Abstract
Mitochondria import more than 1,000 different proteins from the cytosol. The proteins are synthesized as precursors on cytosolic ribosomes and are translocated by protein transport machineries of the mitochondrial membranes. Five main pathways for protein import into mitochondria have been identified. Most pathways use the translocase of the outer mitochondrial membrane (TOM) as the entry gate into mitochondria. Depending on specific signals contained in the precursors, the proteins are subsequently transferred to different intramitochondrial translocases. In this article, we discuss the connection between protein import and mitochondrial membrane architecture. Mitochondria possess two membranes. It is a long-standing question how contact sites between outer and inner membranes are formed and which role the contact sites play in the translocation of precursor proteins. A major translocation contact site is formed between the TOM complex and the presequence translocase of the inner membrane (TIM23 complex), promoting transfer of presequence-carrying preproteins to the mitochondrial inner membrane and matrix. Recent findings led to the identification of contact sites that involve the mitochondrial contact site and cristae organizing system (MICOS) of the inner membrane. MICOS plays a dual role. It is crucial for maintaining the inner membrane cristae architecture and forms contacts sites to the outer membrane that promote translocation of precursor proteins into the intermembrane space and outer membrane of mitochondria. The view is emerging that the mitochondrial protein translocases do not function as independent units, but are embedded in a network of interactions with machineries that control mitochondrial activity and architecture.
Keywords: contact site, membrane architecture, MICOS, mitochondria, protein sorting, protein translocase
Introduction
Mitochondria consist of four compartments: outer membrane, intermembrane space, inner membrane, and matrix (Fig. 1). Mitochondria are known best for their central role in cellular energy metabolism, resulting in the synthesis of ATP by oxidative phosphorylation. However, mitochondria play many more crucial functions in eukaryotic cells, including the metabolism of amino acids and lipids, the biosynthesis of heme and iron-sulfur clusters, the involvement in cellular stress responses and a central role in programmed cell death (apoptosis).1–10
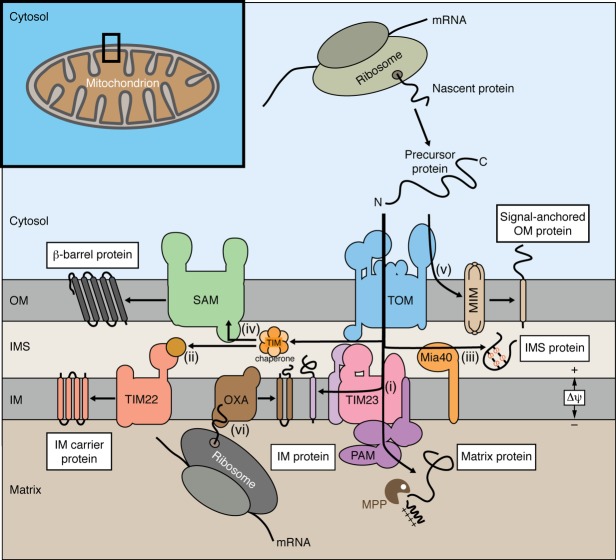
Figure 1.
Biogenesis of mitochondrial proteins. Most mitochondrial proteins are synthesized on cytosolic ribosomes and imported into mitochondria by the translocase of the outer membrane (TOM). (i) Presequence-carrying preproteins are transferred from TOM to the presequence translocase of the inner membrane (TIM23). Hydrophilic preproteins are transported into the matrix with the help of the presequence translocase-associated motor (PAM). Preproteins with hydrophobic sorting signal are arrested in the TIM23 complex and are laterally released into the inner membrane (IM). The presequences are proteolytically removed by the mitochondrial processing peptidase (MPP). (ii) The precursors of multispanning inner membrane proteins like the carrier proteins are imported via TOM, the small TIM chaperones of the intermembrane space (IMS) and the carrier translocase of the inner membrane (TIM22). (iii) IMS proteins with cysteine motifs are imported via TOM and the mitochondrial IMS import and assembly (MIA) machinery. (iv) The precursors of outer membrane (OM) β-barrel proteins use TOM, small TIM chaperones and the sorting and assembly machinery (SAM) for insertion into the outer membrane. (v) For α-helical outer membrane proteins, different pathways have been described. Shown is the import of a precursor protein via the mitochondrial import (MIM) complex. (vi) A small number of hydrophobic proteins are encoded by mtDNA and synthesized in the matrix. These proteins are typically exported into the inner membrane by the cytochrome oxidase activity (OXA) translocase. Δψ, membrane potential across the inner mitochondrial membrane (drives protein import via TIM23 and TIM22).
