Abstract
Small angle solution X-ray and neutron scattering recently resurfaced as powerful tools to address an array of biological problems including folding, intrinsic disorder, conformational transitions, macromolecular crowding, and self or hetero-assembling of biomacromolecules. In addition, small angle solution scattering complements crystallography, nuclear magnetic resonance spectroscopy, and other structural methods to aid in the structure determinations of multidomain or multicomponent proteins or nucleoprotein assemblies. Neutron scattering with hydrogen/deuterium contrast variation, or X-ray scattering with sucrose contrast variation to a certain extent, is a convenient tool for characterizing the organizations of two-component systems such as a nucleoprotein or a lipid-protein assembly. Time-resolved small and wide-angle solution scattering to study biological processes in real time, and the use of localized heavy-atom labeling and anomalous solution scattering for applications as FRET-like molecular rulers, are amongst promising newer developments. Despite the challenges in data analysis and interpretation, these X-ray/neutron solution scattering based approaches hold great promise for understanding a wide variety of complex processes prevalent in the biological milieu.
Keywords: small angle X-ray scattering, anomalous solution scattering, solution neutron scattering, solution structure determination
Introduction
Modern biological applications of small angle X-ray and neutron solution scattering (SAXS/SANS) include low-resolution shape modeling, and the characterizations of protein/RNA folding, intrinsic disorder, conformational transitions, and protein-protein or protein-nucleic acid assembling processes (Fig. 1).1–5 Even though small angle solution scattering (SAS) typically requires very pure and mono-disperse sample, no specialized sample treatments like chemical fixation or crystallization are necessary, making the use of a range of physiologically relevant buffer conditions possible. Furthermore, small sample volume at 1 to 10 mg/mL concentration, rapid data collection at the synchrotron sites (0.5–5 s), practically no size-limitation (depending upon the experimental setup), several dedicated beam-lines at the synchrotron facilities including those with options for automated high-throughput data collection and the availability of user-friendly, automated software for data analysis makes SAXS a very popular technique.5–9 Due to the immense popularity of SAXS, guidelines for publications are actively discussed in the research community.10,11 For the theoretical basis and technical details of small angle scattering, we refer the readers to an excellent, recently published text-book.12 In this review, we will focus our attention on the modern applications of SAXS and SANS in a variety of problems in structural biology.
Figure 1.
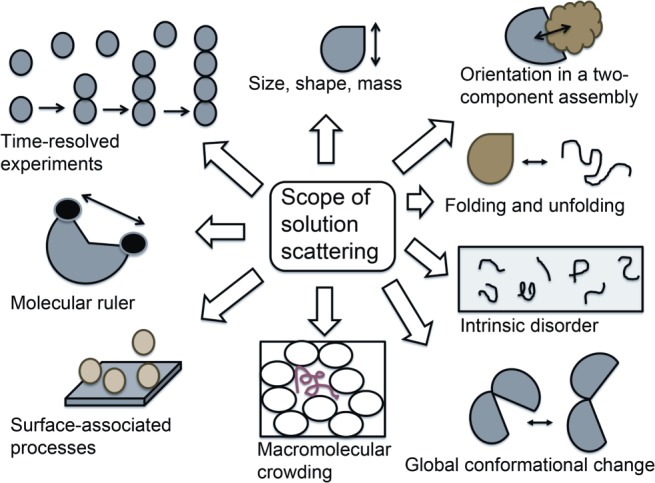
A schematic diagram showing various biological applications of small angle solution scattering technique.
Size and shape of a macromolecule
Analysis of SAS data allows ready estimations of mass, radius of gyration, and maximum diameter of a monodisperse macromolecule in solution.1,2,13 In addition, cross-sectional size and linear mass density (mass per unit length) of a rod-shaped filament and thickness of a disk-shaped particle can be estimated from SAS data.13–15 These quantities and the pair-wise distance distribution profile computed from SAS data are useful for testing various structural hypotheses and can be used as restraints for structural modeling. While small angle X-ray/neutron scattering1–4 provides information about the global size and shape of the macromolecule in solution, wide-angle X-ray scattering (WAXS) data obtained at higher scattering angles contains information about its finer structural features.16 On the other hand, SAXS at ultra-low scattering angles can aid in the elucidation of large-scale, higher-order structures, such as the nucleosome fiber.17
When a crystal structure is not available, SAXS provides a quick way to compute a low-resolution, average molecular shape of a mono-disperse, globular macromolecule from its scattering profile.1,2,18–21 Impositions of anticipated point-group symmetry, connectivity and compactness conditions, and multiple ab initio shape computations with consistent results, aid in obtaining an average, low-resolution shape model.22,23 Due to the inherent ambiguity of SAS-based shape modeling, validations of the model using additional experimental data that are “not seen by the model” are endorsed.12 Comparisons of the SAS-derived shape models with corresponding known structures indicated excellent agreements for a number of cases.24 Recently, methods were developed to model RNA structures using experimental SAXS data in conjunction with the structure prediction algorithms.25,26
SAS as a complementary technique in structural biology
While X-ray crystallography is a routine tool for the atomic resolution structure determinations of bio-molecules held in crystal lattices, SAS can readily reveal their oligomeric states and domain organizations in solution.1,2 Solution scattering profiles are computed from the atomic co-ordinates of available crystal structures for comparisons with the experimental SAXS/WAXS data.27–32 In addition to ab initio shape modeling, SAS can be used in conjunction with the existing substructures to model their overall tertiary or quaternary organizations.33–39 Furthermore, SAS-based shape computations with known partial structures can help to predict the location of a missing component within the structure.40
Despite the tremendous methodological advances in structure determination techniques, many multicomponent or multidomain biological entities continue to pose considerable challenge to structure determination. An effective way to tackle this challenge is to combine information obtained from complementary methods, such as crystallography, nuclear magnetic resonance spectroscopy (NMR), transmission electron microscopy, mass spectrometry and SAS, to learn about the structural organizations of proteins and their assemblies at data-dependent levels of details.41–45 For example, Wang et al., used information about overall shape, relative orientations of the components and the footprints of buried, interface residues obtained from SAXS, NMR and mass spectrometric data to learn about the oligomeric states of a chemokine CCL5.44 Hybrid methods are developed to integrate SAS data with other experimental data for structural modeling.46–49 In particular, NMR with SAS has emerged as a powerful approach to elucidate the structural organizations of various biological entities.44,49–52 Thus, SAS in combination with other complementary techniques can be an effective tool to address structural problems that are not easily amenable to a single technique.
