different Pseudomonas species and strains (Fig. 1B and C). In frame deletion, mutants were generated to investigate the role of rsmY and rsmZ in the regulation of massetolide A biosynthesis. The drop collapse assay, a reliable proxy for detection of massetolide A and other lipopeptide surfactants (de Bruijn et al., 2008; de Bruijn and Raaijmakers, 2009a), showed that mutations in either rsmY or rsmZ alone did not affect massetolide A production (Fig. 2A). However, mutations in both rsmY and rsmZ resulted in loss of massetolide A production which was confirmed by reversed phase-high-performance liquid chromatography (RP-HPLC) (Fig. 2B). Also swarming motility of SS101, a phenotype that depends on massetolide production (de Bruijn et al., 2008), was abolished in the rsmYZ double mutant (Fig. 2C). Mutations in rsmY or rsmZ alone did not affect growth of strain SS101 (Fig. 2D). However, mutations in both rsmY and rsmZ slightly enhanced growth in the early exponential phase but had an adverse effect on growth during the late exponential and stationary phase; similar changes in growth dynamics were observed for the gacS and gacA mutants of strain SS101 (Fig. 2D). These changes in growth dynamics are most likely not related to a lack of massetolide production, because growth of the site-directed massA biosynthesis mutant of SS101 was similar to that of the wild type (de Bruijn and Raaijmakers, 2009a). In summary, these results indicated that both RsmY and RsmZ are an integral component of the GacS/GacA signal transduction cascade and regulate massetolide biosynthesis in P. fluorescens SS101.
Figure 2.
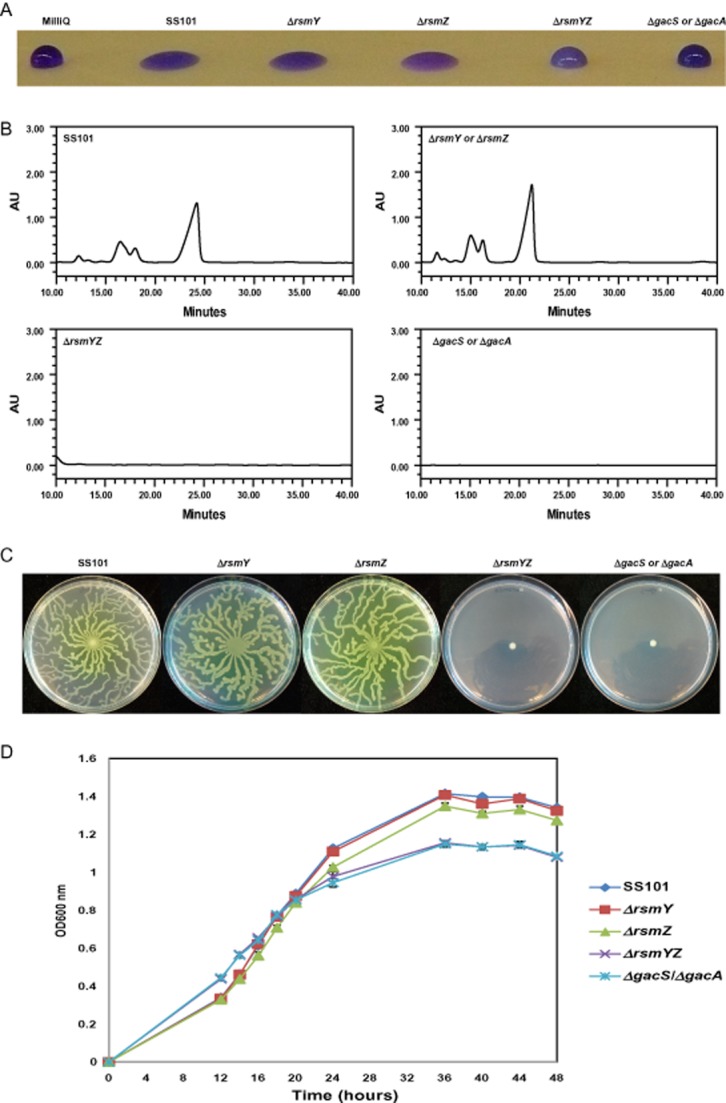
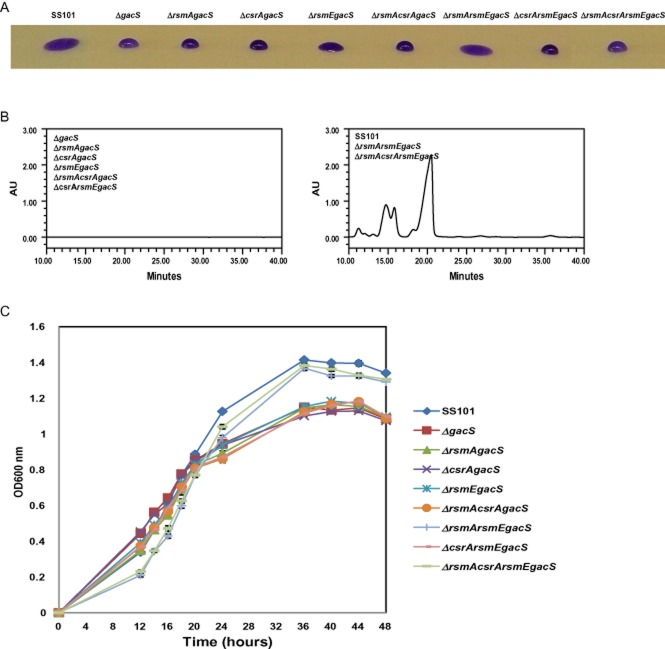
Phenotypic and chemical analyses of P. fluorescens strain SS101 and single or double mutants disrupted in rsmY, rsmZ, gacS or gacA.A. Drop collapse assay with cell cultures of wild-type strain SS101, ΔrsmY, ΔrsmZ, ΔrsmYZ, ΔgacS and ΔgacA mutants. Bacterial cultures grown for 2 days at 25°C on KB agar plates were suspended in sterile water to a final density of 1 × 1010 cells ml−1, and 10-μl droplets were spotted on parafilm, and crystal violet was added to the droplets to facilitate visual assessment. A flat droplet is a highly reliable proxy for the production of the surface-active lipopeptide massetolide A.B. Reversed phase-high-performance liquid chromatography chromatograms of cell-free culture extracts of wild-type strain SS101, ΔrsmY, ΔrsmZ, ΔrsmYZ, ΔgacS and ΔgacA mutants as described in A. The wild-type strain SS101 produces massetolide A (retention time of approximately 23–25 min) and various other derivatives of massetolide A (minor peaks with retention times ranging from 12 to 18 min) which differ from massetolide A in the amino acid composition of the peptide moiety. AU stands for absorbance unit.C. Swarming motility of wild-type strain SS101, ΔrsmY, ΔrsmZ, ΔrsmYZ, ΔgacS and ΔgacA mutants on soft [0.6% (wt/vol)] agar plates. Five microlitres (1 × 1010 cells ml−1) of washed overnight cultures of wild-type SS101 or mutants were spot inoculated in the centre of a soft agar plate and incubated for 48 to 72 h at 25°C.D. Growth of wild-type strain SS101, ΔrsmY, ΔrsmZ, ΔrsmYZ, ΔgacS and ΔgacA mutants in liquid broth at 25°C. At different time points, the optical density of the cell cultures was measured spectrophotometrically (600 nm). Mean values of four biological replicates are given; the error bars represent the standard error of the mean.
