Abstract
The two-component regulatory system, SaeRS, controls expression of important virulence factors, including toxins and invasins, which contribute to the pathogenicity of Staphylococcus aureus. Previously, we conducted a transcriptomics study for identification of SaeRS regulon and found that inactivation of SaeRS dramatically enhances the transcription of a novel transcriptional regulator (SA1804). This led us to question whether SA1804 is involved in bacterial pathogenicity by regulating the expression of virulence factors. To address this question, we created sa1804, saeRS, and sa1804/saeRS double deletion mutants in a USA300 community-acquired MRSA strain, 923, and determined their impact on the pathogenicity. The deletion of sa1804 dramatically increased the cytotoxicity and enhanced the capacity of bacteria to invade into the epithelial cells (A549), whereas the deletion of saeRS eliminated the cytotoxicity and abolished the bacterial ability to invade into the epithelial cells. Moreover, the double deletions of sa1804 and saeRS appeared a similar phenotype with the saeRS null mutation. Furthermore, we determined the regulatory mechanism of SA1804 using qPCR and gel-shift approaches. Our data indicate that the novel virulence repressor SA1804 is dependent on the regulation of SaeRS. This study sheds light on the regulatory mechanism of virulence factors and allows for us further elucidate the molecular pathogenesis of S. aureus.
Keywords: S. aureus, transcriptional regulator SA1804, two-component regulatory system SaeRS, bacterial invasion, cytotoxicity
Introduction
Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA), is a major community and hospital acquired pathogen that can cause superficial infections, including skin and soft tissue lesions (Lowy, 1998), as well as life-threaten infections, such as pneumonia, endocarditis, and toxic shock syndrome (Klevens et al., 2007; Gordon and Lowy, 2008). The ability of this organism to cause such a variety of diseases partially depends on the expression of virulence factors that allow the bacteria to colonize and evade the host immune system.
S. aureus produces various surface-associated proteins to facilitate bacterial colonization and invasion (Foster and Höök, 1998). Fibronectin-binding proteins are the main surface-associated proteins of bacterial cells that function as invasins by assembling the extracellular matrix protein Fn that bridges to the host cell receptors such as α5β1-integrin, which in turn lead to bacterial internalization of host cells (Sinha et al., 1999; Fowler et al., 2000). On the other hand, the exported α-toxin is an important virulence factor of S. aureus. α-toxin can cause cell apoptosis and death via different signaling transduction pathways (Essmann et al., 2003; Liang and Ji, 2007).
Two-component signal transduction regulatory systems (TCSs) have been implicated together with other regulators to mediate the pathogenicity of S. aureus (Novick et al., 1993; Giraudo et al., 1999; Montgomery et al., 2010). The expression of virulence factors is coordinately controlled by TCSs, such as Agr (Novick, 2003), ArlRS (Fournier et al., 2001; Liang et al., 2005), and SaeRS (Giraudo et al., 1999; Liang et al., 2006), and transcriptional regulators, such as SarA (Cheung et al., 1992), Rot (McNamara et al., 2000), and Mgr (Luong et al., 2003). The SaeRS system plays a crucial role in the control of virulence gene expression (Liang et al., 2006; Rogasch et al., 2006) and in biofilm formation (Mrak et al., 2012; Olson et al., 2013). SaeRS not only up-regulates the transcription of hla, hlb, and coa (Giraudo et al., 1999), and nuclease (nuc) production in vitro (Olson et al., 2013), but also controls hla expression in vivo, as the transcriptional level of hla remarkably decreased during infection with sae mutants (Goerke et al., 2001, 2005). Moreover, the SaeRS-dependent and Agr/SarA-independent activation of hla was revealed in exudates accumulated in a guinea pig model of infection, indicating that SaeRS is able to function independently (Goerke et al., 2001, 2005). The disruption of of SaeRS abolished the S. aureus-induced apoptosis and death of epithelial cells, indicating the importance of SaeRS in modulation of bacterial toxicity (Liang et al., 2006). On the other hand, the mutation of sae eliminates the transcription and expression of fnbA and increases expression of CP5 in Newman strain and consequently decreases the internalization of S. aureus by endothelial cells (Steinhuber et al., 2003). We revealed that SaeRS also affects the expression of Efb, a bifunctional protein capable of binding to both extracellular fibrinogen (Peacock et al., 2002) and complement factor C3 (Lee et al., 2004), and the expression of SA1000, a hypothetical fibrinogen-binding protein (Liang et al., 2006). Both Efb and SA1804 contribute to the adherence and internalization of S. aureus by epithelial cells (Liang et al., 2006). Importantly, the role of the SaeRS as a virulence regulator has been demonstrated in several animal models of infection (Benton et al., 2004; Goerke et al., 2005; Liang et al., 2006; Montgomery et al., 2010).