Neutron and X-ray scattering with contrast variation
Small angle neutron scattering or SANS contrast variation experiments reveal the relative dispositions of components within a multicomponent assembly.4,13,53–56 A difference in the neutron scattering length density of hydrogen (H) and deuterium (D) is exploited in a SANS contrast variation experiment to vary the excess scattering density or contrast of the scattering macromolecule.53–57 A typical contrast variation dataset is obtained by collecting SANS data at a number of different H/D ratio, from which individual scattering profile for each component in the assembly as well as the cross-term describing their relative orientations in a two-component assembly can be retrieved.57 In addition, distance between the centers of mass of the two components can be estimated from SANS contrast variation dataset.57 Thus, SANS with H/D contrast variation is very well suited for analyzing relative positioning of the components within an assembly and an induced-fit or a mutual induced fit mode of molecular recognition.
In an alternative contrast matching experiment, neutron scattering contribution from one of the components can be selectively “matched out” or abolished by adjusting the H/D ratio of the buffer. The “matching out” conditions for lipids, proteins, carbohydrates, and DNA/RNA are at about 10 to 14%, 40 to 45%, 47%, and 65 to 72% D2O, respectively, which can be changed by deuteration.1,4,13,57 Neutron scattering with contrast matching is an exceptional tool for observing a selected component in the presence of another “invisible” component in a heteromeric assembly.
Scattering with contrast variation aided in elucidating the organizations of many two-component assemblies, such as nucleoprotein assemblies and protein-protein assemblies with a deuterated protein component.58–64 Owing to the differences in scattering densities between proteins and lipids as well as proteins and detergents, SANS is useful for studying membrane proteins.65,66 Recently, a hybrid strategy to model membrane proteins in lipidic environment by combining SAXS and SANS data was described.67 Thus, the feasibility of deuterium-labeling the macromolecules for changing contrasts and the power of H/D contrast variation allows SANS to characterize a range of two-component assemblies, which is not possible to achieve solely based on SAXS.
Due to a limited number of neutron scattering facilities, a large amount of sample requirement (150 µL or more at 5–10 mg/mL concentration),12 noisier data due to in-coherent scattering and long data collection times (a few hours), resurgence of neutron scattering is gradual. In comparison, SAXS typically requires only 20 to 30 µL of sample and a few seconds of exposure at a powerful synchrotron source. Although less potent than SANS with H/D contrast variation, SAXS with varying amounts of sucrose provides another avenue to protein contrast matching and a limited range of contrast variation.68–72 Moreover, sucrose can reduce the effect of radiation damage at the powerful synchrotron sources.73 However, unaddressed issues such as enhanced viscosity at higher sucrose concentration may limit the utility of these SAXS-based contrast variation experiments and care should be taken to account for these effects.72 High salt conditions are effective for SAXS-based contrast variation experiments, but have limited applicability in biological problems.1,72 Due to the relative ease of obtaining SAXS data in comparison to SANS, a renewed search for additional contrast variation agents for SAXS that are suitable for biological samples will be rewarding to the SAS research community.
Folding, conformational flexibility, and intrinsic disorder
SAS is a powerful global sensor of the folding states and conformational changes in proteins and nucleic acids. A global indication of “folded-ness” of the biomolecule can be obtained from the Kratky plot (I.q2 vs. q, where I is the scattering intensity, and q is the momentum transfer).74 Kratky plots, together with the changes in sizes and pair-distribution function profiles, are used to track folding-unfolding behaviors of biomolecules under varying experimental conditions.74 Furthermore, changes in sizes and pair-distributions functions are model-free indicators of large-scale conformational changes in folded, nonaggregated proteins, which can be combined with shape or structural modeling. Although the applications of SAXS are limited to detections of large movements,75–79 WAXS can sense small amplitude structural changes in proteins.80,81
A large number of naturally occurring proteins are either disordered or contain long stretches of unstructured regions, and are generally classified as intrinsically disordered proteins (IDP).82,83 The disordered regions often fold in the presence of a binding partners(s) and play crucial role in molecular recognition.82,83 Due to a lack of rigid, well-defined structure, characterization of the IDPs pose a challenge to structural biologists. SAS is naturally suitable for studying these IDPs, as the scattering profile represents an ensemble of conformations of the IDP in solution.82 An IDP or a flexible protein can be identified from the global features in the SAS data, such as the Kratky plot, a left-skewed pair-distribution function or a Porod-Debye plot.82,84,85
In addition to the global analysis, a number of different computational methods were developed to generate an ensemble of protein conformers from the SAS data.86–89 These methods typically involve computational generations of multiple conformers followed by their selections based on their compatibility with the experimental SAS data.86–89 Ensemble modeling with SAS data yielded structural information on the spatial occupancy of glycans in glycosylated proteins that are notoriously difficult to crystallize.90 Thus, SAS in combination with suitable computational methods can provide potentially unrestricted access to the conformational spaces of flexible proteins under different conditions, which is a clear advantage over crystallography.
Due to the low information content of SAS and a large number of parameters required for an exhaustive description of an IDP, combining SAS with additional complementary experimental data is a sensible approach, when possible. SAS analysis of IDPs is typically complemented with solution NMR, circular dichroism, dynamic light scattering and other hydrodynamic data analyses.91 Synergy between NMR and SAS were exploited in numerous studies of IDPs, which is recently reviewed elsewhere.92 Bertini et al. used SAXS with NMR on lanthanide-labeled proteins to determine the most abundant conformers of highly flexible calmodulin, which might be needed for a better explanation of its function in solution.93 In a recent article, exhaustive SAXS and NMR based ensemble analyses, together with extensive cross-validations, were used to predict conformations of IDPs relevant to neurodegenerative disorder.94 SAS, particularly in combination with NMR, will continue to play a pivotal role in providing key structural descriptions of IDPs.
Macromolecular crowding
SAS provides an elegant way to examine the important but often neglected effect of macromolecular crowding on proteins.95 WAXS data suggested that concentrated conditions inhibit breathing motions in proteins.96 Furthermore, SAS was found to be a suitable tool to study the effects of crowding on RNA folding.97,98 SAXS-based studies indicated that crowding promotes compactions in the tertiary and quaternary structures of certain modular enzymes and their complexes.99,100 Quite significantly, this compaction correlates with higher enzymatic activity.99,100 On the other hand, a recent, remarkable SANS study on the intrinsically disordered, deuterium-labeled protein N in the presence of two different un-labeled proteins as crowding agents under their “contrast matching” conditions indicated minimal effect of crowding on the protein N.101 Valuable new insights on the effects of crowding on nucleic acids and deuterated proteins, both folded and intrinsically disordered, can be obtained from SANS in the presence of these “invisible” contrast-matched, unlabeled protein crowding agents that realistically mimics the intracellular environment.