Deletion of repressor proteins restores massetolide production
Previous studies with P. protegens CHA0 have shown that Gac/Rsm-mediated regulation of secondary metabolites involves sequestration of the repressor proteins RsmA and RsmE that act post-transcriptionally by binding to the target mRNA (Blumer et al., 1999; Reimmann et al., 2005; Lapouge et al., 2008). Hence, the next step was to determine if these repressor proteins are present in SS101 and if they play a role in Gac/Rsm-mediated regulation of massetolide biosynthesis. In silico analysis of the SS101 genome led to the identification of rsmA (PflSS101_4138), rsmE (PflSS101_3491) and csrA (PflSS101_3653). Phylogenetic analyses showed that they clustered closely with their homologues in other P. fluorescens strains and Pseudomonas species at both DNA and protein levels (Fig. S1). To decipher their role in regulation of massetolide biosynthesis, deletion mutants were made for each of these three repressors in the gacS mutant background of strain SS101. The gacS mutant does not produce massetolide, but according to the regulatory model, a mutation of the repressor proteins would alleviate translational repression and restore production. The results of the drop collapse assay and RP-HPLC analyses showed that a deletion of either rsmA or csrA in the gacS mutant did not restore massetolide production (Fig. 3A and B). Based on the drop collapse assay, a mutation in the rsmE gene partially affected the surface tension (Fig. 3A), but massetolide production was not detectable by RP-HPLC analysis (Fig. 3B). A double mutation in rsmE and rsmA fully restored massetolide production (Fig. 3A and B). A single deletion of either one of the repressor genes did not affect growth as compared with that of the gacS mutant, whereas stacked deletions of rsmA and rsmE in the gacS mutant changed the growth dynamics back to that of the wild type (Fig. 3C). We conclude that Gac/Rsm-mediated regulation of massetolide biosynthesis via rsmY and rsmZ implicates the two small RNA binding proteins RsmA and RsmE, whereas CsrA is not involved.
Figure 3.
Phenotypic and chemical analyses of P. fluorescens strain SS101, ΔgacS mutant and single, double or triple mutants disrupted in rsmA, rsmE and csrA in the ΔgacS background.A. Drop collapse assay with cell suspensions of wild-type SS101, ΔgacS, ΔrsmAgacS, ΔcsrAgacS, ΔrsmEgacS, ΔrsmAcsrAgacS, ΔrsmArsmEgacS, ΔcsrArsmEgacS and ΔrsmAcsrArsmEgacS mutants. Bacterial cultures grown for 2 days at 25°C on KB agar plates were suspended in sterile water to a final density of 1 × 1010 cells ml−1, and 10-μl droplets were spotted on parafilm, and crystal violet was added to the droplets to facilitate visual assessment. A flat droplet is a highly reliable proxy for the production of the surface-active lipopeptide massetolide A.B. Reversed phase-high-performance liquid chromatography chromatograms of cell-free culture extracts of wild-type SS101, ΔrsmAgacS, ΔcsrAgacS, ΔrsmEgacS, ΔrsmAcsrAgacS, ΔrsmArsmEgacS, ΔcsrArsmEgacS and ΔrsmAcsrArsmEgacS mutants as described in A. The wild-type strain SS101 produces massetolide A (retention time of approximately 18–21 min) and various other derivatives of massetolide A (minor peaks with retention times ranging from 12 to 18 min) which differ from massetolide A in the amino acid composition of the peptide moiety. AU stands for absorbance unit. Representative chromatograms of ΔrsmAgacS and ΔrsmArsmEgacS mutants are shown.C. Growth of wild-type SS101, ΔrsmAgacS, ΔcsrAgacS, ΔrsmEgacS, ΔrsmAcsrAgacS, ΔrsmArsmEgacS, ΔcsrArsmEgacS and ΔrsmAcsrArsmEgacS mutants in liquid broth at 25°C. At different time points, the optical density of the cell cultures was measured spectrophotometrically (600 nm). Mean values for four biological replicates are given; the error bars represent the standard errors of the mean.