It has been demonstrated that SaeRS activates the expression of virulence factors through directly binding to the promoter regions of some virulence gene, such as hla, emp, and map/eap by phosphorylated response regulator SaeR (Sun et al., 2010). Our previous transcriptomics study indicated that inactivation of SaeRS dramatically increases the transcription of a novel transcriptional regulator (SA1804). This led us to hypothesize that SA1804 is involved in the pathogenesis of S. aureus. In this study, we created sa1804, saeRS, and sa1804/saeRS double deletion mutants in a USA300 community-acquired MRSA strain, 923, and determined their impact on the pathogenicity.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions
S. aureus strain RN4220 was utilized as the primary recipient for allelic exchange constructs, together with a virulent clinical isolate WCUH29 (NCIMB40771) and USA300 MRSA isolate 923 (Montgomery et al., 2010), as secondary recipients for electroporation. Escherichia coli DH10B (Invitrogen) served as the host for all in vitro recombinant DNA. Bacteria were grown in Tryptic Soy broth (TSB; Difco) and on TSA agar at 37°C with appropriate antibiotics. Bacterial cell cultures were incubated at 37°C with shaking at 200 rpm.
Construction of the saeRS and/or sa1804 In-Frame Deletion Mutants, the saeRS and sa1804 Complementary Strains, and sa1804 Promoter-lux Reporter Fusion
In-frame deletion of saeRS and/or sa1804 was carried out following the pKOR1 allelic exchange protocol as described (Bae and Schneewind, 2006). The deletion in saeRS, sa1804, or saeRS/sa1804 was confirmed diagnostic PCR and DNA sequencing flanking regions of sa1804.
In order to examine whether the expression of saeRS or sa1804 in trans complements the effect of the mutation of endogenous correspondence gene, we constructed complementary plasmids, including pYH4/saeRS and pYH4/sa1804 by cloning the saeRS or sa1804 coding region (which was obtained by PCR using primers listed in Table 1) into the AscI and PmeI sites of pYH4 (Ji et al., 2004). The resulting plasmid was electroporated into the saeRSko and sa1804ko, respectively, resulting in SaeRScom, and SA1804com strains. To confirm the impact of SaeRS on the transcription of sa1804, we created a sa1804 promoter-lux promoter system as described (Yan et al., 2011). The upstream sa1804 promoter region was PCR amplified using the primers listed in Table 1, digested with EcoRI and XmaI (NEB), and cloned into same enzyme sites of pCY1006. The reformed Psa1804-lux reporter fusion was confirmed by diagnostic PCR and electroporated into the saeRSko and parental control. The Psa1804-lux reporter fusion strains were grown in TSB at 37°C with shaking overnight. Bioluminescence intensity and optical density of the cultures were measured at different times of the experiment in duplicate. The relative light units (RLU) were calculated by dividing the average bioluminescence reading by the average OD600nm reading (lum/OD600nm) at each time point. The experiment was repeated three times with separate colonies of each strain.
Table 1.
Primers used in qPCR.
| Primer | Sequence (5′-3′) |
|---|---|
| Sal804proEcoRfor | ATGAATTCTGGAAGTGATGTCGTTATCG |
| Sal804proXmarev | TACCCGGGCATGTTGTCACCGCCTTTC |
| Sal804forPmeI | AGCTTTGTTTAAACTATGAAAACATTAAAAGAGTTGAGG |
| Sal804RevAscI | TTGGCGCGCCTTAAGATGTTTGTTTTTCTTTAAATGC |
| Sa1804Ndefor | TGAAAAAACATATGAAAACATTAAAAGAGT TGA G |
| Sa1804Bamrev | TTGGATCCTTAAGATGTTTGTTTTTCTTTAAA |
| ProfnbBfor | AAGAATTCGCGCTTTATGTCTGATGATTG |
| ProfnbBrev | AACCCGGGCTCCCTTAAATGCAAAATTC |
| SA1000for | GTATCAACGTTTGCCGGTGAATCTC |
| SA1000rev | CAGCTCTTTGTGCTTTACGGTGTGTT |
| Efbfor | GTACAATGATGGTACTTTTAAATATCAATCTAGAC |
| Efbrev | GTTCTTTTTTAATAGTTGCATCAGTTTTCGCT |
| Hlafor | CAACTGATAAAAAAGTAGGCTGGAAAGTGAT |
| Hlarev | CTGGTGAAAACCCTGAAGATAATAGAG |
| FnbB for | GCAGTGAGCGACCATACAACAGTT |
| FnbBrev | CAATCACGCCATAATTACCGTGACCA |
| gyrBfor | GGTGGCGACTTTGATCTAGC |
| gyrBrev | TTATACAACGGTGGCTGTGC |
RNA Purification and Real-Time RT-PCR Analysis
Overnight cultures of S. aureus were inoculated in 5% in TSB medium and grown to the mid-exponential (3 h) phase of growth. Cells were harvested by centrifugation; RNA was isolated using the RNAPrep Kit (Promega, MI) as described (Lei et al., 2014). Contaminating DNA was removed with a DNA-free Kit (Ambion). The first strand cDNA was synthesized using reverse transcriptase with the SuperScript III Platinum Two-Step qRT-PCR Kit (Life Science Technology). For each RNA sample, duplicate reactions of reverse transcription were performed, as well as a control without reverse transcriptase, in order to determine the levels of DNA contamination. PCR reactions were set up in triplicate by using the SYBR Green PCR Master Mix (Bio-Rad). Real-time sequence-specific detection and relative quantization were performed with the Stratagene Mx3000P Real Time PCR System. Gene-specific primers were designed to yield ∼100 bp of specific products (Table 1). Relative quantification of the product will be calculated using the Comparative CT method, as described for the Stratagene Mx3000P system. The housekeeping gene gyrB was used as an endogenous control. All samples were analyzed in triplicate and normalized against gyrB gene expression. The experiments were at least repeated twice and analyzed for correlation to the microarray results.
