Abstract
2-Ketoisocaproate (KIC), the last intermediate in l-leucine biosynthesis, has various medical and industrial applications. After deletion of the ilvE gene for transaminase B in l-leucine production strains of Corynebacterium glutamicum, KIC became the major product, however, the strains were auxotrophic for l-isoleucine. To avoid auxotrophy, reduction of IlvE activity by exchanging the ATG start codon of ilvE by GTG was tested instead of an ilvE deletion. The resulting strains were indeed able to grow in glucose minimal medium without amino acid supplementation, but at the cost of lowered growth rates and KIC production parameters. The best production performance was obtained with strain MV-KICF1, which carried besides the ilvE start codon exchange three copies of a gene for a feedback-resistant 2-isopropylmalate synthase, one copy of a gene for a feedback-resistant acetohydroxyacid synthase and deletions of ltbR and iolR encoding transcriptional regulators. In the presence of 1 mM l-isoleucine, MV-KICF1 accumulated 47 mM KIC (6.1 g l−1) with a yield of 0.20 mol/mol glucose and a volumetric productivity of 1.41 mmol KIC l−1 h−1. Since MV-KICF1 is plasmid free and lacks heterologous genes, it is an interesting strain for industrial application and as platform for the production of KIC-derived compounds, such as 3-methyl-1-butanol.
Introduction
Corynebacterium glutamicum is the major host for biotechnological production of amino acids, the most important ones being the flavor enhancer l-glutamate and the feed additive l-lysine. In the past decades, C. glutamicum strains have been developed for the production of various other commercially interesting compounds (Becker and Wittmann, 2012), including organic acids (Okino et al., 2008; Litsanov et al., 2012a,b; Wieschalka et al., 2013), diamines (Mimitsuka et al., 2007; Kind and Wittmann, 2011; Schneider and Wendisch, 2011) or alcohols (Inui et al., 2004; Smith et al., 2010; Blombach et al., 2011). Besides small molecules, also heterologous proteins can be efficiently produced with this Gram-positive bacterium (Scheele et al., 2013, and references therein). Thus, C. glutamicum has become a production platform in white biotechnology. Three monographs (Eggeling and Bott, 2005; Burkovski, 2008; Yukawa and Inui, 2013) document the rapidly increasing knowledge on this species, which is based on the genome sequence (Ikeda and Nakagawa, 2003; Kalinowski et al., 2003) and efficient techniques for its genetic engineering (Kirchner and Tauch, 2003).
The spectrum of amino acids produced with C. glutamicum includes the essential branched-chain amino acids (BCAAs) l-valine, l-isoleucine and l-leucine, which are produced in quantities of up to 5000 tons per year in a steadily growing market (Becker and Wittmann, 2012). They have different applications in the food, feed and pharmaceutical industry (Park and Lee, 2010). The biosynthesis pathways of the BCAAs in C. glutamicum are overlapping and partly share the same precursors and enzymes (Fig. 1). The direct precursors of l-valine, l-isoleucine and l-leucine are 2-ketoisovalerate (KIV), 2-keto-3-methylvalerate (KMV), and 2-ketoisocaproate (KIC) respectively. These keto acids are predominantly transaminated to the respective amino acids by the transaminase IlvE (Radmacher et al., 2002; Marienhagen et al., 2005). Similar to their corresponding amino acids, KIV, KMV and KIC have a variety of applications in the medical, biological and food area, since they play an important role in living organisms as regulatory factors in metabolism and key intermediates in biosynthesis (Krause et al., 2010; Zhu et al., 2011; Bückle-Vallant et al., 2014). They are used, for example, in the therapy of chronic kidney disease patients (Aparicio et al., 2012). Similar to l-leucine, KIC has anti-catabolic properties through inhibition of muscle proteolysis and provokes enhancement of protein synthesis, especially in the skeletal muscle (Escobar et al., 2010; Zanchi et al., 2011). Additionally, an insulin-releasing action of KIC (Heissig et al., 2005) and an inhibitory effect on glucagon release (Leclercq-Meyer et al., 1979) were discussed. It has been shown that KIC can also serve as a basis for the production of the biofuel isopentanol (Cann and Liao, 2010).
Figure 1.
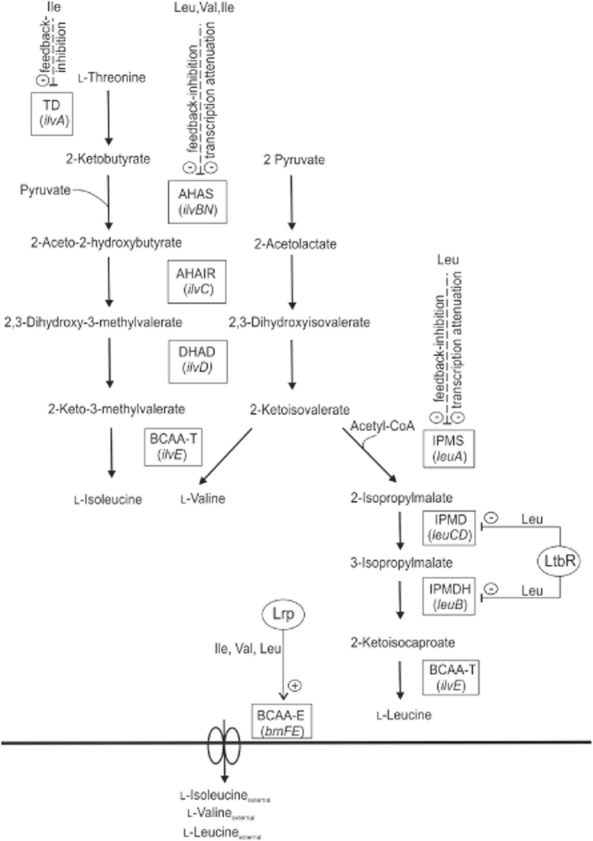
Biosynthesis pathways and their control by various regulatory mechanisms of the three branched-chain amino acids and the respective keto acids in C. glutamicum. Enzymes and their corresponding genes are shown in boxes. Lines with ‘+’ indicate activation of gene expression; ‘-’ indicates repression of gene expression (solid lines) or transcription attenuation or feedback inhibition (dashed lines). ‘Leu’, ‘Val’ and ‘Ile’ indicate the presence of l-leucine, l-valine and l-isoleucine respectively. Not shown is the avtA gene encoding the branched-chain amino acid transaminase AvtA, which predominantly transaminates 2-ketoisovalerate to l-valine. Abbreviations: AHAIR, acetohydroxyacid isomeroreductase; AHAS, acetohydroxyacid synthase; BCAA-E, branched-chain amino acid exporter (BrnFE); BCAA-T, branched-chain amino acid transaminase IlvE; DHAD, dihydroxyacid dehydratase; IPMD, 3-isopropylmalate dehydratase; IPMDH, 3-isopropylmalate dehydrogenase; IPMS, 2-isopropylmalate synthase; Lrp, leucine-responsive regulatory protein; LtbR, leucine and tryptophane biosynthesis regulator; TD, threonine dehydratase (threonine ammonia-lyase).