SAXS-based molecular ruler
SAS provides global information on size and shape but no local structural information on a particular site can be generally obtained. However, for certain biological applications, such site-specific details can be very instructive. One way to obtain local information from SAXS is to label the protein with electron-rich elements.102,103 DNA coupled to gold nanocrystal as a probe was successfully used to measure the length of DNA using scattering interference.104 In another recent SAXS study, gold cluster-labeled DNA was used to track conformational changes induced by a DNA mismatch repair protein, leading to new insights into the repair mechanism.105 Under the experimental conditions of low concentrations, scattering from the electron-dense gold-component dominated the total scattering.105 Grishaev et al. determined the structural organization of lead-substituted calmodulin-peptide complex using a combination of sucrose contrast matching, conventional SAXS/WAXS and NMR residual dipolar coupling measurements.70 Scattering contribution from the protein parts was contrast matched by sucrose, leading to the heavy-atom dominated scattering profile, which was critical for the correct positioning of structural components.70 Unlike the Förster resonance energy transfer or FRET-based molecular rulers that are restricted around the Förster distance of the available donor-acceptor pairs, SAXS-based molecular rulers can be potentially used to measure a much larger range of distances.104 Therefore, use of labeled samples for SAXS analyses of localized conformational changes, molecular recognition processes and multimeric organizations, especially coupled with time-resolved SAXS experiments,104 electron microscopy,104 and contrast matching,70 will almost certainly become more common in future.
Anomalous solution X-ray scattering
Anomalous scattering properties of metal ions, which are routinely exploited in multiwavelength anomalous dispersion phasing methods in macromolecular crystallography, has been used to a limited extent in SAXS.106–109 Strong anomalous solution X-ray scattering (ASAXS) of terbeium at the LIII edge was exploited to measure the mean distance between the terbiums substituting the calcium binding sites within parvalbumin and the center of mass of the protein, which was consistent with the crystallographic results.107 Makowski et al. recently used anomalous contribution from iron in the solution scattering data at the Fe K edge to determine the distance between centers of mass of hemoglobin/myoglobin and the metal ion.109 Anomalous contribution due to the metal label to the total scattering is typically quite small, requiring very careful measurements at a tunable-wavelength, powerful synchrotron X-ray source. Although small anomalous contribution due to the cross-term between the label and the protein segment is usable, scattering contribution due to the interference within the anomalous scattering metal component itself is negligible.109 Theoretical calculations predict that the use of metal clusters, such as a gold nanocrystal, may alleviate this problem for ASAXS studies.110 With further explorations of dissimilar anomalous scattering atomic clusters as labels to measure specific distances, and concomitant development of theories,110 applications of ASAXS-based molecular rulers to probe intricate conformational changes and molecular recognition processes may become routine.
Mixed systems
Sample heterogeneity pose challenges to routine SAS data analysis that generally assumes stable molecular species of one kind in dilute, noninteracting solution state. However, many interesting biological phenomena take place in a mixture of interacting particles in solution, which might include weak and transient interactions. SAXS is one of the very few structural techniques available to study these mixed systems.
For a separable mixture, such as a monomer with a stable dimer, SAXS at a powerful synchrotron source coupled with in-line size-exclusion column chromatography (SAXS-SEC) allows an elegant way to study individual entities while being eluted.111,112 In an extension of this coupled approach, scattering from a low-affinity complex of actin with a peptide ligand was measured using SAXS-SEC in a buffer saturated with this peptide ligand, while effectively excluding any scattering from the higher aggregates.113,114 Sokolova et al. modeled intermediate filament assembling pathway from SAXS data by finding out conditions dominated by each of the constituents.115 However, separation of individual entities in a mixture may not be possible or desirable in all cases.
Computational approaches were developed for extracting individual scattering profiles of the constituents from concentration-dependent SAXS dataset obtained from mixed systems involving weak homo- or hetero-oligomers co-existing with monomers.116–118 These approaches are original in a sense that they allow derivations of low-resolution shape models as well as the association constants and assembling pathway from the same experiment.116–118 Petoukhov et al. recently published a method for composition analysis and shape modeling of oligomeric assemblies from condition-dependent scattering curves for a variety of user-defined mixed system scenarios.119 However, conformational changes upon binding can make simple interpretations of SAS data obtained from mixed systems difficult. Above computational approaches involving equilibrium mixtures can be potentially used in conjunction with time series of SAXS to provide a rich variety of information on the mixed system.116
Time-resolved SAS to track biological processes
Time-resolved SAXS, SANS, and WAXS studies can provide direct mechanistic insights on the course of a biological reaction/process in terms of its constituent components, including the pathway intermediates. Data collection capability up to about 100 ps time resolution at the modern, powerful synchrotron sources equipped with better detectors and better methods to trigger an event make it possible to observe the time progressions of SAXS/WAXS profiles of a variety of evolving systems.120–124 SANS provides another avenue for time-resolved studies, albeit at a longer time scale, which can be judiciously combined with contrast matching for a two-component system.125,126 The target biological process can be initiated by a number of ways, such as laser light for a light-initiated process (pump-probe) or by using a mixing device or by quickly removing one component from the mixture, depending upon the nature and time-scale of the process under investigation.120–124 Thus, time-resolved SAS is a valuable tool for investigating a range of processes including the slow assembling (second or longer time-scale) as well as fast folding kinetics (second to 100 µs) and ultra-fast protein movements in the subnanosecond region.12 In recent times, 10 to 100 fs X-ray laser pulses are opening up the first-time possibility to observe even faster, light-triggered processes, such as a “protein-quake.”124,127
Time-resolved SAS is suitable for probing many biological processes, such as capsid maturation and fibrillation pathways that are difficult to study by traditional means. Examples include movements in light-driven proton pumps,128 microtubule formation,129 allostery and structural dynamics in hemoglobin,120,130 fibrillation pathway in insulin amyloid and alpha-synuclein,131,132 viral capsid maturation,133–135 protein folding,136–138 and RNA folding.139,140 In a seminal study by Vestergaard et al., insulin fibrillation process was modeled using time-resolved SAXS data, leading to an elongation pathway for amyloid fibril formation from a helical nucleus.131 In another study, global size information obtained from time-resolved SAS was effectively complemented with local solvent accessibility data from time-resolved hydroxyl radical footprinting to provide unique insights into the RNA folding pathway.141 Recently, Chen et al. performed time-resolved SAXS under protein contrast matching conditions to track salt-induced DNA unwrapping in the nucleosome core particle.72 An ability to probe unrestricted movements of label-free biomolecules in near-physiological conditions at a wide range of temporal resolutions is a huge advantage of time-resolved SAS. Furthermore, time-resolved SAXS/WAXS on proteins with site-specific heavy atom labeling can be used to track local changes, while simultaneously monitoring the global changes in the protein.142
Concluding remarks
Advances in SAS methods opened up the exciting opportunities to learn about size, shape, folding, recognition, flexibility and disorder of soluble single/multidomain proteins, membrane proteins, glycoproteins, intrinsically disordered proteins, DNA/RNA, and their assemblies (Fig. 1). Feasibility of time-resolved SAXS/WAXS studies of biological processes, contrast variation for multicomponent assemblies, convenient applications of SAXS or ASAXS on heavy-atom labeled samples as molecular rulers and appropriate combinations of above are some of the key advantages and future promises of SAS technique. Furthermore, grazing incidence solution scattering and related reflectivity techniques are suitable for studying membrane-associated proteins in near-natural environment.143 Although SAS is essentially a low-information technique, combining SAS with complementary techniques can circumvent this limitation in many cases.47–50 However, much work remains in method developments for data acquisition, analysis and model validation.144–146 In the coming years, modern X-ray sources and methodological advances will probably open up unprecedented ways to reconstruct solution structures of biomacromolecules, and will conceivably reveal unforeseen wealth of information on protein motion, which will be a giant step forward.114,124,147,148
Glossary
Abbreviations:
- ASAX
anomalous solution X-ray scattering
- FRET
Förster resonance energy transfer
- IDP
Intrinsically disordered protein
- NMR
Nuclear magnetic resonance
- SANS
small angle neutron scattering
- SAS
small angle scattering
- SAXS
small angle X-ray scattering
- SAXS-SEC
small angle X-ray scattering–size exclusion chromatography
- WAXS
wide angle X-ray scattering.