Cell Culture and Epithelial Cell Invasion Assay
A549 human lung epithelial cells (ATCC CCL 185) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Invitrogen). Cultures of A549 cells were maintained in a medium containing penicillin (5 μg/ml) and streptomycin (100 μg/ml; Sigma). Bacterial invasion assyas were performed in RPMI 1640 medium with supplement of 10% FBS (RPMI-FBS) as described (Yang and Ji, 2014). Briefly, one day prior to infection, approximately 2 × 105 cells were seeded in each well of 24-well plates, and incubated overnight at 37°C in a CO2 incubator. Monolayers of A549 cells (2 × 105 cells/well) were infected by adding 0.5 ml RPMI containing approximately 5 × 105 cfu of bacteria followed a centrifugation at 100 g for 5 min, and incubated for 2 h at 37°C in 5% CO2. The bacteria outside of the monolayer cells were killed by adding1 ml of RPMI-10% FCS containing 100 μg/ml gentamicin and 5 μg/ml lysostaphin to invasion wells. We used 0.025% TritonX-100 for better cell lysis. The numbers of bacterial cfu released from the lysed epithelial cells were determined by plating of diluted lysates on TSA-agar plates. Each experiment was repeated three times and statistically analyzed by TTEST, using Microsoft Excel software. P-values of < 0.05 were considered significant.
Cytotoxicity Assays
All cells were grown in 96-well plates to 70% confluence. Inhibitors were diluted serially in complete medium and applied to the cells. The supernatants were collected from the overnight cultures, sterilized by filtration, and applied to the epithelial cells, and the treated cells were incubated at 37°C with 5% CO2 overnight for 18 h. At the end of the experiment, cell viability was determined using the CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay (Promega) as manufacturer’s instructions. Each experiment was repeated three times and statistically analyzed by TTEST, using Microsoft Excel software. P-values of < 0.05 were considered significant.
Cloning, Expression, and Purification of the SA1804-HIS Tagged Fusion Protein in Escherichia coli
The sa1804 gene was obtained by PCR amplification using the premiers listed in Table 1, cloned into pET28a, and resulted in pSA1804-28a. Plasmid pSA1804-28a were introduced into a BL21(DE3) strain. The resulting strains were grown in LB medium at room temperature; the expression of SA1804 was induced when the culture media reached OD600nm equal to 0.6 by addition of 1 mM IPTG (isopropyl-b-d-thiogalactoside) and incubation pursued for 4 h. The SA1804-his tagged protein was purified using Ni-NTA agarose column (Novagen) as described (Yan et al., 2011).
Electrophoretic Mobility Shift Assay
To determining whether SA1804 directly regulates hla, sa1000, and/or efb transcription, a DIG-labeled probe of hla promoter region was utilized for as described (Liang et al., 2011). The dual 5’-biotin labeled promoter fragments of sa1000 and efb were obtained by high-fidelity PCR and gel-purified using a NucleoSpin Gel Clean-up kit (Macherey–Nagel). The purified biotin-labeled probes were utilized for the electrophoretic mobility shift assay (EMSA) using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific). All samples contained 1X LightShift Binding Buffer, 50 ng/μl Poly (dI∙dC), and 2.5% glycerol. The labeled probe, SA1804-His, non-labeled specific probe, BSA, and non-specific non-labeled probe were all added to concentrations as outlined in the figure and ultrapure water was added so that all reaction volumes totaled 20 μl. The reactions were incubated at room temperature for 20 min followed by addition of 5 μl of 5X loading buffer to each reaction. 20 μl of each reaction were loaded into the wells of a pre-run 8% TBE native polyacrylamide gel and electrophoresed at 75 V for three hours at 4°C. The samples were transferred to nylon membrane and processed as outlined in the manufacturer’s protocol. BioMax Light Film (Kodak) was used to detect the chemiluminescent reaction.