KIV, KMV and KIC are mainly produced by chemical synthesis using harsh reaction conditions and multiple purification steps resulting in plenty of waste (Cooper et al., 2010). The biotechnological production of these keto acids is thus an interesting alternative. Besides a biotransformation process with Rhodococcus opacus using l-leucine as substrate for KIC formation (Zhu et al., 2011), fermentative processes with glucose as substrate have recently been described for the production of KIV (Krause et al., 2010) and KIC (Bückle-Vallant et al., 2014), showing that deletion of ilvE in certain engineered C. glutamicum strains results in KIC formation. Whereas these strains contained plasmids and in part heterologous genes, the C. glutamicum KIC production strains developed in our work are plasmid free and lack heterologous genes.
Results and discussion
Initial studies on KIC production using plasmid-containing strains of C. glutamicum
Based on recently developed efficient production strains of C. glutamicum ATCC 13032 (Abe et al., 1967) for l-leucine (Vogt et al., 2014), we intended to modify these strains for the production of KIC. The conversion of KIC to l-leucine is catalysed by the transaminase IlvE, which also converts KIV to l-valine and KMV to l-isoleucine using l-glutamate as amino donor (Radmacher et al., 2002; Marienhagen et al., 2005). An ilvE deletion has been reported to cause auxotrophy for l-leucine and l-isoleucine, but not for l-valine, since the transaminase AvtA also effectively converts KIV to l-valine using l-alanine as amino donor (Marienhagen et al., 2005). According to this knowledge, deletion of ilvE in l-leucine production strains should lead to the accumulation of KIC and potentially also KMV. In a first series of experiments, we deleted ilvE in the wild-type C. glutamicum ATCC 13032 and transformed the ΔilvE mutant with plasmid pAN6-leuA_B018, carrying an IPTG-inducible leuA allele encoding a feedback-resistant 2-isopropylmalate synthase (IPMS) (Vogt et al., 2014). 2-Isopropylmalate synthase of C. glutamicum is strongly inhibited by l-leucine with a Ki of 0.4 mM (Pátek et al., 1994) and the presence of a feedback-resistant variant is the key for l-leucine overproduction (Vogt et al., 2014). The ΔilvE mutant and the ΔilvE strain with plasmid pAN6-leuA_B018 were cultivated in 500 ml baffled Erlenmeyer flasks with 50 ml CGXII minimal medium (Keilhauer et al., 1993) with 4% (w/v) glucose, 1 mM l-leucine and 1 mM l-isoleucine at 30°C and 120 rpm on a rotary shaker. Keto acids and amino acids were quantified by high-performance liquid chromatography as described (Vogt et al., 2014). Chromosomal in-frame deletions and integrations of DNA fragments were performed by two-step homologous recombination using the vector pK19mobsacB (Schäfer et al., 1994) and a method described previously (Niebisch and Bott, 2001).
Corynebacterium glutamicum ΔilvE exhibited a growth rate of 0.38 ± 0.01 h−1 and excreted up to 5 mM KIV but no detectable concentrations of KIC (detection limit < 0.1 mM), whereas C. glutamicum ΔilvE pAN6-leuA_B018 showed a growth rate of 0.30 ± 0.01 h−1 and accumulated 37 ± 0.7 mM KIC in the supernatant when induced with 0.1 mM IPTG, confirming that overexpression of the leuA allele encoding the feedback-resistant IPMS increased metabolic flux into the leucine pathway (Fig. 1). Surprisingly, C. glutamicum ΔilvE carrying pAN6-leuA_B018 also accumulated l-leucine (12.3 ± 0.4 mM) and in fact was only auxotrophic for l-isoleucine, but not for l-leucine. A possible limitation of l-valine due to a high metabolic flux from KIV towards KIC was excluded for this strain since additional supplementation of l-valine did not improve growth (data not shown). Accumulation of l-leucine was also reported for other KIC-producing ΔilvE strains and explained by the activity of unspecific transaminases (e.g. AlaT or AvtA) using KIC as substrate when it is present in high concentrations (Bückle-Vallant et al., 2014). Consequently, supplementation of the medium with l-leucine was omitted in the following cultivations. The formation of l-leucine as by-product additionally necessitates the presence of feedback-resistant IPMS for KIC overproduction. The results described above demonstrated that our previously described l-leucine producers (Vogt et al., 2014) can serve as basis for the construction of KIC production strains.