References
- Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- Whitten AE, Trewhella J. Small-angle scattering and neutron contrast variation for studying bio-molecular complexes. Methods Mol Biol. 2009;544:307–323. doi: 10.1007/978-1-59745-483-4_20. [DOI] [PubMed] [Google Scholar]
- Graewert MA, Svergun DI. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS) Curr Opin Struct Biol. 2013;23:748–754. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystrallogr. 2006;39:277–286. [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, Tsutakawa SE, Jenney FE, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH. Small angle X-ray scattering as a complementary tool for high-throughput structural studies. Biopolymers. 2011;95:517–530. doi: 10.1002/bip.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel A, Liu P, Weiss TM, Niebuhr M, Tsuruta H. An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. J Synchrotron Radiat. 2012;19:431–434. doi: 10.1107/S0909049512008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Guss JM, Svergun DI, Trewhella J. Publication guidelines for structural modeling of small-angle scattering data from biomolecules in solution. Acta Crystallogr D. 2012;68:620–626. doi: 10.1107/S0907444912012073. [DOI] [PubMed] [Google Scholar]
- Jacques DA, Trewhella J. Small-angle scattering for structural biology-expanding the frontier while avoiding the pitfalls. Protein Sci. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Koch MHJ, Timmins PA, May RP. Small angle X-ray and neutron scattering from solutions of biological macromolecules. UK: Oxford University Press; 2013. [Google Scholar]
- Perkins SJ. Structural studies of proteins by high-flux X-ray and neutron solution scattering. Biochem J. 1988;254:313–327. doi: 10.1042/bj2540313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiyagarajan P, Burkoth TS, Urban V, Seifert S, Benzinger TLS, Morgan DM, Gordon D, Meredith SC, Lynn DG. pH dependent self assembly of β-amyloid (10–35) and β-amyloid(10–35)-PEG3000. J Appl Crystallogr. 2000;33:535–539. [Google Scholar]
- Qian S, Dean R, Urban VS, Chaudhuri BN. The internal organization of mycobacterial partition assembly: does the DNA wrap a protein core? PLoS One. 2012;7:e52690. doi: 10.1371/journal.pone.0052690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti RF, Rodi DJ, Mirza A, Irving TC, Kondrashkina E, Makowski L. High-resolution wide-angle X-ray scattering of protein solutions: effect of beam dose on protein integrity. J Synchrotron Radiat. 2003;10:398–404. doi: 10.1107/s0909049503016583. [DOI] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón P, Díaz JF, Morán F, Andreu JM. Reconstruction of protein form with X-ray solution scattering and a genetic algorithm. J Mol Biol. 2000;299:1289–1302. doi: 10.1006/jmbi.2000.3784. [DOI] [PubMed] [Google Scholar]
- Walther D, Cohen FE, Doniach S. Reconstruction of low-resolution three-dimensional density maps from one-dimensional small-angle X-ray solution scattering data for biomolecules. J Appl Crystallogr. 2000;33:350–363. [Google Scholar]
- Vigil D, Gallagher SC, Trewhella J, Garcia AE. Functional dynamics of the hydrophobic cleft in the N-domain of calmodulin. Biophys J. 2001;80:2082–2092. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Koch MH. Advances in structure analysis using small-angle scattering in solution. Curr Opin Struct Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- Durchschlag H, Zipper P, Krebs AA. Comparison of protein models obtained by small-angle X-ray scattering and crystallography. J Appl Crystallogr. 2007;40:1123–1134. [Google Scholar]
- Yang S, Parisien M, Major F, Roux B. RNA structure determination using SAXS data. J Phys Chem B. 2010;114:10039–10048. doi: 10.1021/jp1057308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J Mol Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitevin F, Orland H, Doniach S, Koehl P, Delarue M. AQUASAXS: a web server for computation and fitting of SAXS profiles with non-uniformally hydrated atomic models. Nucleic Acids Res. 2011;39:W184–W189. doi: 10.1093/nar/gkr430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen J, Makowski L, Sosnick Tobin R, Freed KF. Modeling the hydration layer around proteins: applications to small- and wide-angle X-ray scattering. Biophys J. 2011;101:2061–2069. doi: 10.1016/j.bpj.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Morris RJ, Hexemer A, Grandison S, Zwart PH. Computation of small-angle scattering profiles with three- dimensional Zernike polynomials. Acta Crystallogr A. 2012;68:278–285. doi: 10.1107/S010876731104788X. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modelling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Monie TP, Allain FH, Matthews S, Curry S, Svergun DI. Conformation of polypyrimidine tract binding protein in solution. Structure. 2006;14:1021–1027. doi: 10.1016/j.str.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Förster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. J Mol Biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons C, D'Abramo M, Svergun DI, Orozco M, Bernadó P, Fernández-Recio J. Structural characterization of protein-protein complexes by integrating computational docking with small-angle scattering data. J Mol Biol. 2010;403:217–230. doi: 10.1016/j.jmb.2010.08.029. [DOI] [PubMed] [Google Scholar]