Results
Transcriptional Repression of sa1804 By SaeRS
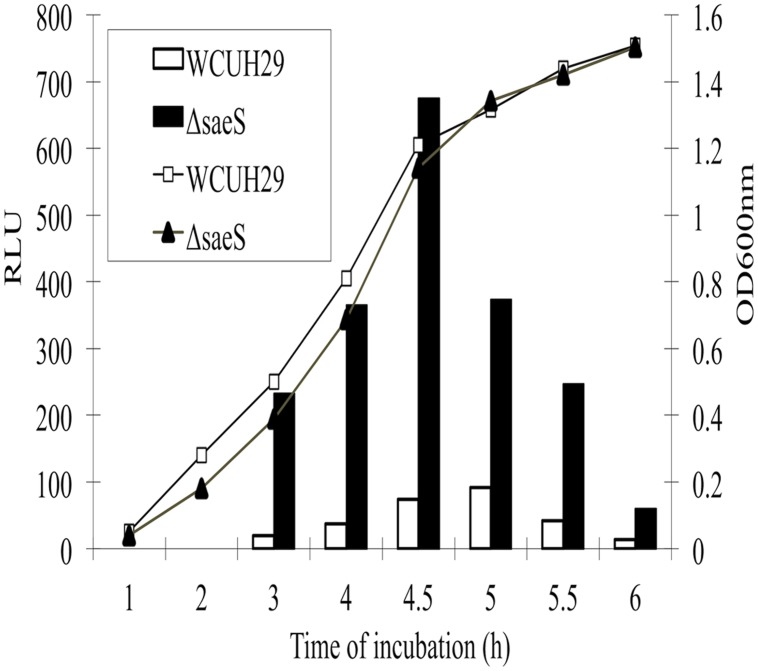
Our previous microarray analysis showed that the mutation of saeS significantly induced the transcription of a putative regulator gene (sa1804), suggesting that SaeR might be a repressor of SA1804 (Liang et al., 2006). To further confirm whether SaeRS system negatively mediates the transcription of sa1804, we conducted a Psa1804-lux reporter experiment. We detected the sa1804 transcription levels by kinetically measuring the intensity of bioluminescence signal during growth over time. The transcription level of sa1804 strikingly increased from the mid-log phase of growth and peaked at the early stationary phase of growth in the saeRS null mutant compared to the control strain (Figure 1). This suggests that SaeRS system transcriptionally modulates the expression of SA1804.
FIGURE 1.
Effect of the deletion of saeS on the sa1804 promoter transcription activity. Both bioluminescence signals and cell growth were monitored at different phases of growth at 37°C by measuring the light intensity with a Chiron luminometer and optical density at 600 nm (OD600) with a SpectraMax plus Spectrophotometer. Relative light unit (RLU) was calculated with the bioluminescence intensity divided by the optical density in the same time of culture. The experiments were repeated at least three times. Solid column represents the saeS deletion mutant and open column represents the parental wild type strain.
The Deletion Mutation of sa1804 Enhances the Capacity of S. aureus to Invade Into Epithelial Cells in a SaeRS-Dependent Manner
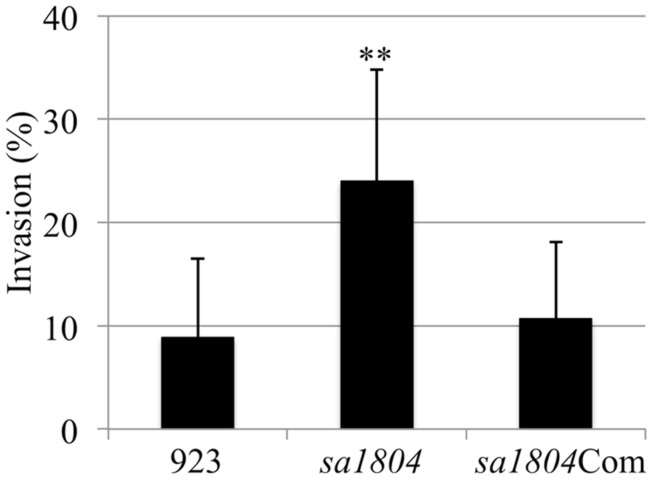
Our previous studies demonstrated that the SaeRS is a major regulator of bacterial adhesion and invasion and SaeRS represses sa1804 transcription. This led us to question the impact of SA1804 on the capacity of bacteria to adhere to and invade into the host cells. To address this question, first we constructed a sa1804 null mutant using a USA 300 human clinical S. aureus isolate 923. Then, we examined the impact of the null mutation of SA1804 on invasion using a human lung epithelial cell line (A549). The results showed that the mutation of sa1804 significantly elevated the capacity of bacteria to invade into the epithelial cells (Figure 2). To confirm the role of Sa1804, we constructed sa1804 complementary strain, and determined whether the expression of Sa1804 in trans complements the effect of depletion of SA1804. The results showed that the complementation of Sa1804 alleviated the bacterial ability to invade epithelial cells (Figure 2).