Deletion of ilvE in plasmid-free l-leucine production strains
Analogous to our strategy used for l-leucine strain development (Vogt et al., 2014), we intended to construct KIC production strains devoid of plasmids, heterologous genes and auxotrophies. Depending on the composition of the medium used in the fermentation process, auxotrophies can necessitate the addition of supplements, increasing the costs of the fermentation process. Plasmids usually necessitate the addition of antibiotics to the medium, which is undesirable for production strains applied in the food and feed industry and can be prohibited by regulatory authorities (Tauch et al., 2002). Moreover, the absence of plasmids, antibiotic resistance markers and heterologous genes often results in more stable producer strains (Pátek, 2007). The use of heterologous genes is also an undesired trait for strains used in the food and feed industry. In a first attempt to construct a plasmid-free KIC producer, we deleted the ilvE gene in the previously constructed l-leucine producer MV-Leu20 (Table 1; Vogt et al., 2014), which contains a deletion of the ltbR gene, encoding a repressor of the l-leucine biosynthesis genes, and a replacement of the wild-type leuA gene by the feedback-resistant variant leuA_B018 under control of the strong tuf promoter (Vogt et al., 2014). In shake flask cultivations with CGXII medium containing 4% (w/v) glucose, MV-Leu20 accumulated about 20 mM l-leucine. When cultivated in the same medium supplemented with 1 mM l-isoleucine, the strain MV-Leu20 ΔilvE accumulated 18.0 ± 1.6 mM KIC in the supernatant and formed as by-products 5.6 ± 0.3 mM l-leucine, 2.1 ± 0.5 mM KIV and 7.3 ± 1.7 mM KMV. Since KIV and KMV are substrates of the transaminase IlvE (Marienhagen et al., 2005), the ilvE deletion leads to an accumulation of these keto acids. The presumably low concentrations of l-isoleucine and l-valine in strain MV-Leu20 ΔilvE may also contribute to overproduction of KIV and KMV by reducing the feedback-inhibition of threonine dehydratase (encoded by ilvA) by l-isoleucine (Möckel et al., 1992) and of acetohydroxyacid synthase (encoded by ilvBN) by l-valine and l-isoleucine (Eggeling et al., 1987).
Table 1.
| Strain or plasmid | Relevant characteristicsc | Source or reference |
|---|---|---|
| C. glutamicum strains | ||
| Wild type | ATCC 13032, biotin-auxotrophic | Abe and colleagues (1967) |
| ΔilvE | ATCC 13032 derivative with in-frame deletion of ilvE | Marienhagen and colleagues (2005) |
| MV-Leu20 | Rationally designed C. glutamicum l-leucine producer (ΔltbR ΔleuA::Ptuf-leuA_B018) | Vogt and colleagues (2014) |
| MV-Leu20 ΔilvE | MV-Leu20 derivative with in-frame deletion of ilvE | This study |
| SH-KIC20 | MV-Leu20 derivative with chromosomal replacement of ATG start codon of ilvE by GTG start codon | This study |
| MV-LeuF1 | Rationally designed C. glutamicum l-leucine producer (ΔltbR::Ptuf-leuA_B018 ΔleuA::Ptuf-leuA_B018 IR(cg1121/1122)::Ptuf-leuA_B018 ΔiolR ilvN_fbr) | Vogt and colleagues (2014) |
| MV-KICF1 | MV-LeuF1 derivative with chromosomal replacement of ATG start codon of ilvE by GTG start codon | This study |
| ΔilvE Δcg0018 | ΔilvE derivative with cg0018 in-frame deletion | This study |
| ΔilvE Δcg1121 | ΔilvE derivative with cg1121 in-frame deletion | This study |
| ΔilvE Δcg1219 | ΔilvE derivative with cg1219 in-frame deletion | This study |
| ΔilvE Δcg1419 | ΔilvE derivative with cg1419 in-frame deletion | This study |
| ΔilvE Δcg1658 | ΔilvE derivative with cg1658 in-frame deletion | This study |
| ΔilvE Δcg2557 | ΔilvE derivative with cg2557 in-frame deletion | This study |
| ΔilvE Δcg2676 | ΔilvE derivative with cg2676 in-frame deletion | This study |
| ΔilvE Δcg3334 | ΔilvE derivative with cg3334 in-frame deletion | This study |
| ΔilvE Δcg1121::cg1121 | ΔilvE Δcg1121 derivative with re-integrated gene cg1121 into its wild-type locus | This study |
| E. coli strains | ||
| DH5α | F− Φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | Invitrogen (Karlsruhe, Germany) |
| Plasmids | ||
| pAN6 | Kanr; E. coli/C. glutamicum shuttle vector for inducible gene expression (Ptac, lacIq, pBL1 pUC18 oriVE.coli, pBL1 oriVC.glutamicum) | Frunzke and colleagues (2008) |
| pAN6-leuA_B018 | Kanr; pAN6 derivative containing leuA allele coding for feedback-resistant 2-isopropylmalate synthase under control of the tac promoter | Vogt and colleagues (2014) |
| pAN6-leuA_B018-cg1121 | Kanr; pAN6-leuA_B018 derivative carrying additional gene coding for cg1121 along with its upstream (94 bp) and downstream (305 bp) regions | This study |
| pK19mobsacB | Kanr; vector for allelic exchange in C. glutamicum (pK18 oriVE.coli sacB lacZα) | Schäfer and colleagues (1994) |
| pK19mobsacB-ΔilvE | Kanr, pK19mobsacB derivative for in-frame deletion of gene ilvE | Marienhagen and colleagues (2005) |
| pK19mobsacB-GTG-ilvE | Kanr, pK19mobsacB derivative for replacement of ATG start codon of ilvE by GTG | This study |
| pK19mobsacB-Δcg0018 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg0018 | This study |
| pK19mobsacB-Δcg1121 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg1121 | This study |
| pK19mobsacB-Δcg1219 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg1219 | This study |
| pK19mobsacB-Δcg1419 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg1419 | This study |
| pK19mobsacB-Δcg1658 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg1658 | This study |
| pK19mobsacB-Δcg2557 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg2557 | This study |
| pK19mobsacB-Δcg2676 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg2676 | This study |
| pK19mobsacB-Δcg3334 | Kanr, pK19mobsacB derivative for in-frame deletion of gene coding for Cg3334 | This study |
| pK19mobsacB-cg1121 | Kanr, pK19mobsacB derivative for re-integration of gene coding for Cg1121 into its wild-type locus | This study |
All constructed plasmids as well as chromosomal deletions and integrations in engineered strains were verified by DNA sequencing.