Potential targets of the RsmA/RsmE repressor proteins in P. fluorescens SS101
To determine the potential targets of the RsmA and RsmE repressor proteins, we conducted a whole genome search for putative Rsm binding sites at or near the 5′ untranslated leader mRNA by using the conserved motif 5′-A/U CANGGANGU/A-3′ (N is any nucleotide) (Lapouge et al., 2008). A total of 17 genes were found with this conserved motif located in the ribosome binding site (RBS) (Table 2). For six of these 17 genes, transcription was significantly downregulated in the gacS/gacA mutants and also in the rsmYZ double mutant (Table 2). These six genes included: PflSS101_0554 with unknown function; gcd (PflSS101_1096) encoding the quinoprotein glucose dehydrogenase; ompA (PflSS101_1239); aprA (PflSS101_2560), which encodes an extracellular protease; PflSS101_2598, a gene predicted to encode a formyl-transferase domain/enoyl-CoA hydratase/isomerase family protein; and massAR (PflSS101_3396), the LuxR-type transcriptional regulatory gene located upstream of the massA biosynthesis gene and essential for massetolide biosynthesis (de Bruijn and Raaijmakers, 2009a,b). There was no GacA box sequence upstream of massA, massBC or massBCR (LuxR type regulator downstream of massBC). Alignment of the 5′ untranslated leader regions of these six putative target genes, with hcnA and aprA of P. protegens CHA0 and P. aeruginosa PAO1 as references, revealed the position of the consensus motif close to the RBS (Fig. 4A). When the alignment for massAR was performed with genes of several closely related LuxR-type transcriptional regulator genes flanking other lipopeptide biosynthesis genes in different Pseudomonas species and strains, similar consensus motifs were found (Fig. 4B). Based on these findings, we postulate that (i) the LuxR-type transcriptional regulator MassAR is the most likely target of the RsmA and RsmE repressor proteins in Gac/Rsm-mediated regulation of massetolide biosynthesis in P. fluorescens SS101; and (ii) lipopeptide biosynthesis in other Pseudomonas species is most likely regulated in a similar manner.
Table 2.
Predicted target genes of the RsmA and RsmE repressor proteins in P. fluorescens SS101
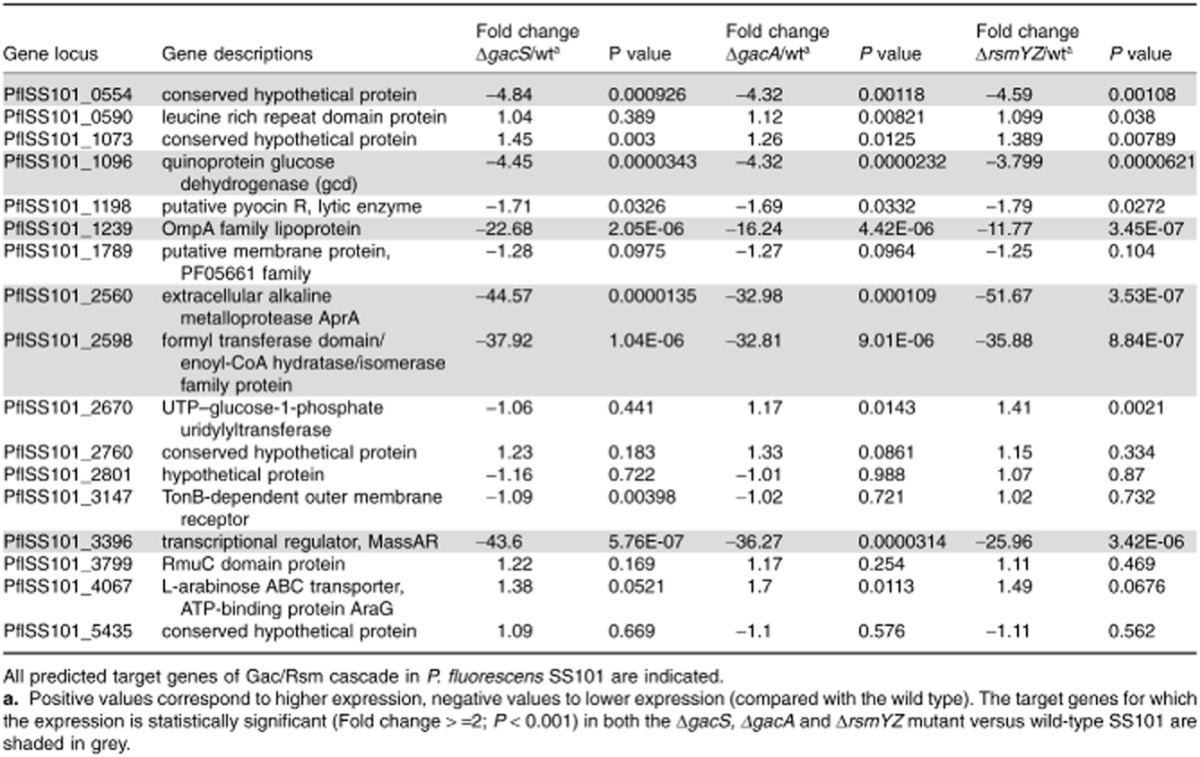
Figure 4.