FIGURE 2.
Effect of the deletion of sa1804 on the bacterial invasion of the human lung epithelial cell line (A549). Intracellular invasion was calculated as described in Section “Materials and Methods." Data are the means ± SE of the means from at least three repeated experiments. 923: wild type CA-MRSA control; sa1804: sa1804 deletion mutant; sa1804Com: sa1804 complementary strain. The symbol **represents P < 0.01 between wild type control and sa1804 knockout mutant.
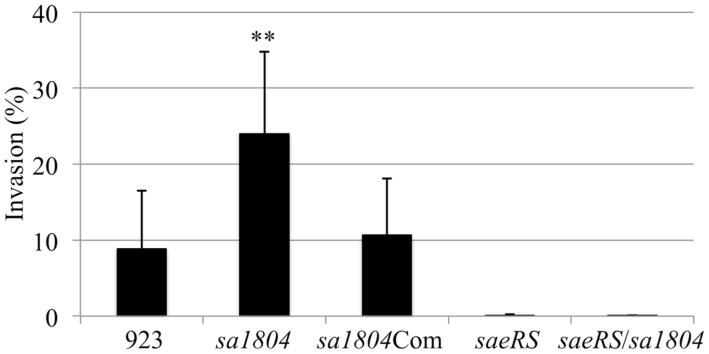
To elucidate whether SA1804 functions through the regulation of SaeRS, we generated saeRS and saeRS/ sa1804 double knockout mutants and examined their impact on bacterial invasion. Consistent with our previous findings the deletion of saeRS loci totally eliminated the bacterial capacity of invading of epithelial cells (Figure 3). The deletion mutation of saeRS eliminated the effect of SA1804 on the invasion ability of in the sa1804 null mutant (Figure 3). The above data indicate that the involvement of SA1804 in regulation of bacterial invasion is dependent on SaeRS.
FIGURE 3.
Effect of the deletion of saeRS, sa1804, or saeRS/sa1804 on the bacterial invasion of the human lung epithelial cell line (A549). Intracellular invasion was calculated as described in Section “Materials and Methods." Data are the means ± SE of the means from at least three repeated experiments. 923: wild type CA-MRSA control; sa1804: sa1804 deletion mutant; sa1804Com: sa1804 complementary strain; saeRS: saeRS deletion mutant; saeRS/sa1804: saeRS and sa1804 double deletion mutant. The symbol **represents P < 0.01 between wild type control and sa1804 knockout mutant.
The Deletion Mutation of sa1804 Increases the Cytotoxicity of S. aureus to Epithelial Cells in a SaeRS-Dependent Manner
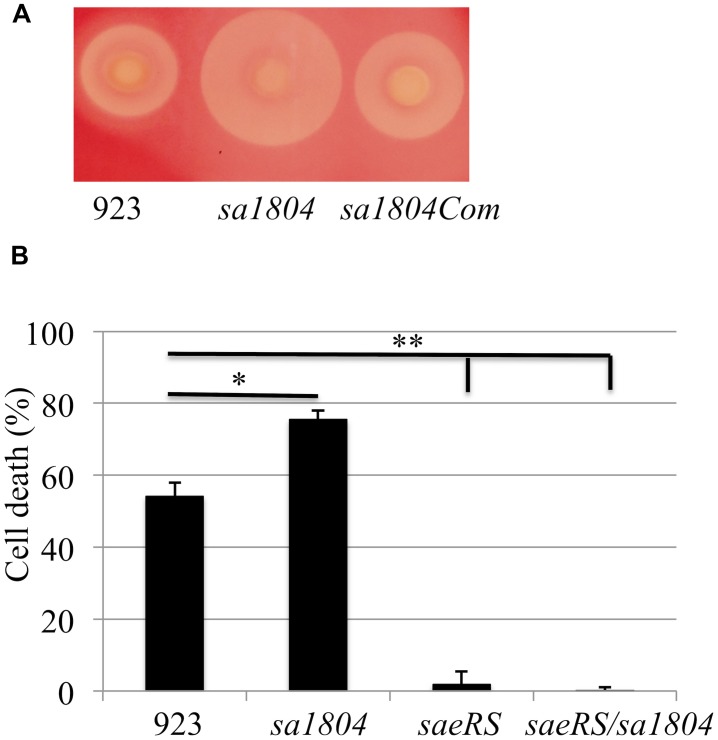
To further determine the role of SA1804 in pathogenicity of S. aureus, we performed hemolysis and cytotoxicity assays by measuring LDH as described (Liang et al., 2006). The deletion of sa1804 increased the hemolysis on a sheep blood agar plate, whereas the expression of SA1804 in trans complemented the hemolytic activity of S. aureus 923 isolate (Figure 4A). We found that the supernatant of the wild type strain, 923, caused 54% of A549 cells died 16 h after exposure, whereas 75% of the cells died after exposing to the supernatant from the sa1804 knockout mutant culture (Figure 4B). Deletion of saeRS abolished the toxicity of the supernatant of the culture, whereas knockout of saeRS had no impact of the deleted sa1804 on cytotoxicity. This further indicates that the regulator SA1804 plays a role in the regulation of virulence factors in a saeRS dependent manner.