Plasmid constructions were performed in E. coli DH5α. Description of plasmid constructions and used DNA oligonucleotides (Table S1) can be found in the Supporting Information.
Kanr, kanamycin resistance
As mentioned above, l-leucine formation in the absence of the transaminase IlvE is presumably due to high cytoplasmic KIC concentrations, allowing its conversion by other transaminases such as AvtA that have weak affinities for KIC (Marienhagen et al., 2005). To test this assumption, we measured the cytoplasmic KIC concentrations in the wild type and strain MV-Leu20 ΔilvE. Cells were grown in CGXII medium with 4% (w/v) glucose, harvested in the early exponential phase (optical density at 600 nm (OD600 = 5), and cytoplasmic concentrations were determined as described (Paczia et al., 2012). The internal KIC concentration of the wild type was below the detection limit of 5 μM, whereas MV-Leu20 ΔilvE accumulated approximately 2.8 mM KIC inside the cell, corresponding to a more than 500-fold increase.
Exchange of the ilvE start codon in plasmid-free l-leucine production strains
To avoid the l-isoleucine auxotrophy of strain MV-Leu20 ΔilvE, we intended to reduce IlvE activity to a value that was high enough to provide l-isoleucine for growth, but low enough to allow KIC overproduction. For this purpose, the ATG start codon of ilvE was exchanged against GTG, which should decrease the translation rate (Becker et al., 2010) of the ilvE transcript and thereby reduce the specific IlvE activity. The start codon exchange was performed in the l-leucine producers MV-Leu20 and MV-LeuF1 (Table 1). The latter strain contains (i) three copies of the leuA_B018 gene in the chromosome under control of the tuf promoter, two of them replacing ltbR and the native leuA gene, (ii) a deletion of iolR (Klaffl et al., 2013) for enhanced glucose uptake and (iii) a feedback-resistant acetohydroxyacid synthase encoded by ilvN_fbr (Vogt et al., 2014). The strains SH-KIC20 (from MV-Leu20) and MV-KICF1 (from MV-LeuF1) resulting from the ilvE start codon exchange (Table 1) were cultivated in CGXII medium with 4% (w/v) glucose to test for KIC accumulation in the supernatant (Fig. 2).
Figure 2.
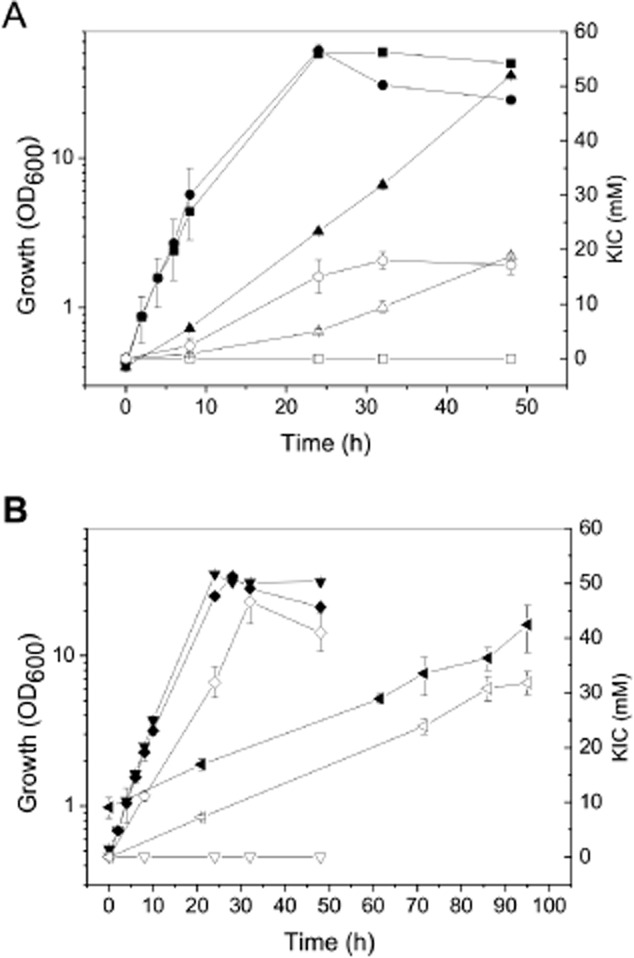
Growth and KIC formation of different C. glutamicum strains in shake flasks with CGXII minimal medium containing 4% (w/v) glucose.A. MV-Leu20 without supplements (growth, ▪; KIC, □), MV-Leu20 ΔilvE supplemented with 1 mM l-isoleucine (growth, •; KIC, ○) and SH-KIC20 without supplements (growth, ▴; KIC, △).B. MV-LeuF1 without supplements (growth, ▾; KIC, ▽), MV-KICF1 without supplements (growth, ◂; KIC, ◃) and MV-KICF1 with 1 mM l-isoleucine (growth, ♦; KIC, ⋄). The data represent mean values and standard deviations obtained from three independent cultivations.
Without supplementation of l-isoleucine, SH-KIC20 showed a strongly reduced growth rate of 0.08 ± 0.01 h−1 compared with the ancestor strain MV-Leu20 (0.31 ± 0.01 h−1). This phenotype suggests that the ilvE start codon exchange reduced the availability of BCAAs and consequently the growth rate. Strain SH-KIC20 accumulated about 19 mM KIC in 49 h, which correlates with the KIC concentration produced by MV-Leu20 ΔilvE. The slower growth of SH-KIC20 led to a lowered volumetric productivity of 0.38 mmol l−1 h−1 in comparison to MV-Leu20 ΔilvE (approximately 0.8 mmol l−1 h−1). Strain MV-KICF1 showed a growth rate of only 0.03 ± 0.01 h−1, which is 85% lower than the one of the parent strain (μ = 0.20 ± 0.01 h−1), and yielded a maximal KIC concentration of 33 mM after 95 h (Table 2). Supplementation of the medium with 1 mM l-isoleucine enabled MV-KICF1 to reach the same growth rate (μ = 0.21 ± 0.01 h−1) as its parent MV-LeuF1 and to form 47 ±q 4 mM KIC (6.1 g l−1) after 32 h with a yield of 0.20 ± 0.02 mol KIC per mol of glucose and a productivity of 1.41 ± 0.13 mmol KIC l−1 h−1 (Table 2). Both SH-KIC20 and MV-KICF1 formed KIV, KMV and l-leucine as by-products (Table 2).