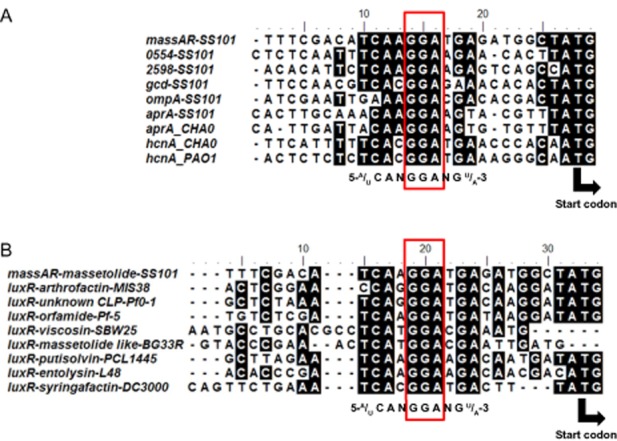
A. Alignment of the upstream regions of five putative target genes of the RsmA and RsmE repressor proteins of P. fluorescens SS101. The aprA and hcnA genes of P. protegens CHA0 and P. aeruginosa PAO1 were used as references. The translation initiation ATG codon is shown at the 3′ end. B. Alignment of the regions upstream of LuxR-type transcriptional regulatory genes that flank different lipopeptide biosynthesis gene clusters in Pseudomonas fluorescens SS101, Pseudomonas sp. MIS38, P. fluorescens Pf0-1, P. protegens Pf-5, P. fluorescens SBW25, P. synxantha BG33R, P. putida PCL1445, P. entomophila L48 and Pseudomonas syringae pv. tomato DC3000. The translation initiation ATG codon is shown at the 3′ end.
Other genes of the Rsm regulon in P. fluorescens SS101
To explore the potential roles of rsmY and rsmZ in global gene regulation in strain SS101, we conducted a genome-wide microarray analysis on the rsmYZ double mutant and the wild-type strain, both sampled in the mid-exponential growth phase (OD600 ∼ 0.6). In rsmYZ, the expression of rsmY and rsmZ was reduced 89 and 82-fold, respectively, due to the deletion of the corresponding genes. Various other significant changes in gene expression were observed with 121 and 272 genes significantly (fold change > 2.0; P < 0.001) up- and downregulated respectively (Table S4; Table S5). Next to the genes involved in massetolide biosynthesis, the chitinase encoding gene chiC (PflSS101_3606) and a gene predicted to encode a bacterioferritin family protein (PflSS101_0584) were significantly downregulated in the rsmYZ mutant. Moreover, 19 genes (PflSS101_5338–5358) homologous to the HSI-I type VI secretion system of P. aeruginosa (Mougous et al., 2006) were downregulated (Fig. 5A). Another type VI secretion system HSI-II was not differentially regulated in the rsmYZ mutant. The putative functions of these type VI secretion systems in SS101, including a role in antibacterial activity or in plant-growth promotion (Decoin et al., 2014), are yet unknown.
Figure 5.
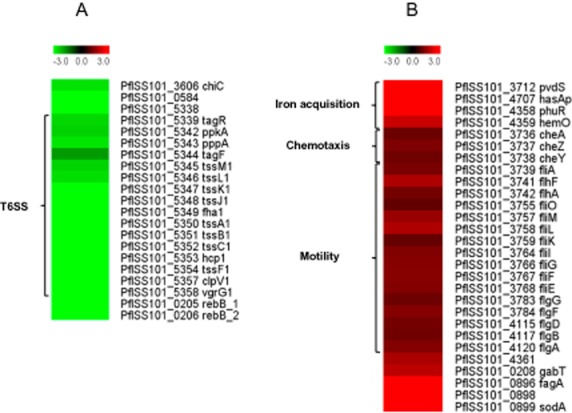
Whole genome transcriptome analysis of P. fluorescens SS101 and the ΔrsmYZ mutant. Heat maps showing significant log2-fold changes (P < 0.001) in the expression of genes in the ΔrsmYZ versus wild-type cells. Wild-type SS101 and the ΔrsmYZ mutant were grown in liquid KB at 25°C to an optical cell density of OD600 = 0.6. The fold changes shown here represent averages of three biological replicates. A represents known genes that were downregulated in the ΔrsmYZ mutant, whereas B represents known genes upregulated in the ΔrsmYZ mutant versus wild-type SS101. For a list of all genes differentially regulated in the ΔrsmYZ mutant versus wild-type SS101, we refer to Tables S2 and S3.
Transcriptomic analysis also revealed that rebB_1 (PflSS101_0205) and rebB_2 (PflSS101_0206) were downregulated more than 44-fold and 93-fold, respectively, in the rsmYZ mutant (Table S3). For certain endosymbionts, such as Caedibacter in Paramecium, these genes have been reported to encode insoluble proteins referred to as refractile bodies (R bodies) (Schrallhammer et al., 2012). It has been noted that R bodies unwind under certain conditions and are associated with toxicity, i.e. the ability to kill symbiont-free competitors. For free-living bacteria, including P. fluorescens SS101, the functions of these R bodies are not known yet. Given that not all downregulated genes in rsmYZ double mutant harbour the conserved motif 5′-A/U CANGGANGU/A-3′ in the ribosome-binding site (data not shown), we postulate that the altered expression of these genes might be due to indirect regulation by the Rsm regulon as was reported for P. aeruginosa (Brencic and Lory, 2009).
Genes upregulated in the rsmYZ mutant represent genes involved in iron acquisition, chemotaxis and cell motility (Fig. 5B). Also, gabT (PflSS101_0208), which is involved in γ-aminobutyric acid utilization, was upregulated in the rsmYZ mutant. Upregulation was also found for three genes of the fagA-fumC-orfX-sodA operon (PflSS101_0896, 0898, 0899) (Fig. 5B), which functions in oxidative stress adaptation in P. aeruginosa (Polack et al., 1996; Hassett et al., 1997a,b).