FIGURE 4.
Effect of SA1804 on bacterial hemolysis and on the bacterial cytotoxicity. (A) Hemolytic analysis on Sheep Blood Agar. (B) Effect of supernatants of overnight cultures on cell death. The supernatants were collected from overnight cultures and sterilized by filtration. Monolayers of A549 cells (2 × 105 cells/well) were exposed to the supernatant of the cultures. Cell viability was measured at 18 h after treatment and is expressed as an average of at least three experiments ± SD. 923: wild type CA-MRSA control; sa1804: sa1804 deletion mutant; sa1804Com: sa1804 complementary strain; saeRS: saeRS deletion mutant; saeRS/sa1804: saeRS and sa1804 double deletion mutant. The symbol *represents P < 0.05 between wild type control and sa1804 knockout mutant; the symbol **represents P < 0.01 between wild type control and saeRS knockout or saeRS/sa1804 double knockout mutant.
The Putative Transcriptional Regulator (SA1804) Negatively Regulates the Expression of efb, sa1000 and hla in S. aureus
To elucidate potential molecular mechanism behind the impact of SA1804 on bacterial invasion and cytotoxicity, we examined the impact of SA1804 on transcriptions of several potential target genes using qPCR approach. Total RNA was purified from the mid-log phase of cultures of the sa1804 null mutant, saeRS null mutant, saeRS/sa1804 double knockout mutant, and parental control. The transcription levels of selected genes were detected by qPCR. The housekeeping gene gyrB was used as an internal control. No difference of gyrB gene transcription level was revealed between the sa1804 and/or saeRS null mutant and it parental control. However, the transcriptional levels of hla, efb, and sa1000 increased approximately threefold in the sa1804 null mutant than that parental control (Table 2). In contrast, the deletion of saeRS remarkably decreased these genes transcription. Taken together, the above data demonstrated that the putative regulator SA1804 transcriptionally represses the expression of hla, efb, and sa1000 in S. aureus 923 isolate.
Table 2.
qPCR analysis of expression of genes regulated by SaeRS.
| N315 gene | Description | Fold change a in sa1804ko | Fold change a in saeRSko | Fold change in sa1804/saeRSko |
|---|---|---|---|---|
| hla | Alpha-hemolysin | 3.0 | -56.49 | -40.93 |
| sal 000 | Hypothetical fibrinogen-binding protein | 2.9 | -20.38 | -20.97 |
| efb | Extracellular fibrinogen-binding protein | 3.86 | -12.91 | -11.0 |
| fnbB | Fibronectin-binding protein B | -0.96 | -1.7 | -1.33 |
aNormalized values in the knockout mutant over values in the wild type 923 strain. Negative number denotes down-regulated in mutant strain.
The Putative Regulator Sa1804 Binds the hla Promoter Region in vitro
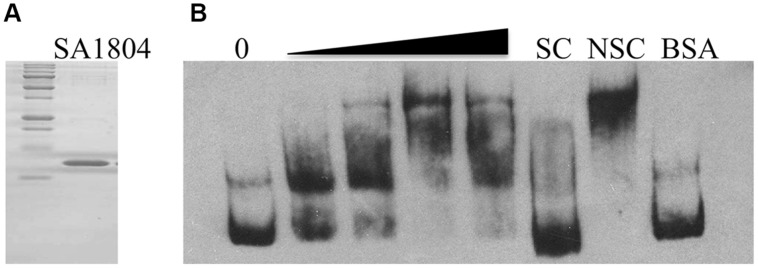
To elucidate the potential mechanism of regulation, we examined whether SA1804 directly or indirectly regulates the transcription of the hla, efb, and sa1000 genes using gel-shift assays. We cloned, expressed, and purified SA1804-his tag protein as described (Figure 5A). BSA protein was used as a control. The promoter regions of hla, efb, and sa1000 genes were obtained by PCR, labeled, and purified using PCR cleanup kits. Unlabeled probe was utilized as specific competitor (SC) accordingly. Apparent hla probe-SA1804 complex formed in a dose-dependent manner (Figure 5B). However, no hla probe-SA1804 complex was observed for the efb and sa1000 promoters (data not shown). These suggest that SA1804 may directly mediate hla transcription by binding to hla promoter region, but indirectly regulate efb and sa1000 transcription.
FIGURE 5.