Table 2.
| Parameter | MV-KICF1 + 1 mM l-isoleucine | MV-KICF1 without l-isoleucine | SH-KIC20d without l-isoleucine |
|---|---|---|---|
| Growth rate (h−1) | 0.21 ± 0.01 | 0.03 ± 0.01 | 0.08 ± 0.01 |
| KIC (mM) | 46.7 ± 4.1 | 31.8 ± 2.1 | 18.8 ± 0.67 |
| By-productsc: | |||
| KIV (mM) | 13.3 ± 2.2 | 19.0 ± 4.1 | 2.6 ± 0.3 |
| KMV (mM) | 8.8 ± 1.3 | 4.9 ± 0.6 | 8.7 ± 0.1 |
| l-leucine (mM) | 3.0 ± 0.2 | 10.3 ± 3.1 | 4.8 ± 0.2 |
| Molar product yield (mol KIC per mol glucose) | 0.204 ± 0.018 | 0.143 ± 0.010 | 0.084 ± 0.001 |
| Volumetric productivity (mmol KIC l−1 h−1) | 1.41 ± 0.13 | 0.34 ± 0.02 | 0.38 ± 0.02 |
Cultivations were performed in 500 ml baffled shake flasks containing 50 ml CGXII minimal medium with 4% (w/v) glucose. Supplementation of l-isoleucine is indicated.
Mean values and standard deviations from three independent cultivations are shown.
Concentrations of l-valine and l-isoleucine were below 2 mM.
Cultivation of SH-KIC20 supplemented with 1 mM l-isoleucine was not tested.
The results described above demonstrate that the start codon exchange for reduction of IlvE activity was successful and allowed growth and KIC accumulation without supplementation of BCAAs; however, the production parameters were lower compared with supplementation with 1 mM l-isoleucine (Table 2). A successful industrial application depends on high product yields combined with sufficient cell growth, resulting in competitive productivity. Therefore, the addition of l-isoleucine is still important to improve growth of the constructed strains to reach better productivity values. As an alternative approach to adjust the IlvE activity to an optimal value for prototrophic growth with simultaneous KIC production, ilvE gene expression could be fine-tuned by testing promoters with varying strength (Vašicová et al., 1999; Hammer et al., 2006).
Recently, Bückle-Vallant and colleagues (2014) described a plasmid-based C. glutamicum strain for the production of KIC. This strain is characterized by deletions of the genes ilvE, ltbR, prpC1 and prpC2, an exchange of the two gltA promoters (van Ooyen et al., 2011) by the mutated dapA promoter L1 (Vašicová et al., 1999) to reduce citrate synthase activity (van Ooyen et al., 2012), and plasmid-based overexpression of ilvBNCD and a leuA allele of Escherichia coli encoding a feedback-resistant IPMS. This strain accumulated up to 71 mM KIC when cultivated with glucose plus acetate as carbon sources and supplemented with 2 mM each of l-isoleucine and l-valine. Under cultivation conditions comparable to ours, i.e. without acetate, this strain reached KIC titres (54 ± 4 mM) and yields (0.22 mol per mol of glucose) in a similar range as strain MV-KICF1 when supplemented with l-isoleucine. In comparison, the biotransformation with Rhodococcus opacus transcribed by Zhu and colleagues (2011) reached about 10 mM KIC using 39 mM l-leucine as substrate.
2-Ketoisocaproate transport
When MV-KICF1 was batch cultivated in a bioreactor as described (Vogt et al., 2014) using a medium supplemented with 1 mM l-isoleucine, comparable KIC titres as in shake flasks of about 50 mM were achieved (data not shown). Interestingly, additional feeding of glucose in fed-batch experiments did not further increase the KIC concentrations and led to an arrest of cell growth and glucose consumption (data not shown). A possible explanation is a block of the l-leucine biosynthesis pathway by elevated KIC concentrations, as Bückle-Vallant and colleagues (2014) found competitive inhibition of IPMS by KIC and non-competitive inhibition of 3-isopropylmalate dehydratase by KIC. Additionally, a competitive inhibition of acetohydroxyacid synthase by KIV has been reported by Krause and colleagues (2010). To avoid a reduced flux into the leucine synthesis pathway by competitive inhibition of IPMS by KIC, the concentration of KIV within the cell needs to be increased and/or the concentration of KIC within the cell should be decreased. These concentrations are determined on one hand by the rates of synthesis and further metabolic conversion and on the other hand by the rates of export to and import from the supernatant.
Knowledge on KIC transport is very limited. Obviously, as shown by our studies and that of Bückle-Vallant and colleagues (2014), KIC can leave the cell, and previous studies by Groeger and Sahm (1987) demonstrated that KIC can enter the cell. As previously described for l-isoleucine (Zittrich and Krämer, 1994), passive diffusion, carrier-mediated uptake and carrier-mediated excretion must be considered as possibilities for an amphiphilic solute like KIC to cross the cytoplasmic membrane. Due to its similarity to l-isoleucine, it seems likely that also KIC is able to diffuse across the membrane. However, a necessity to possess carriers for KIC import or KIC export is not obvious, in contrast to the advantage of having importers for amino acids and exporters for non-catabolizable amino acids. In C. glutamicum, the export of BCAAs and methionine is catalysed by the exporter BrnFE (Kennerknecht et al., 2002; Trötschel et al., 2005; Xie et al., 2012), but evidence is available that BrnFE is not involved in KIC export (Radespiel, 2010). The identification of transporters can be beneficial for biotechnological processes to increase productivity since transport of desired products into the medium often represents a bottleneck. For example, export was identified as a limiting factor for l-isoleucine production with C. glutamicum (Morbach et al., 1996), and the production of this BCAA was improved by overexpression of the respective transporter encoded by brnFE (Kennerknecht et al., 2002; Xie et al., 2012).