Comparison of the Rsm regulon and the Gac regulon of P. fluorescens SS101
Many of the genes differentially regulated in the rsmYZ mutant of strain SS101 have also been reported previously to be differentially expressed in Gac mutants of other Pseudomonas species and strains (Brencic et al., 2009; Hassan et al., 2010; Cheng et al., 2013; Wang et al., 2013). In P. aeruginosa, the GacS/GacA transduction system acts exclusively through its control over the transcription of rsmY and rsmZ (Brencic et al., 2009). However, the possibility that the system directly regulates other genes cannot be excluded for other Pseudomonas species and strains. For instance, in L. pneumophila, LetA (orthologue of GacS) regulates expression of flagellar genes by a mechanism that appears to be independent of RsmY and RsmZ (Sahr et al., 2009). In our study, comparative analyses of the Gac regulon and Rsm regulon of P. fluorescens SS101 were conducted according to Sahr and colleagues (2009). Briefly, we made a direct comparison (fold change > 2.0, P value < 0.05) of the gene expression pattern of ΔgacA and ΔrsmYZ. Additionally, we analysed genes differentially expressed in either ΔgacA/wt or in ΔrsmYZ/wt. Collectively, these analyses resulted in five genes differentially expressed in the ΔgacA mutant and 11 genes differentially expressed in the ΔrsmYZ mutant. One of the five genes (PflSS101_2039) that was differentially expressed in the ΔgacA mutant is located directly downstream of gacA. Hence, its differential expression is most likely due to a polar effect of the gac mutation. Therefore, this gene was excluded from the comparison. In summary, the expression of four and 11 genes varied in ΔgacA and ΔrsmYZ mutants respectively. One of these four genes is related to iron uptake, one is involved in amino acid transport and metabolism, and two genes are predicted to encode a hypothetical protein. The 11 genes uniquely expressed in the rsmYZ mutant (Table S6) were all significantly upregulated. One gene, encoding a secondary thiamine-phosphate synthase enzyme, showed the most increased expression (nine-fold change), but its function in strain SS101 is not known yet. In summary, this analysis suggests that most, not all, of the genes controlled by GacS/GacA two-component system are controlled via RsmY/RsmZ.
Conclusions
Through in silico analyses of the genome of the rhizobacterium P. fluorescens SS101, 16 small RNAs were identified. Subsequent experiments revealed, for the first time, that the Rsm signal transduction pathway plays a critical role in the regulation of massetolide biosynthesis, a cyclic lipopeptide important for biofilm formation, swarming motility, antimicrobial activity and induction of systemic resistance in plants. We showed that the effects of the two sRNAs RsmY and RsmZ are channeled through the RsmA and RsmE repressor proteins, and we predicted that the LuxR-type transcriptional regulator MassAR is one of the targets of these repressor proteins in strain SS101. To date, most information on the Rsm regulon in Pseudomonas species comes from studies on P. aeruginosa and P. protegens. Here, new information is provided that the Rsm system regulates lipopeptide biosynthesis in P. fluorescens SS101 and possibly other Pseudomonas species. Our study also provided, for the first time, a whole genome comparison of the Rsm and Gac regulons in a Pseudomonas species other than P. aeruginosa. The results of these analyses revealed that most but not all of the genes controlled by RsmY/RsmZ are also controlled by the GacS/GacA two-component system, whereas in P. aeruginosa, the Gac regulon controls downstream genes exclusively through the sRNAs RsmY and RsmZ.
Experimental procedures
Bioinformatic prediction of sRNAs in P. fluorescens SS101 genome
sRNA searches were performed by blast and yass (Noe and Kucherov, 2005) against the Rfam database (http://rfam.janelia.org/), as well as by erpin (Gautheret and Lambert, 2001), infernal (Nawrocki et al., 2009) and darn (Zytnicki et al., 2008), which are included in the rnaspace package (Cros et al., 2011).
Bacterial strains and cultural conditions
Bacterial strains used in this study are listed in Table 3. Pseudomonas fluorescens strains were cultured in liquid King's medium B (KB) (King et al., 1954) at 25°C. The gacS and gacA plasposon mutants were obtained with plasmid pTnModOKm (Dennis and Zylstra, 1998). Escherichia coli strain DH5α was used as a host for the plasmids used for site-directed mutagenesis. Escherichia coli strains were grown on Luria–Bertani (LB) plates or in LB broth (Bertani, 1951) amended with the appropriate antibiotics.
Table 3.
Bacterial strains and mutants used in this study
| Strain | Relative characteristics | Reference source |
|---|---|---|
| Pseudomonas fluorescens | ||
| SS101 | Wild type, Rifr | de Souza et al., 2003 |
| ΔgacS | Plasposon mutant, Kmr | This study |
| ΔgacA | Plasposon mutant, Kmr | This study |
| ΔrsmY | rsmY deletion mutant | This study |
| ΔrsmZ | rsmZ deletion mutant | This study |
| ΔrsmYZ | rsmY rsmZ deletion mutant | This study |
| ΔrsmAgacS | rsmA deletion mutant in the ΔgacS background | This study |
| ΔcsrAgacS | csrA deletion mutant in the ΔgacS background | This study |
| ΔrsmEgacS | rsmE deletion mutant in the ΔgacS background | This study |
| ΔrsmAcsrAgacS | rsmA csrA deletion mutant in the ΔgacS background | This study |
| ΔrsmArsmEgacS | rsmA rsmE deletion mutant in the ΔgacS background | This study |
| ΔcsrArsmEgacS | csrA rsmE deletion mutant in the ΔgacS background | This study |
| ΔrsmAcsrArsmEgacS | rsmA csrA rsmE deletion mutant in the ΔgacS background | This study |
Rifr: Rifampin resistance; Kmr: Kanamycin resistance.