(A) SDS-PAGE analysis of the purity of purified recombinant SA1804; (B) Electrophoretic mobility shift analysis of the hla promoter regulated by SA1804. The promoter region of the hla gene was obtained by PCR, purified, and labeled with Digoxigenin. The mobility of the labeled promoter fragment without addition of SA1804 is shown in the first lane. Different amounts of SA1804 protein (25, 100, 250, 500 ng) were incubated with each labeled hla promoter probe in a 15 μl reaction volume. Specific competitor (SC) control: incubation in the presence of 100-fold excess unlabeled SC. Non-specific competitor (NSC) control: incubation in the presence of 100-fold excess unlabeled non-specific internal gene probe. BSA (1 μg) was used as non-specific binding control.
Discussion
In this study, we are the first demonstrating that SA1804 is a repressor of several virulence factors, including hla, efb, and sa1000 in a CA-MRSA USA300 isolate 923. Moreover, our data suggest that SA1804 indirectly mediates the transcription of efb and sa1000, whereas directly controls the transcription of hla through binding to the promoter region of hla. Furthermore, our data clearly indicate the role of SA1804 in manipulation of the pathogenicity of S. aureus, as the null mutation of sa1804 enhanced the capacity of bacteria internalizing into the epithelial cells and the cytotoxicity. In addition, we further confirmed that SaeRS negatively regulates the transcription of the novel transcriptional regulator SA1804. Our data suggest that the regulatory function of SA1804 is dependent on the activation of SaeRS, as the disruption of SaeRS eliminated the influence of SA1804 on the transcription of selected virulence factors, including hla, efb, and sa1000, as well as the bacterial invasion ability and cytotoxicity.
Our sa1804 promoter-lux reporter fusion experiment showed that the inactivation of SaeRS remarkably elevated the transcription of sa1804, which is consistent with our microarray analysis result (Liang et al., 2006). Together with previous findings in different S. aureus isolates (Giraudo et al., 1999; Liang et al., 2006; Rogasch et al., 2006), our qPCR analysis further confirmed the effect of SaeRS on positive regulation of key virulence factors, including hla, efb, fnbB, and sa1000 in CA-MRSA USA300 isolate 923. Moreover, our invasion and cytotoxicity assays demonstrated that SaeRS plays a key role in the pathogenesis of S. aureus, which is highly supportive for the importance of SaeRS in pathogenicity in animal models of infection (Liang et al., 2006; Montgomery et al., 2010). Previous reports indicate that SaeRS tightly controls the expression of fnb (Giraudo et al., 1999) since no mRNA of fnbA and fnbB was detectable in the saeS mutant strain (Steinhuber et al., 2003). In this study, we found that the deletion mutation of saeRS had a weak effect on the transcription of fnbB. This discrepancy is likely due to the difference of genetic background among different S. aureus isolates.
It was well established that SaeRS is a major regulator of hla expression in S. aureus (Liang et al., 2006; Montgomery et al., 2010); SaeR can directly regulate the hla transcription by binding to its promoter region (Cho et al., 2012; Olson et al., 2013). Although our gel-shift result indicated that the novel transcriptional regulator, SA1804, is able to bind to the hla promoter region, our data clearly demonstrated that SaeRS plays a predominant role in the regulation of the hla transcription in S. aureus.
Acknowledgments
This project is partially supported by grant AI 057451 from the NIH and USDA Hatch Formula Fund.
REFERENCES
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55 58–63 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Benton B. M., Zhang J., Bond S., Pope C., Christian T., Lee L., et al. (2004). Large-scale identification of genes required for full virulence of Staphylococcus aureus. J. Bacteriol. 186 8478–8489 10.1128/JB.186.24.8478-8489.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. L., Koomey J. M., Butler C. A., Projan S. J., Fischetti V. A. (1992). Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U.S.A. 89 6462–6466 10.1073/pnas.89.14.6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Jeong D. W., Li C., Bae T. (2012). Organizational requirements of the SaeR binding sites for a functional P1 promoter of the sae operon in Staphylococcus aureus. J. Bacteriol. 194 2865–2876 10.1128/JB.06771-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann F., Bantel H., Totzke G., Engels I., Sinha B., Schulze-Osthoff K., et al. (2003). Staphylococcus aureus alpha-toxin-induced cell death: predominant necrosis despite apoptotic caspase activation. Cell Death Differ. 10 1260–1272 10.1038/sj.cdd.4401301 [DOI] [PubMed] [Google Scholar]
- Foster T. J., Höök M. (1998). Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6 484–488 10.1016/S0966-842X(98)01400-0 [DOI] [PubMed] [Google Scholar]
- Fournier B., Klier A., Rapoport G. (2001). The two-component system ArlS–ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41 247–261 10.1046/j.1365-2958.2001.02515.x [DOI] [PubMed] [Google Scholar]
- Fowler T., Wann E. R., Joh D., Johansson S., Foster T. J., Höök M. (2000). Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell [veerupbeta]1 integrins. Eur. J. Cell Biol. 79 672–679 10.1078/0171-9335-00104 [DOI] [PubMed] [Google Scholar]
- Giraudo A. T., Calzolari A., Cataldi A. A., Bogni C., Nagel R. (1999). The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177 15–22 10.1111/j.1574-6968.1999.tb13707.x [DOI] [PubMed] [Google Scholar]