In order to test if a KIC exporter is present in C. glutamicum that might be useful to improve KIC overproduction, we searched for genes showing increased mRNA levels during KIC production by performing comparative transcriptome analyses using DNA microarrays as described previously (Vogt et al., 2014). KIC producer strain MV-Leu20 ΔilvE was compared with the wild type to determine differentially expressed genes, resulting in a list of eight candidate transporter genes (Table 3). Each of these genes was deleted in the C. glutamicum ΔilvE background, and the resulting double deletion mutants were transformed with pAN6-leuA_B018. When cultivated in glucose minimal medium with 0.1 mM IPTG, one of the eight strains, which contained a deletion of cg1121 (annotated as putative permease of the major facilitator superfamily), showed a reduced growth rate (0.23 ± 0.01 h−1) and a reduced maximal KIC titre (22.4 mM) compared with the reference strain ΔilvE pAN6-leuA_B018 (0.30 ± 0.01 h−1, 37 mM KIC). The phenotype could be complemented by reintegration of cg1121 into the genome of strain ΔilvE Δcg1121 or by plasmid-borne expression of cg1121 (Fig. 3). However, the specific KIC export rates (determined as described by Kennerknecht et al., 2002) of strain ΔilvE Δcg1121 (6.5 ± 1.0 nmol min−1 gCDW−1) were not significantly reduced compared with that of the reference strain ΔilvE (8.0 ± 1.5 nmol min−1 gCDW−1) and the cytoplasmic KIC concentrations of the two strains at an OD600 of 5 were comparable. Therefore, the role of Cg1121 for growth and KIC production remains unclear and needs further investigations.
Table 3.
Putative transporter genes showing increased expression in a KIC producer
| Gene | Annotation | mRNA ratioa (MV-Leu20 ΔilvE/ Wild type | TMHb |
|---|---|---|---|
| cg0018 | putative membrane protein, conserved | 3.5 | 9 |
| cg1121 | putative permease of the major facilitator superfamily | 2.2 | 7 |
| cg1219 | putative membrane protein | 3.5 | 10 |
| cg1419 | putative Na+-dependent transporter, bile acid:Na+ symporter BASS family | 7.2 | 8 |
| cg1658 | putative permease of the major facilitator superfamily | 35.6 | 12 |
| cg2557 | putative secondary Na+/bile acid symporter, bile acid:Na+ symporter BASS family | 2.5 | 8 |
| cg2676 | putative ABC-type dipeptide/oligopeptide/nickel transport system, permease component | 2.1 | 6 |
| cg3334 | putative arabinose efflux permease, MFS type | 2.0 | 12 |
Transcriptome analyses of KIC producer MV-Leu20 ΔilvE in comparison to the wild type were performed using DNA microarrays as described (Vogt et al., 2014). Candidate transporter genes were chosen based on an mRNA ratio (MV-Leu20 ΔilvE/wild type) of > 2, an annotation as (putative) membrane or transporter proteins and the prediction of multiple transmembrane helices in the encoded proteins. Data represent mean values of at least two (maximum four) evaluable microarray experiments (P-value < 0.05).
TMH, number of transmembrane helices predicted with the sosui engine version 1.11.
Figure 3.
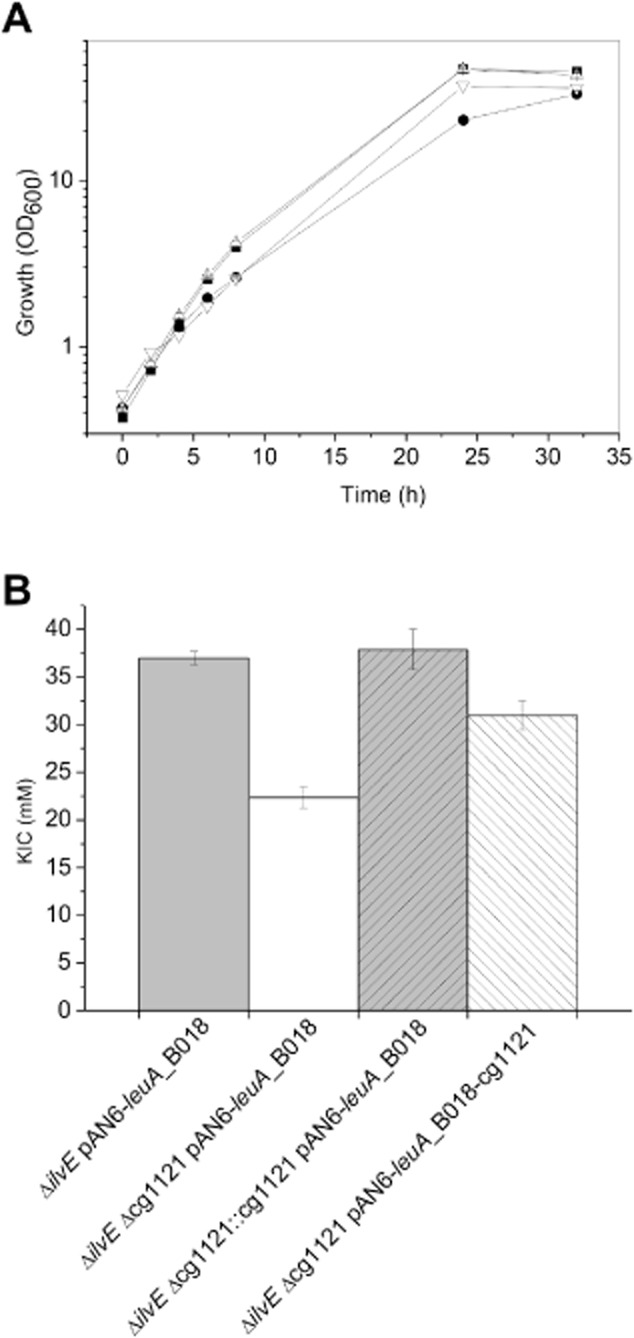
Complementation of the effects on growth and KIC accumulation caused by deletion of cg1121 in strain C. glutamicum ΔilvE carrying pAN6-leuA_B018.A. Growth of strains ΔilvE with pAN6-leuA_B018 (▪), ΔilvE Δcg1121 with pAN6-leuA_B018 (•), ΔilvE Δcg1121::cg1121 with pAN6-leuA_B018 (△) and ΔilvE Δcg1121 with pAN6-leuA_B018-cg1121 (▽) are shown.B. Maximal KIC concentrations reached after 32 h cultivation in 500 ml baffled shake flasks with 50 ml CGXII minimal medium containing 4% (w/v) glucose and 0.1 mM IPTG at 30°C and 120 rpm on a rotary shaker. The deletion of cg1121 was complemented either by genomic reintegration of cg1121 (ΔilvE Δcg1121::cg1121 pAN6-leuA_B018) or by plasmid-borne expression of cg1121 (ΔilvE Δcg1121 pAN6-leuA_B018-cg1121). The data represent mean values and standard deviations obtained from three independent cultivations.