Bacterial mutagenesis
Site-directed mutagenesis of the two small RNAs and three repressor protein genes was performed with the pEX18Tc suicide vector as described by de Bruijn and colleagues (de Bruijn et al., 2008). The primers used are listed in Table S7. For each mutant construct, two fragments were amplified: Up and down fragments. In the first-round polymerase chain reaction (PCR), the up and down fragments were amplified respectively. The first round PCR was performed with Pfu polymerase (Promega). The program used for the PCR consisting 1 min denaturation at 95°C, followed by 30 cycles of 95°C 1 min, Tm 30 s and 72°C 2 min. The last step of the PCR was 72°C for 7 min. All fragments were separated on a 1% (wt/vol) agarose gel and purified with an Illustra GFX PCR DNA and Gel Band Purification Kit. The second round PCR was performed by mixing equimolar amounts of the up and down fragments as templates, up forward and down reverse primers were added in the Pfu PCR reaction system. All fragments were separated on a 1% agarose gel, and bands of the right size were purified with a Qiagen kit. The fragments were digested with EcoRI and HindIII and cloned into pEX18Tc. Escherichia coli DH5α was transformed with pEX18TC-rsmY, pEX18TC-rsmZ, pEX18TC-rsmA, pEX18TC-csrA or pEX18TC-rsmE plasmids by heat shock transformation according to method of Inoue and colleagues (Inoue et al., 1990), and transformed colonies were selected on LB supplemented with 25 μg ml−1 tetracycline (Sigma). Integration of the inserts was verified by restriction analysis of the plasmids. The plasmid inserts were verified by sequencing (Macrogen, Amsterdam, the Netherlands). The correct pEX18Tc-rsmY and pEX18Tc-rsmZ constructs were subsequently electroporated into P. fluorescens SS101; pEX18Tc-rsmA, pEX18Tc-csrA and pEX18Tc-rsmE constructs were transformed into the ΔgacS mutant. Electrocompetent cells were obtained according to the method of Choi and colleagues (2006), and electroporation occurred at 2.4 kV and 200 μF. After incubation in SOC medium [2% Bacto tryptone (Difco), 0.5% Bacto yeast extract (Difco), 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose (pH 7)] for 2 h at 25°C, the cells were plated on KB supplemented with tetracycline (25 μg ml−1) and rifampin (50 μg ml−1). The single crossover colonies obtained were grown in LB overnight at 25°C and plated on LB supplemented 5% sucrose to accomplish the double crossover. The plates were incubated at 25°C for at least 48 h, and colonies were re-streaked on LB supplemented with tetracycline (25 μg ml−1) and on LB supplemented with 5% sucrose. Colonies that grew on LB with sucrose, but not on LB with tetracycline, were selected and subjected to colony PCR to confirm the deletion of the genes.
Lipopeptide extraction and RP-HPLC separation
Massetolide extractions and RP-HPLC analysis were conducted according to the methods described previously (de Bruijn et al., 2008; de Bruijn and Raaijmakers, 2009a). Briefly, Pseudomonas strains were grown on Pseudomonas agar plates (Pseudomonas agar 38 g l−1, glycerol 10 g l−1) for 48 h at 25°C. The cells were suspended in sterile de-mineralized water (∼ 40 ml per plate), transferred to 50 ml tubes, shaken vigorously for 2 min and then centrifuged (30 min, 6000 rpm, 4°C). The culture supernatant was transferred to a new tube and acidified to pH 2.0 with 9% HCl. The precipitate was obtained by centrifugation (30 min, 6000 rpm, 4°C) and washed three times with acidified dH2O (pH 2.0). The precipitate was re-suspended in 5 ml dH2O and the pH adjusted to 8.0 with 0.2 M NaOH; the precipitate dissolves. The solution was centrifuged (30 min, 6000 rpm, 4°C) and the supernatant transferred to a new tube and subjected to lyophilization. Analytical HPLC separations were carried out on 5 μm C18 column (Waters Symmetry column, Waters, Etten-Leur, Netherlands), a 55 min linear gradient of 0% to 100% acetonitrile + 0.1% (v/v) trifluoroacetic acid with a flow rate of 0.5 ml min−1. Detection was performed with a photodiode array detector (Waters) at wavelengths from 200 to 450 nm.
Swarming motility
Swarming motility assays of the bacterial strains and mutants were conducted according to the method described previously (de Bruijn and Raaijmakers, 2009a). Swarming motility of wild type strain SS101 and the mutants was assessed on soft (0.6% wt/vol) standard succinate agar medium (SSM) consisting of 32.8 mM K2HPO4, 22 mM KH2PO4, 7.6 mM (NH4)2SO4, 0.8 mM MgSO4 and 34 mM succinic acid and adjusted to pH 7 with NaOH. After autoclaving, the medium was cooled down in a water bath to 55°C and kept at 55°C for 1 h. Twenty millilitres of SSM was pipetted into a 9 cm diameter petri dish, and the plates were kept for 24 h at room temperature (20°C) prior to the swarming assay. For all swarming assays, the same conditions (agar temperature and volume, time period of storage of the poured plates) were kept constant to maximize reproducibility. Overnight cultures of wild-type SS101, mutants, were washed three times with 0.9% NaCl, and 5 μl of the washed cell suspension (1 × 1010 cells ml−1) was spot inoculated in the centre of the soft SSM agar plate and incubated for 48–72 h at 25°C.