- Goerke C., Fluckiger U., Steinhuber A., Bisanzio V., Ulrich M., Bischoff M., et al. (2005). The role of Staphylococcus aureus global regulators sae and [veerupsigma]B in virulence gene expression during device-related infection. Infect. Immun. 73 3415–3421 10.1128/IAI.73.6.3415-3421.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C., Fluckiger U., Steinhuber A., Zimmerli W., Wolz C. (2001). Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of [veerupalpha]-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40 1439–1447 10.1046/j.1365-2958.2001.02494.x [DOI] [PubMed] [Google Scholar]
- Gordon R. J., Lowy F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46 S350–S359 10.1086/533591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Yin D., Fox B., Holmes D., Payne D., Rosenberg M. (2004). Validation of antibiotic mechanism of action by regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231 177–184 10.1016/S0378-1097(03)00931-5 [DOI] [PubMed] [Google Scholar]
- Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298 1763–1771 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Lee L., Liang X., Hook M., Brown E. (2004). Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrogen-binding protein (Efb). J. Biol. Chem. 279 50710–50716 10.1074/jbc.M408570200 [DOI] [PubMed] [Google Scholar]
- Lei T., Becker A., Ji Y. (2014). Transcriptomic analysis of Staphylococcus aureus using microarray and advanced next-generation RNA-seq technologies. Methods Mol. Biol. 1085 213–229 10.1007/978-1-62703-664-1_13 [DOI] [PubMed] [Google Scholar]
- Liang X., Hall J. W., Yang J., Yan M., Doll K., Bey R., et al. (2011). Identification of single nucleotide polymorphisms associated with hyperproduction of alpha-toxin in Staphylococcus aureus. PLoS ONE 6:e18428 10.1371/journal.pone.0018428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Ji Y. (2007). Involvement of alpha5beta1-integrin and TNF-alpha in Staphylococcus aureus alpha-toxin-induced death of epithelial cells. Cell Microbiol. 9 1809–1821 10.1111/j.1462-5822.2007.00917.x [DOI] [PubMed] [Google Scholar]
- Liang X., Yu C., Sun J., Liu H., Landwehr C., Holmes D., et al. (2006). Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74 4655–4665 10.1128/IAI.00322-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zheng L., Landwehr C., Lunsford D., Holmes D., Ji Y. (2005). Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187 5486–5492 10.1128/JB.187.15.5486-5492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339 520–532 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Luong T. T., Newell S., Lee C. Y. (2003). Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185 3703–3710 10.1128/JB.185.13.3703-3710.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P., Milligan-Monroe K., Khalili S., Proctor R. A. (2000). Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 1823 3197–3203 10.1128/JB.182.11.3197-3203.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery C. P., Boyle-Vavra S., Daum R. S. (2010). Importance of the global regulators agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE 5:e15177 10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak L. N., Zielinska A. K., Beenken K. E., Mrak I. N., Atwood D. N., Griffin L. M., et al. (2012). saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS ONE 7:e38453 10.1371/journal.pone.0038453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. (2003). Autoinduction and signal transduction in the regulation of Staphylococcal virulence. Mol. Microbiol. 48 1429–1449 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of Staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. E., Nygaard T. K., Ackermann L., Watkins R. L., Zurek O. W., Pallister K. B., et al. (2013). Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect. Immun. 81 1316–1324 10.1128/IAI.01242-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock S. J., Moore C. E., Justice A., Kantzanou M., Story L., Mackie K., et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70 4987–4996 10.1128/IAI.70.9.4987-4996.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogasch K., Rühmling V., Pané-Farré J., Höper D., Weinberg C., Fuchs S., et al. (2006). Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188 7742–7758 10.1128/JB.00555-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B., François P. P., Nüße O., Foti M., Hartford O. M., Vaudaux P., et al. (1999). Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin-[veerupalpha]5[veerupbeta]1. Cell Microbiol. 1 101–117 10.1046/j.1462-5822.1999.00011.x [DOI] [PubMed] [Google Scholar]
- Steinhuber A., Goerke C., Bayer M. G., Döring G., Wolz C. (2003). Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185 6278–6286 10.1128/JB.185.21.6278-6286.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Li C., Jeong D., Sohn C., He C., Bae T. (2010) In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J. Bacteriol. 192 2111–2127 10.1128/JB.01524-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Yu C., Yang J., Ji Y. (2011). The essential two-component system YhcSR is involved in regulation of the nitrate respiratory pathway of Staphylococcus aureus. J. Bacteriol. 193 1799–1805 10.1128/JB.01511-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ji Y. (2014). Investigation of Staphylococcus aureus adhesion and invasion of host cells. Methods Mol. Biol. 1085 187–194 10.1007/978-1-62703-664-1_11 [DOI] [PubMed] [Google Scholar]