- Zheng W, Tekpinar M. Accurate flexible fitting of high-resolution protein structures to small-angle X-ray scattering data using a coarse-grained model with implicit hydration shell. Biophys J. 2011;101:2981–2991. doi: 10.1016/j.bpj.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinozzi F, Beltramini M. Quafit: a novel method for the quaternary structure determination from small-angle scattering data. Biophys J. 2012;103:511–521. doi: 10.1016/j.bpj.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Eady NA, Brown KA, Svergun DI. Addition of missing loops and domains to protein models by x-ray solution scattering. Biophys J. 2002;83:3113–3125. doi: 10.1016/S0006-3495(02)75315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Tsuruta H, Chiu W. Scaling structure factor amplitudes in electron cryomicroscopy using X-ray solution scattering. J Struct Biol. 1999;128:51–57. doi: 10.1006/jsbi.1999.4173. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, Grossmann JG, Liu H, Putz NS, Roberts GC, Volkmann N, Hanein D, Barsukov IL, Critchley DR. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc Natl Acad Sci USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Watson C, Sharp JS, Handel TM, Prestegard JH. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure. 2011;19:1138–1148. doi: 10.1016/j.str.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowieson NP, King G, Cookson D, Ross I, Huber T, Hume DA, Kobe B, Martin JL. Cortactin adopts a globular conformation and bundles actin into sheets. J Biol Chem. 2008;283:16187–16193. doi: 10.1074/jbc.M708917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Rossi A, Avila-Sakar A, Kim SJ, Velázquez-Muriel J, Strop P, Liang H, Krukenberg KA, Liao M, Kim HM, Sobhanifar S, Dötsch V, Rajpal A, Pons J, Agard DA, Cheng Y, Sali A. A method for integrative structure determination of protein-protein complexes. Bioinformatics. 2012;28:3282–3289. doi: 10.1093/bioinformatics/bts628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Schneidman D, Velázquez-Muriel J, Sali A. Putting the pieces together: integrative structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Bonvin AM. On the usefulness of ion-mobility mass spectrometry and SAXS data in scoring docking decoys. Acta Crystallogr D. 2013;69:683–694. doi: 10.1107/S0907444913007063. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Clore GM. Using small angle solution scattering data in Xplor-NIH structure calculations. Prog Nucl Magn Reson Spectrosc. 2014;80:1–11. doi: 10.1016/j.pnmrs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multidomain protein structures by combination of solution small-angle X-ray scattering and NMR data. J Am Chem Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- Hennig J, Wang I, Sonntag M, Gabel F, Sattler M. Combining NMR and small angle X-ray and neutron scattering in the structural analysis of a ternary protein-RNA complex. J Biomol NMR. 2013;56:17–30. doi: 10.1007/s10858-013-9719-9. [DOI] [PubMed] [Google Scholar]
- Lapinaite A, Simon B, Skjaerven L, Rakwalska-Bange M, Gabel F, Carlomagno T. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013;502:519–523. doi: 10.1038/nature12581. [DOI] [PubMed] [Google Scholar]
- Engelman DM, Moore PB. Determination of quaternary structure by small angle neutron scattering. Annu Rev Biophys Bioeng. 1975;4:219–241. doi: 10.1146/annurev.bb.04.060175.001251. [DOI] [PubMed] [Google Scholar]
- Grossmann JG, Callaghan AJ, Marcaida MJ, Luisi BF, Alcock FH, Tokatlidis K, Moulin M, Haertlein M, Timmins P. Complementing structural information of modular proteins with small angle neutron scattering and contrast variation. Eur Biophys J. 2008;37:603–611. doi: 10.1007/s00249-008-0278-z. [DOI] [PubMed] [Google Scholar]
- Neylon C. Small angle neutron and X-ray scattering in structural biology: recent examples from the literature. Eur Biophys J. 2008;37:531–541. doi: 10.1007/s00249-008-0259-2. [DOI] [PubMed] [Google Scholar]
- Stuhrmann HB. Small-angle scattering and its interplay with crystallography, contrast variation in SAXS and SANS. Acta Crystallogr A. 2008;64:181–191. doi: 10.1107/S0108767307046569. [DOI] [PubMed] [Google Scholar]
- Whitten AE, Cai S, Trewhella J. MULCh: ModULes for the analysis of small-angle neutron contrast variation data from biomolecular complexes. J Appl Crystallogr. 2008;41:222–226. [Google Scholar]
- DiCapua E, Schnarr M, Timmins PA. The location of DNA in complexes of recA protein with double-stranded DNA. A neutron scattering study. Biochemistry. 1989;28:3287–3292. doi: 10.1021/bi00434a025. [DOI] [PubMed] [Google Scholar]
- Svergun DI, Nierhaus KH. A map of protein-rRNA distribution in the 70 S Escherichia coli ribosome. J Biol Chem. 2000;275:14432–14439. doi: 10.1074/jbc.275.19.14432. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Grishaev A, Whitten AH, Tsigelny I, Taylor P, Trewhella J. Small-angle solution scattering of the neuroligin/β-neurexin complex reveals its synaptic arrangement. Structure. 2007;15:693–705. doi: 10.1016/j.str.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten AE, Jacques DA, Hamouda B, Hanley T, King GF, Guss MJ, Trewhella J, Langley DB. The structure of the Sda-KinA complex suggests an allosteric mechanism of histidine kinase inhibition. J Mol Biol. 2007;368:407–420. doi: 10.1016/j.jmb.2007.01.064. [DOI] [PubMed] [Google Scholar]
- Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proc Natl Acad Sci USA. 2008;105:18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri BN, Gupta S, Urban VS, Chance MR, D'Mello R, Smith L, Lyons K, Gee J. A combined global and local approach to elucidate spatial organization of the Mycobacterial ParB-parS partition assembly. Biochemistry. 2011;50:1799–1807. doi: 10.1021/bi1016759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mély Y, Svergun DI, Moras D. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- Clifton LA, Neylon C, Lakey JH. Examining protein-lipid complexes using neutron scattering. Methods Mol Biol. 2013;974:119–150. doi: 10.1007/978-1-62703-275-9_7. [DOI] [PubMed] [Google Scholar]
- Gabel F, Lensink MF, Clantin B, Jacob-Dubuisson F, Villeret V, Ebel C. Probing the conformation of FhaC with small-angle neutron scattering and molecular modeling. Biophys J. 2014;107:185–196. doi: 10.1016/j.bpj.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynde SA, Skar-Gislinge N, Pedersen MC, Midtgaard SR, Simonsen JB, Schweins R, Mortensen K, Arleth L. Small-angle scattering gives direct structural information about a membrane protein inside a lipid environment. Acta Crystallogr D. 2014;70:371–383. doi: 10.1107/S1399004713028344. [DOI] [PubMed] [Google Scholar]
- Inoko Y, Yamamoto M, Fujiwara S, Ueki T. X-ray scattering study of the shape of the DNA region in nucleosome core particle with synchrotron radiation. J Biochem. 1992;111:310–316. doi: 10.1093/oxfordjournals.jbchem.a123755. [DOI] [PubMed] [Google Scholar]