Acknowledgments
We thank Dr Nicole Paczia (IBG-1: Biotechnology, Forschungszentrum Jülich, Germany) for her help with the measurements of cytoplasmic 2-ketoisocaproate concentrations.
Conflict of interest
None declared.
Supporting Information
Table S1. DNA oligonucleotides used in this study.
References
- Abe S, Takayama KI. Kinoshita S. Taxonomical studies on glutamic acid-producing bacteria. J Gen Appl Microbiol. 1967;13:279–301. [Google Scholar]
- Aparicio M, Bellizzi V, Chauveau P, Cupisti A, Ecder T, Fouque D, et al. Keto acid therapy in predialysis chronic kidney disease patients: final consensus. J Ren Nutr. 2012;22:S22–S24. doi: 10.1053/j.jrn.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Becker J. Wittmann C. Systems and synthetic metabolic engineering for amino acid production – the heartbeat of industrial strain development. Curr Opin Biotechnol. 2012;23:718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- Becker J, Buschke N, Bücker R. Wittmann C. Systems level engineering of Corynebacterium glutamicum – reprogramming translational efficiency for superior production. Eng Life Sci. 2010;10:430–438. [Google Scholar]
- Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF. Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovski A. Corynebacteria: Genomics and Molecular Biology. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- Bückle-Vallant V, Krause FS, Messerschmidt S. Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisocaproate production. Appl Microbiol Biotechnol. 2014;98:297–311. doi: 10.1007/s00253-013-5310-2. [DOI] [PubMed] [Google Scholar]
- Cann AF. Liao JC. Pentanol isomer synthesis in engineered microorganisms. Appl Microbiol Biotechnol. 2010;85:893–899. doi: 10.1007/s00253-009-2262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJ, Ginos JZ. Meister A. Synthesis and properties of the α-keto acids. Chem Rev. 1983;83:321–358. [Google Scholar]
- Eggeling L. Bott M. Handbook of. Boca Raton, FL, USA: Taylor & Francis; 2005. Corynebacterium glutamicum. [Google Scholar]
- Eggeling I, Cordes C, Eggeling L. Sahm H. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to l-isoleucine. Appl Microbiol Biotechnol. 1987;25:346–351. [Google Scholar]
- Escobar J, Frank JW, Suryawan A, Nguyen HV, Van Horn CG, Hutson SM. Davis TA. Leucine and α-ketoisocaproic acid, but not norleucine, stimulate skeletal muscle protein synthesis in neonatal pigs. J Nutr. 2010;140:1418–1424. doi: 10.3945/jn.110.123042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frunzke J, Engels V, Hasenbein S, Gätgens C. Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol. 2008;67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger U. Sahm H. Microbial production of l-leucine from α-ketoisocaproate by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1987;25:352–356. [Google Scholar]
- Hammer K, Mijakovic I. Jensen PR. Synthetic promoter libraries – tuning of gene expression. Trends Biotechnol. 2006;24:53–55. doi: 10.1016/j.tibtech.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Heissig H, Urban KA, Hastedt K, Zunkler BJ. Panten U. Mechanism of the insulin-releasing action of α-ketoisocaproate and related α-keto acid anions. Mol Pharmacol. 2005;68:1097–1105. doi: 10.1124/mol.105.015388. [DOI] [PubMed] [Google Scholar]
- Ikeda M. Nakagawa S. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol. 2003;62:99–109. doi: 10.1007/s00253-003-1328-1. [DOI] [PubMed] [Google Scholar]
- Inui M, Kawaguchi H, Murakami S, Vertes AA. Yukawa H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol. 2004;8:243–254. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- Keilhauer C, Eggeling L. Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerknecht N, Sahm H, Yen MR, Patek M, Saier Jr MH., Jr Eggeling L. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol. 2002;184:3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind S. Wittmann C. Bio-based production of the platform chemical 1,5-diaminopentane. Appl Microbiol Biotechnol. 2011;91:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- Kirchner O. Tauch A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J Biotechnol. 2003;104:287–299. doi: 10.1016/s0168-1656(03)00148-2. [DOI] [PubMed] [Google Scholar]
- Klaffl S, Brocker M, Kalinowski J, Eikmanns BJ. Bott M. Complex regulation of the phosphoenolpyruvate carboxykinase gene pck and characterization of its GntR-type regulator IolR as a repressor of myo-inositol utilization genes in Corynebacterium glutamicum. J Bacteriol. 2013;195:4283–4296. doi: 10.1128/JB.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause FS, Blombach B. Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq-Meyer V, Marchand J, Leclercq R. Malaisse WJ. Interactions of α-ketoisocaproate, glucose and arginine in the secretion of glucagon and insulin from the perfused rat pancreas. Diabetologia. 1979;17:121–126. doi: 10.1007/BF01222213. [DOI] [PubMed] [Google Scholar]
- Litsanov B, Brocker M. Bott M. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol. 2012a;78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Kabus A, Brocker M. Bott M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2012b;5:116–128. doi: 10.1111/j.1751-7915.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienhagen J, Kennerknecht N, Sahm H. Eggeling L. Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J Bacteriol. 2005;187:7639–7646. doi: 10.1128/JB.187.22.7639-7646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitsuka T, Sawai H, Hatsu M. Yamada K. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem. 2007;71:2130–2135. doi: 10.1271/bbb.60699. [DOI] [PubMed] [Google Scholar]