Transcriptional profiling
Wild-type SS101,the ΔgacA and the ΔrsmYZ mutant were grown in King's medium B in 24-well plates, and harvested for RNA isolation at the mid-exponential growth stage (OD600 = 0.6). Cells of these strains were collected in triplicates. Total RNA was extracted with Trizol reagent (Invitrogen) and further purified with the NucleoSpin RNA kit (Macherey-Nagel). A tiling microarray for P. fluorescens SS101 was developed in the MicroArray Department (MAD), University of Amsterdam (UvA), Amsterdam, the Netherlands. In total, 134 276 probes (60 mer) were designed with, in general, a gap of 32 nucleotides between adjacent probes on the same strand and an overlap by 14 nucleotides when regarding both strands. In addition, 5000 custom negative control probes were hybridized, and used as an internal control to validate the designed probes in a comparative genomic hybridization experiment of four arrays. Probes were annotated and assembled into probe sets for known genes based on location information retrieved from the Pathosystems Resource Integration Center (http://patricbrc.org). Probes outside of known genes were labelled as InterGenic Region. Complementary DNA (cDNA) labelling was conducted as described previously (52). Briefly, cDNA was synthesized in presence of Cy3-dUTP (Cy3) for the test samples and with Cy5-dUTP (Cy5) for the common reference. The common reference was made by an equimolar pool of the test samples (3 μg per sample). Five micrograms of total RNA per reaction was used and yielded 1.5–2.5 μg cDNA for each sample with more than 16 pmol of Cy3 or Cy5 dye per microgram. Hybridizations were performed according to Pennings and colleagues (Pennings et al., 2011). Slides were washed according to the procedures described in the Nimblegen Arrays User's Guide – Gene Expression Arrays Version 5.0 and scanned in an ozone-free room with a Agilent DNA microarray scanner G2565CA (Agilent Technologies). Feature extraction was performed with NimbleScan v2.5 (Roche Nimblegen). Data pre-processing consisted of log2-transformation of the raw probe-intensity data, followed by a within slide Lowess normalization. Thus, normalized sample (Cy3) channel intensities were summarized into probe sets values and normalized between arrays using the Robust Multi-Array Analysis algorithm (Irizarry, et al. 2003). All results described were found to be significant using a false discovery rate of less than 5%. The Arraystar 12 software (DNASTAR, Madison, Wisconsin, USA) was used for analysing the pre-normalized array data. Statistical analyses were carried out with the normalized data using a moderated t-test to determine differential transcript abundance. Genes with a fold change > 2 and P-value < 0.05 were considered to be differentially regulated.
Acknowledgments
We thank the MAD lab: Dutch Genomics Service & Support Provider for conducting the microarray analysis. This publication is No.5680 of the Netherlands Institute of Ecology (NIOO-KNAW).
Conflict of interest
The authors of this manuscript have no conflicts of interest to declare.
Supporting Information
Fig. S1. Phylogenetic analyses of RsmA/CsrA-like proteins in different Pseudomonas species and strains. The phylogenetic tree is based on amino acid sequences of RsmA, RsmE and CsrA from 23 bacterial genomes, and was generated by neighbor-joining (NJ) (Saitou and Nei, 1987) in MEGA 6 (Tamura et al., 2013). The evolutionary distances were computed using Jones–Taylor–Thornton (JTT) model. The variation rate among sites was modelled with a gamma distribution. Bootstrap values (1000 repetitions) are shown on branches. Rsm proteins from P. fluorescens strain SS101 are indicated in bold.
Table S1. tRNA and rRNA in SS101.
Table S2. Whole genome transcriptome analysis of ΔgacS/wt.
Table S3. Whole genome transcriptome analysis of ΔgacA/wt.
Table S4. Whole genome transcriptome analysis of ΔrsmYZ/wt, up-regulated genes with P < 0.001, fold change > 2.
Table S5. Whole genome transcriptome analysis of ΔrsmYZ/wt, down-regulated genes with P < 0.001, fold change > 2.
Table S6. Unique expression genes in ΔgacA and ΔrsmYZ mutants.
Table S7. Primers used for in frame deletion mutagenesis of the small RNAs and repressor proteins.
References
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer C, Heeb S, Pessi G. Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A. Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL. Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn I. Raaijmakers JM. Regulation of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens by the ClpP protease. J Bacteriol. 2009a;191:1910–1923. doi: 10.1128/JB.01558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn I. Raaijmakers JM. Diversity and functional analysis of LuxR-type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl Environ Microbiol. 2009b;75:4753–4761. doi: 10.1128/AEM.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn I, de Kock MJD, de Waard P, van Beek TA. Raaijmakers JM. Massetolide a biosynthesis in Pseudomonas fluorescens. J Bacteriol. 2008;190:2777–2789. doi: 10.1128/JB.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, de Bruijn I, van der Voort M, Loper JE. Raaijmakers JM. The Gac regulon of Pseudomonas fluorescens SBW25. Environ Microbiol Rep. 2013;5:608–619. doi: 10.1111/1758-2229.12061. [DOI] [PubMed] [Google Scholar]
- Choi KH, Kumar A. Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Cros MJ, de Monte A, Mariette J, Bardou P, Grenier-Boley B, Gautheret D, et al. RNAspace.org: an integrated environment for the prediction, annotation, and analysis of ncRNA. RNA. 2011;17:1947–1956. doi: 10.1261/rna.2844911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoin V, Barbey C, Bergeau D, Latour X, Feuilloley MGJ, Orange N. Merieau A. A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS ONE. 2014;9:e89411. doi: 10.1371/journal.pone.0089411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JJ. Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JC. Brown JW. The RNase P family. RNA Biol. 2009;6:362–369. doi: 10.4161/rna.6.4.9241. [DOI] [PubMed] [Google Scholar]
- Fortune DR, Suyemoto M. Altier C. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangipani E, Visaggio D, Heeb S, Kaever V, Camara M, Visca P. Imperi F. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ Microbiol. 2014;16:676–688. doi: 10.1111/1462-2920.12164. [DOI] [PubMed] [Google Scholar]