- Bu Z, Engelman DM. A method for determining transmembrane helix association and orientation in detergent micelles using small angle x-ray scattering. Biophys J. 1999;77:1064–1073. doi: 10.1016/S0006-3495(99)76956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishaev A, Anthis NJ, Clore GM. Contrast-matched small-angle X-ray scattering from a heavy-atom-labeled protein in structure determination: application to a lead-substituted calmodulin-peptide complex. J Am Chem Soc. 2012;134:14686–14689. doi: 10.1021/ja306359z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Contreras LM, Smith D, Qu G, Huang T, Spruce LA, Seeholzer SH, Belfort M, Van Duyne GD. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res. 2014;42:5347–5360. doi: 10.1093/nar/gku140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tokuda JM, Topping T, Sutton JL, Meisburger SP, Pabit SA, Gloss LM, Pollack L. Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Res. 2014;42:8767–8776. doi: 10.1093/nar/gku562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwamoto S, Akiyama S, Fujisawa TJ. Radiation damage to a protein solution, detected by synchrotron X-ray small-angle scattering: dose-related considerations and suppression by cryoprotectants. Synchrotron Radiat. 2004;11:462–468. doi: 10.1107/S0909049504019272. [DOI] [PubMed] [Google Scholar]
- Doniach S. Changes in biomolecular conformation seen by small angle X-ray scattering. Chem Rev. 2001;101:1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- Trewhella J, Krueger JK. Small-angle solution scattering reveals information on conformational dynamics in calcium-binding proteins and in their interactions with regulatory targets. Methods Mol Biol. 2002;173:137–159. doi: 10.1385/1-59259-184-1:137. [DOI] [PubMed] [Google Scholar]
- Ashish, Juncadella IJ, Garg R, Boone CD, Anguita J, Krueger JK. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J Biol Chem. 2008;283:2761–2772. doi: 10.1074/jbc.M708325200. [DOI] [PubMed] [Google Scholar]
- Durand D, Vivès C, Cannella D, Pérez J, Pebay-Peyroula E, Vachette P, Fieschi F. NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J Struct Biol. 2010;169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Chaudhuri BN, Dean R. The evidence of large-scale DNA-induced compaction in the mycobacterial chromosomal ParB. J Mol Biol. 2011;413:901–907. doi: 10.1016/j.jmb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti RF, Rodi DJ, Gore DB, Makowski L. Wide-angle X-ray solution scattering as a probe of ligand-induced conformational changes in proteins. Chem Biol. 2004;11:1431–1443. doi: 10.1016/j.chembiol.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Makowski L, Bardhan J, Gore D, Lal J, Mandava S, Park S, Rodi DJ, Ho NT, Ho C, Fischetti RF. WAXS studies of the structural diversity of hemoglobin in solution. J Mol Biol. 2011;408:909–921. doi: 10.1016/j.jmb.2011.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadó P, Svergun DI. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol Biosyst. 2012;8:151–167. doi: 10.1039/c1mb05275f. [DOI] [PubMed] [Google Scholar]
- Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel F. Small angle neutron scattering for the structural study of intrinsically disordered proteins in solution: a practical guide. Methods Mol Biol. 2012;896:123–135. doi: 10.1007/978-1-4614-3704-8_8. [DOI] [PubMed] [Google Scholar]
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Park S, Makowski L, Roux B. A rapid coarse residue-based computational method for X-ray solution scattering characterization of protein folds and multiple conformational states of large protein complexes. Biophys J. 2009;96:4449–4463. doi: 10.1016/j.bpj.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Różycki B, Kim YC, Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. 2011;19:109–116. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Weinkam P, Sali A, Lee KK. All-atom ensemble modeling to analyze small-angle x-ray scattering of glycosylated proteins. Structure. 2013;21:321–331. doi: 10.1016/j.str.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Receveur-Bréchot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Sibille N, Bernado P. Structural characterization of intrinsically disordered proteins by the combined use of NMR and SAXS. Biochem Soc Trans. 2012;40:955–962. doi: 10.1042/BST20120149. [DOI] [PubMed] [Google Scholar]
- Bertini I, Giachetti A, Luchinat C, Parigi G, Petoukhov MV, Pierattelli R, Ravera E, Svergun DI. Conformational space of flexible biological macromolecules from average data. J Am Chem Soc. 2010;132:13553–13558. doi: 10.1021/ja1063923. [DOI] [PubMed] [Google Scholar]
- Schwalbe M, Ozenne V, Bibow S, Jaremko M, Jaremko L, Gajda M, Jensen MR, Biernat J, Becker S, Mandelkow E, Zweckstetter M, Blackledge M. Predictive atomic resolution descriptions of intrinsically disordered hTau40 and α-synuclein in solution from NMR and small angle scattering. Structure. 2014;22:238–249. doi: 10.1016/j.str.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Minton AP. How can biochemical reactions within cells differ from those in test tubes? J Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- Makowski L, Rodi DJ, Mandava S, Minh DD, Gore DB, Fischetti RF. Molecular crowding inhibits intramolecular breathing motions in proteins. J Mol Biol. 2008;375:529–546. doi: 10.1016/j.jmb.2007.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc. 2010;132:8690–6896. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D, Roh JH, Behrouzi R, Briber RM, Woodson SA. Crowders perturb the entropy of RNA energy landscapes to favor folding. J Am Chem Soc. 2013;135:10055–10063. doi: 10.1021/ja4030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akabayov SR, Akabayov B, Richardson CC, Wagner G. Molecular crowding enhanced ATPase activity of the RNA helicase eIF4A correlates with compaction of its quaternary structure and association with eIF4G. J Am Chem Soc. 2013;135:10040–10047. doi: 10.1021/ja404404h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akabayov B, Akabayov SR, Lee SJ, Wagner G, Richardson CC. Impact of macromolecular crowding on DNA replication. Nat Commun. 2013;4:1615. doi: 10.1038/ncomms2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg DP, Argyle B. Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophys J. 2014;106:905–914. doi: 10.1016/j.bpj.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein BK, Feigin LA, Lvov YM, Gvozdev RI, Marakushev SA, Likhtenshtein GI. Determination of the distance between heavy-atom markers in haemoglobin and histidine decarboxylase in solution by small-angle x-ray scattering. FEBS Lett. 1980;116:107–110. doi: 10.1016/0014-5793(80)80539-4. [DOI] [PubMed] [Google Scholar]