- Morbach S, Sahm H. Eggeling L. l-Isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl Environ Microbiol. 1996;62:4345–4351. doi: 10.1128/aem.62.12.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckel B, Eggeling L. Sahm H. Functional and structural analyses of threonine dehydratase from Corynebacterium glutamicum. J Bacteriol. 1992;174:8065–8072. doi: 10.1128/jb.174.24.8065-8072.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebisch A. Bott M. Molecular analysis of the cytochrome bc1aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch Microbiol. 2001;175:282–294. doi: 10.1007/s002030100262. [DOI] [PubMed] [Google Scholar]
- Okino S, Noburyu R, Suda M, Jojima T, Inui M. Yukawa H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008;81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- van Ooyen J, Emer D, Bussmann M, Bott M, Eikmanns BJ. Eggeling L. Citrate synthase in Corynebacterium glutamicum is encoded by two gltA transcripts which are controlled by RamA, RamB, and GlxR. J Biotechnol. 2011;154:140–148. doi: 10.1016/j.jbiotec.2010.07.004. [DOI] [PubMed] [Google Scholar]
- van Ooyen J, Noack S, Bott M, Reth A. Eggeling L. Improved l-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol Bioeng. 2012;109:2070–2081. doi: 10.1002/bit.24486. [DOI] [PubMed] [Google Scholar]
- Paczia N, Nilgen A, Lehmann T, Gätgens J, Wiechert W. Noack S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb Cell Fact. 2012;11:122. doi: 10.1186/1475-2859-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH. Lee SY. Fermentative production of branched chain amino acids: a focus on metabolic engineering. Appl Microbiol Biotechnol. 2010;85:491–506. doi: 10.1007/s00253-009-2307-y. [DOI] [PubMed] [Google Scholar]
- Pátek M. Branched-chain amino acids. In: Wendisch VF, editor. Amino Acid Biosynthesis – Pathways, Regulation and Metabolic Engineering. Berlin, Germany: Springer; 2007. pp. 129–162. [Google Scholar]
- Pátek M, Krumbach K, Eggeling L. Sahm H. Leucine synthesis in Corynebacterium glutamicum: enzyme activities, structure of leuA, and effect of leuA inactivation on lysine synthesis. Appl Environ Microbiol. 1994;60:133–140. doi: 10.1128/aem.60.1.133-140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radespiel T. 2010. Dissertation abstract. URL http://kups.ub.uni-koeln.de/id/eprint/3156.
- Radmacher E, Vaitsikova A, Burger U, Krumbach K, Sahm H. Eggeling L. Linking central metabolism with increased pathway flux: l-valine accumulation by Corynebacterium glutamicum. Appl Environ Microbiol. 2002;68:2246–2250. doi: 10.1128/AEM.68.5.2246-2250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G. Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Scheele S, Oertel D, Bongaerts J, Evers S, Hellmuth H, Maurer KH, et al. Secretory production of an FAD cofactor-containing cytosolic enzyme (sorbitol-xylitol oxidase from Streptomyces coelicolor) using the twin-arginine translocation (Tat) pathway of Corynebacterium glutamicum. Microb Biotechnol. 2013;6:202–206. doi: 10.1111/1751-7915.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. Wendisch VF. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol. 2011;91:17–30. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- Smith KM, Cho KM. Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Götker S, Pühler A, Kalinowski J. Thierbach G. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. J Biotechnol. 2002;99:79–91. doi: 10.1016/s0168-1656(02)00159-1. [DOI] [PubMed] [Google Scholar]
- Trötschel C, Deutenberg D, Bathe B, Burkovski A. Krämer R. Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol. 2005;187:3786–3794. doi: 10.1128/JB.187.11.3786-3794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vašicová P, Pátek M, Nešvera J, Sahm H. Eikmanns B. Analysis of the Corynebacterium glutamicum dapA promoter. J Bacteriol. 1999;181:6188–6191. doi: 10.1128/jb.181.19.6188-6191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M, Haas S, Klaffl S, Polen T, Eggeling L, van Ooyen J. Bott M. Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for l-leucine overproduction. Metab Eng. 2014;22:40–52. doi: 10.1016/j.ymben.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Wieschalka S, Blombach B, Bott M. Eikmanns BJ. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol. 2013;6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Xu L, Shi J, Xu Q. Chen N. Effect of transport proteins on l-isoleucine production with the l-isoleucine-producing strain Corynebacterium glutamicum YILW. J Ind Microbiol Biotechnol. 2012;39:1549–1556. doi: 10.1007/s10295-012-1155-4. [DOI] [PubMed] [Google Scholar]
- Yukawa H. Inui M. Corynebacterium glutamicum: Biology and Biotechnology. Heidelberg, Germany: Springer; 2013. [Google Scholar]
- Zanchi NE, Gerlinger-Romero F, Guimaraes-Ferreira L, de Siqueira Filho MA, Felitti V, Lira FS, et al. HMB supplementation: clinical and athletic performance-related effects and mechanisms of action. Amino Acids. 2011;40:1015–1025. doi: 10.1007/s00726-010-0678-0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li J, Liu L, Du G. Chen J. Production of α-ketoisocaproate via free-whole-cell biotransformation by Rhodococcus opacus DSM 43250 with l-leucine as the substrate. Enzyme Microb Technol. 2011;49:321–325. doi: 10.1016/j.enzmictec.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Zittrich S. Krämer R. Quantitative discrimination of carrier-mediated excretion of isoleucine from uptake and diffusion in Corynebacterium glutamicum. J Bacteriol. 1994;176:6892–6899. doi: 10.1128/jb.176.22.6892-6899.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. DNA oligonucleotides used in this study.