- Gautheret D. Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol. 2001;313:1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T. Haas D. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics. 2008;9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, et al. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LD, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ Microbiol. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- Hassett DJ, Howell ML, Sokol PA, Vasil ML. Dean GE. Fumarase C activity is elevated in response to iron deprivation and in mucoid, alginate-producing Pseudomonas aeruginosa: cloning and characterization of fumC and purification of native fumC. J Bacteriol. 1997a;179:1442–1451. doi: 10.1128/jb.179.5.1442-1451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DJ, Howell ML, Ochsner UA, Vasil ML, Johnson Z. Dean GE. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J Bacteriol. 1997b;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Blumer C. Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002a;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Blumer C. Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002b;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nojima H. Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U. Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kay E, Dubuis C. Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci USA. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, et al. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK. Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- Koch HG, Hengelage T, Neumann-Haefelin C, MacFarlane J, Hoffschulte HK, Schimz KL, et al. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol Biol Cell. 1999;10:2163–2173. doi: 10.1091/mbc.10.7.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P. Haas D. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol. 2007;66:341–356. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FHT. Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cui YY, Mukherjee A. Chatterjee AK. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol. 1998;29:219–234. doi: 10.1046/j.1365-2958.1998.00924.x. [DOI] [PubMed] [Google Scholar]
- Livny J, Fogel MA, Davis BM. Waldor MK. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res. 2005;33:4096–4105. doi: 10.1093/nar/gki715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Brencic A, Lory S. Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK. Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Mazzola M, de Bruijn I, Cohen MF. Raaijmakers JM. Protozoan-induced regulation of cyclic lipopeptide biosynthesis is an effective predation defense mechanism for Pseudomonas fluorescens. Appl Environ Microbiol. 2009;75:6804–6811. doi: 10.1128/AEM.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel JE, Ha T, Govers F. Raaijmakers JM. Cellular responses of the late blight pathogen Phytophthora infestans to cyclic lipopeptide surfactants and their dependence on G proteins. Appl Environ Microbiol. 2009;75:4950–4957. doi: 10.1128/AEM.00241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel JE, de Vos RCH, Dekkers E, Pineda A, Guillod L, Bouwmeester K, et al. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012;160:2173–2188. doi: 10.1104/pp.112.207324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL. Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe L. Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33:W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings JL, Rodenburg W, Imholz S, Koster MP, van Oostrom CT, Breit TM, et al. Gene expression profiling in a mouse model identifies fetal liver- and placenta-derived potential biomarkers for Down Syndrome screening. PLoS ONE. 2011;6:e18866. doi: 10.1371/journal.pone.0018866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C. Felden B. Proteins that interact with bacterial small RNA regulators. FEMS Microbiol Rev. 2007;31:614–625. doi: 10.1111/j.1574-6976.2007.00079.x. [DOI] [PubMed] [Google Scholar]
- Polack B, Dacheux D, Delic-Attree I, Toussaint B. Vignais PM. The Pseudomonas aeruginosa fumC and sodA genes belong to an iron-responsive operon. Biochem Biophys Res Commun. 1996;226:555–560. doi: 10.1006/bbrc.1996.1393. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, de Bruijn I, Nybroe O. Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- Reimmann C, Valverde C, Kay E. Haas D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in Biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol. 2005;187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad MA, Larsen N, Samuelsson T. Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C. Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009;72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JAM, Mendieta MES, et al. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci USA. 2010;107:15223–15228. doi: 10.1073/pnas.1007805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrallhammer M, Galati S, Altenbuchner J, Schweikert M, Gortz HD. Petroni G. Tracing the role of R-bodies in the killer trait: absence of toxicity of R-body producing recombinant E. coli on paramecia. Eur J Protistol. 2012;48:290–296. doi: 10.1016/j.ejop.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Sharma CM. Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E. Haas D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl Microbiol Biotechnol. 2011;91:63–79. doi: 10.1007/s00253-011-3332-1. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, et al. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- de Souza JT, de Boer M, de Waard P, van Beek TA. Raaijmakers JM. Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl Environ Microbiol. 2003;69:7161–7172. doi: 10.1128/AEM.69.12.7161-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar J. Gunasekaran P. Computational small RNA prediction in bacteria. Bioinform Biol Insights. 2013;7:83–95. doi: 10.4137/BBI.S11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Ficke A, Asiimwe T, Hofte M. Raaijmakers JM. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytologist. 2007;175:731–742. doi: 10.1111/j.1469-8137.2007.02138.x. [DOI] [PubMed] [Google Scholar]
- Vallet-Gely I, Novikov A, Augusto L, Liehl P, Bolbach G, Pechy-Tarr M, et al. Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl Environ Microbiol. 2010;76:910–921. doi: 10.1128/AEM.02112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde C, Heeb S, Keel C. Haas D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol. 2003;50:1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- Wang D, Lee SH, Seeve C, Yu JM, Pierson LS., 3rd Pierson EA. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30–84. Microbiologyopen. 2013;2:505–524. doi: 10.1002/mbo3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a regulator of transcription. Mol Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA. Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, et al. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci USA. 2013;110:E3487–E3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zytnicki M, Gaspin C. Schiex T. DARN! A weighted constraint solver for RNA motif localization. Constraints. 2008;13:91–109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic analyses of RsmA/CsrA-like proteins in different Pseudomonas species and strains. The phylogenetic tree is based on amino acid sequences of RsmA, RsmE and CsrA from 23 bacterial genomes, and was generated by neighbor-joining (NJ) (Saitou and Nei, 1987) in MEGA 6 (Tamura et al., 2013). The evolutionary distances were computed using Jones–Taylor–Thornton (JTT) model. The variation rate among sites was modelled with a gamma distribution. Bootstrap values (1000 repetitions) are shown on branches. Rsm proteins from P. fluorescens strain SS101 are indicated in bold.
Table S1. tRNA and rRNA in SS101.
Table S2. Whole genome transcriptome analysis of ΔgacS/wt.
Table S3. Whole genome transcriptome analysis of ΔgacA/wt.
Table S4. Whole genome transcriptome analysis of ΔrsmYZ/wt, up-regulated genes with P < 0.001, fold change > 2.
Table S5. Whole genome transcriptome analysis of ΔrsmYZ/wt, down-regulated genes with P < 0.001, fold change > 2.
Table S6. Unique expression genes in ΔgacA and ΔrsmYZ mutants.
Table S7. Primers used for in frame deletion mutagenesis of the small RNAs and repressor proteins.