- Kataoka M, Nakasako M, Tokunaga F. Structural information on proteins obtainable from small-angle X-ray scattering with heavy-atom labeling. Application to solubilized bacteriorhodopsin. J Appl Crystallogr. 1988;21:355–362. [Google Scholar]
- Mathew-Fenn RS, Das R, Silverman JA, Walker PA, Harbury PA. A molecular ruler for measuring quantitative distance distributions. PLoS One. 2008;3:e3229. doi: 10.1371/journal.pone.0003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Tsai CL, Claridge SA, Mendillo ML, Smith JM, Williams GJ, Mastroianni AJ, Alivisatos AP, Putnam CD, Kolodner RD, Tainer JA. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci USA. 2013;110:17308–17313. doi: 10.1073/pnas.1308595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhrmann HB, Notbohm H. Configuration of the four iron atoms in dissolved human hemoglobin as studied by anomalous dispersion. Proc Natl Acad Sci USA. 1981;78:6216–6220. doi: 10.1073/pnas.78.10.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miake-Lye RC, Doniach S, Hodgson KO. Anomalous x-ray scattering from terbium-labeled parvalbumin in solution. Biophys J. 1983;41:287–292. doi: 10.1016/S0006-3495(83)84440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Mills TT, Kwok LW, Maskel GS, Millett IS, Doniach S, Finkelstein KD, Herschlag D, Pollack L. Counterion distribution around DNA probed by solution X-ray scattering. Phys Rev Lett. 2003;90:188103. doi: 10.1103/PhysRevLett.90.188103. [DOI] [PubMed] [Google Scholar]
- Makowski L, Bardhan J, Gore D, Rodi DJ, Fischetti RF. Multiwavelength anomalous diffraction using medium-angle X-ray solution scattering (MADMAX) Biophys J. 2012;102:927–933. doi: 10.1016/j.bpj.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield VJ, Scott DJ. Anomalous small angle X-ray scattering simulations: Proof of concept for distance measurements for nanoparticle-labelled biomacromolecules in solution. PLoS One. 2014;9:e95664. doi: 10.1371/journal.pone.0095664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew E, Mirza A, Menhart N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J Synchrotron Radiat. 2004;11:314–318. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Inoko Y. Size-exclusion chromatography combined with small-angle X-ray scattering optics. J Chromatogr A. 2009;1216:7461–7465. doi: 10.1016/j.chroma.2009.02.053. [DOI] [PubMed] [Google Scholar]
- Didry D, Cantrelle FX, Husson C, Roblin P, Moorthy AM, Perez J, Le Clainche C, Hertzog M, Guittet E, Carlier MF, van Heijenoort C, Renault L. How a single residue in individual b-thymosin/WH2 domains controls their functions in actin assembly. EMBO J. 2011;31:1000–1013. doi: 10.1038/emboj.2011.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez J, Nishino Y. Advances in X-ray scattering: from solution SAXS to achievements with coherent beams. Curr Opin Struct Biol. 2012;22:670–678. doi: 10.1016/j.sbi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Sokolova AV, Kreplak L, Wedig T, Mücke N, Svergun DI, Herrmann H, Aebi U, Strelkov SV. Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc Natl Acad Sci USA. 2006;103:16206–16211. doi: 10.1073/pnas.0603629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson TE, Craig BA, Kondrashkina E, Bailey-Kellogg C, Friedman AM. Analysis of self-associating proteins by singular value decomposition of solution scattering data. Biophys J. 2008;94:4906–4923. doi: 10.1529/biophysj.107.113167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel J, Bernadó P, Svergun DI, Tauler R, Pons M. Low-resolution structures of transient protein-protein complexes using small-angle X-ray scattering. J Am Chem Soc. 2009;131:4378–4386. doi: 10.1021/ja808490b. [DOI] [PubMed] [Google Scholar]
- Chandola H, Williamson TE, Craig BA, Friedman AM, Bailey-Kellogg C. Stoichiometries and affinities of interacting proteins from concentration series of solution scattering data: decomposition by least squares and quadratic optimization. J Appl Crystallogr. 2014;47:899–914. doi: 10.1107/S1600576714005913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Billas IM, Takacs M, Graewert MA, Moras D, Svergun DI. Reconstruction of quaternary structure from X-ray scattering by equilibrium mixtures of biological macromolecules. Biochemistry. 2013;52:6844–6855. doi: 10.1021/bi400731u. [DOI] [PubMed] [Google Scholar]
- Cammarata M, Levantino M, Schotte F, Anfinrud PA, Ewald F, Choi J, Cupane A, Wulff M, Ihee H. Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering. Nat Methods. 2008;5:881–886. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MH, Toft KN, David G, Havelund S, Pérez J, Vestergaard B. Time-resolved SAXS measurements facilitated by online HPLC buffer exchange. J Synchrotron Radiat. 2010;17:769–773. doi: 10.1107/S0909049510030372. [DOI] [PubMed] [Google Scholar]
- Graceffa R, Nobrega RP, Barrea RA, Kathuria SV, Chakravarthy S, Bilsel O, Irving TC. Sub-millisecond time-resolved SAXS using a continuous-flow mixer and X-ray microbeam. J Synchrotron Radiat. 2013;20:820–825. doi: 10.1107/S0909049513021833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria SV, Guo L, Graceffa R, Barrea R, Nobrega RP, Matthews CR, Irving TC, Bilsel O. Minireview: structural insights into early folding events using continuous-flow time-resolved small-angle X-ray scattering. Biopolymers. 2011;95:550–558. doi: 10.1002/bip.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutze R, Moffat K. Time-resolved structural studies at synchrotrons and X-ray free electron lasers: opportunities and challenges. Curr Opin Struct Biol. 2012;22:651–659. doi: 10.1016/j.sbi.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösslea M, Manakovaa E, Holzingerb J, Vanataluc K, Mayb RP, Heumanna H. Time-resolved small-angle neutron scattering of proteins in solution. Physica B. 2000;276:532–533. [Google Scholar]
- Perevozchikova T, Stanley CB, McWilliams-Koeppen HP, Rowe EL, Berthelier V. Investigating the structural impact of the glutamine repeat in huntingtin assembly. Biophys J. 2014;107:411–421. doi: 10.1016/j.bpj.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnlund D, Johansson LC, Wickstrand C, Barty A, Williams GJ, Malmerberg E, Davidsson J, Milathianaki D, DePonte DP, Shoeman RL, Wang D, James D, Katona G, Westenhoff S, White TA, Aquila A, Bari S, Berntsen P, Bogan M, van Driel TB, Doak RB, Kjær KS, Frank M, Fromme R, Grotjohann I, Henning R, Hunter MS, Kirian RA, Kosheleva I, Kupitz C, Liang M, Martin AV, Nielsen MM, Messerschmidt M, Seibert MM, Sjöhamn J, Stellato F, Weierstall U, Zatsepin NA, Spence JC, Fromme P, Schlichting I, Boutet S, Groenhof G, Chapman HN, Neutze R. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat Methods. 2014;11:923–926. doi: 10.1038/nmeth.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Malmerberg E, Westenhoff S, Katona G, Cammarata M, Wöhri AB, Johansson LC, Ewald F, Eklund M, Wulff M, Davidsson J, Neutze R. Structural dynamics of light-driven proton pumps. Structure. 2009;17:1265–1275. doi: 10.1016/j.str.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Diaz JF, Andreu JM, Diakun G, Towns-Andrews E, Bordas J. Structural intermediates in the assembly of taxoid-induced microtubules and GDP-tubulin double rings: time-resolved X-ray scattering. Biophys J. 1996;70:2408–2420. doi: 10.1016/S0006-3495(96)79809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Muniyappan S, Oang KY, Kim JG, Nozawa S, Sato T, Koshihara SY, Henning R, Kosheleva I, Ki H, Kim Y, Kim TW, Kim J, Adachi S, Ihee H. Direct observation of cooperative protein structural dynamics of homodimeric hemoglobin from 100 ps to 10 ms with pump- probe X-ray solution scattering. J Am Chem Soc. 2012;134:7001–7008. doi: 10.1021/ja210856v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard B, Groenning M, Roessle M, Kastrup JS, van de Weert M, Flink JM, Frokjaer S, Gajhede M, Svergun DI. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SB, Macchi F, Raccosta S, Langkilde AE, Giehm L, Kyrsting A, Svane AS, Manno M, Christiansen G, Nielsen NC, Oddershede L, Vestergaard B, Otzen DE. Wildtype and A30P mutant alpha-synuclein form different fibril structures. PLoS One. 2013;8:e67713. doi: 10.1371/journal.pone.0067713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canady MA, Tsuruta H, Johnson JE. Analysis of rapid, large-scale protein quaternary structural changes: time-resolved X-ray solution scattering of Nudaurelia capensis omega virus (NomegaV) maturation. J Mol Biol. 2001;311:803–814. doi: 10.1006/jmbi.2001.4896. [DOI] [PubMed] [Google Scholar]
- Lee KK, Tsuruta H, Hendrix RW, Duda RL, Johnson JE. Cooperative reorganization of a 420 subunit virus capsid. J Mol Biol. 2005;352:723–735. doi: 10.1016/j.jmb.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Matsui T, Tsuruta H, Johnson JE. Balanced electrostatic and structural forces guide the large conformational change associated with maturation of T = 4 virus. Biophys J. 2010;98:1337–1343. doi: 10.1016/j.bpj.2009.12.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack L, Tate MW, Finnefrock AC, Kalidas C, Trotter S, Darnton NC, Lurio L, Austin RH, Batt CA, Gruner SM, Mochrie SG. Time resolved collapse of a folding protein observed with small angle x-ray scattering. Phys Rev Lett. 2001;86:4962–4965. doi: 10.1103/PhysRevLett.86.4962. [DOI] [PubMed] [Google Scholar]
- Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Conformational landscape of cytochrome c folding studied by microsecond-resolved small-angle X-ray scattering. Proc Natl Acad Sci USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa T, Kimura T, Ishimori K, Morishima I, Matsui T, Ikeda-Saito M, Takahashi S, Akiyama S, Fujisawa T. Time-resolved small-angle X-ray scattering investigation of the folding dynamics of heme oxygenase: implication of the scaling relationship for the submillisecond intermediates of protein folding. J Mol Biol. 2006;357:997–1008. doi: 10.1016/j.jmb.2005.12.089. [DOI] [PubMed] [Google Scholar]
- Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SG, Pande V, Doniach S, Herschlag D, Pollack L. Rapid compaction during RNA folding. Proc Natl Acad Sci USA. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SG, Thiyagarajan P, Doniach S, Pollack L, Herschlag D. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J Mol Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- Andersson M, Vincent J, van der Spoel D, Davidsson J, Neutze R. A proposed time-resolved X-ray scattering approach to track local and global conformational changes in membrane transport proteins. Structure. 2008;16:21–28. doi: 10.1016/j.str.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Abuillan W, Vorobiev A, Hartel A, Jones NG, Engstler M, Tanaka M. Quantitative determination of the lateral density and intermolecular correlation between proteins anchored on the membrane surfaces using grazing incidence small-angle X-ray scattering and grazing incidence X-ray fluorescence. J Chem Phys. 2012;137:204907. doi: 10.1063/1.4767569. [DOI] [PubMed] [Google Scholar]
- Meisburger SP, Warkentin M, Chen H, Hopkins JB, Gillilan RE, Pollack L, Thorne RE. Breaking the radiation damage limit with Cryo-SAXS. Biophys J. 2013;104:227–236. doi: 10.1016/j.bpj.2012.11.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PB. The effects of thermal disorder on the solution-scattering profiles of macromolecules. Biophys J. 2014;106:1489–1496. doi: 10.1016/j.bpj.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldin DK, Poon HC, Shneerson VL, Howells M, Chapman HN, Kirian RA, Schmidt KE, Spence JCH. Beyond small-angle x-ray scattering: exploiting angular correlations. Phys Rev B. 2010;81:174105. [Google Scholar]
- Liu H, Poon BK, Saldin DK, Spence JC, Zwart PH. Three-dimensional single-particle imaging using angular correlations from X-ray laser data. Acta Crystallogr A. 2013;69:365–373. doi: 10.1107/S0108767313006016. [DOI] [PubMed] [Google Scholar]