Proteomic studies revealed that mitochondria contain more than 1,000 different proteins.11–16 Although mitochondria carry a complete genetic system in the matrix, only ∼1% of mitochondrial proteins are encoded by the mitochondrial genome. Most mitochondrial-encoded proteins are strongly hydrophobic subunits of the oxidative phosphorylation machinery of the inner membrane. They are synthesized on mitochondrial ribosomes and are typically exported into the inner membrane by the OXA (cytochrome oxidase activity) translocase [Fig. 1, pathway (vi)].17,18
The vast majority of mitochondrial proteins are encoded by nuclear genes and synthesized on cytosolic ribosomes. The precursor proteins carry targeting signals that direct the precursors to receptors on the mitochondrial surface, followed by translocation into mitochondria. Experimental systems to study mitochondrial protein import mostly use precursor proteins that can be imported in a post-translational manner. However, it has been shown that cytosolic ribosomes associate with mitochondria and various precursor proteins can initiate translocation into mitochondria while part of the precursor is still being synthesized on ribosomes (cotranslational import).19–27 It is likely that both post-translational and cotranslational mechanisms are important for the biogenesis of mitochondrial proteins.
From electron micrographic pictures of mitochondria, sites of close contact between outer and inner mitochondrial membranes have been known for decades.19,28–34 The molecular nature and function of membrane contact sites for mitochondrial activity and biogenesis are the subject of ongoing research. In the current view, mitochondria do not possess one defined, permanent contact site structure but contain multiple dynamic contact sites that involve a variety of protein complexes from outer and inner membranes. Here, we first provide an overview on the five main protein import pathways into mitochondria and then discuss how preprotein translocases are involved in forming different types of contact sites. Recent findings indicate that contact sites are involved in at least three protein import pathways and stimulate the import of different classes of precursor proteins by promoting an efficient cooperation of translocases.
Five Mitochondrial Protein Import Pathways—An Overview
Depending on their intramitochondrial destination, mitochondrial precursor proteins contain distinct types of targeting and intramitochondrial sorting signals. Currently, at least five major classes of precursor proteins can be distinguished that use different pathways of translocation into mitochondria (Fig. 1). The protein translocase of the outer membrane (TOM) is the major mitochondrial entry site used by the majority of import pathways.
The classical presequence pathway is used by ∼60% of all mitochondrial precursor proteins.35 The preproteins are synthesized with amino-terminal presequences that form positively charged amphipathic α-helices.36 Upon recognition by Tom receptors, the preproteins are translocated through an outer membrane channel formed by Tom40 and are transferred to the presequence translocase of the inner membrane (TIM23 complex) [Fig. 1, pathway (i)].37–39 Hydrophilic preproteins are imported into the matrix with the help of the presequence translocase-associated motor (PAM).40–44 Preproteins destined for the inner membrane contain a hydrophobic sorting signal behind the positively charged matrix targeting signal. These preproteins are laterally released into the lipid phase of the inner membrane.45,46 For both, matrix-translocated and inner membrane-sorted preproteins, the positively charged matrix targeting signal is proteolytically removed by the mitochondrial processing peptidase (MPP).47–49
The carrier pathway imports the precursors of noncleavable multispanning inner membrane proteins. The precursors contain internal targeting signals that are not removed during import but remain part of the mature mitochondrial protein.50 The hydrophobic metabolite carriers of the inner membrane are the major substrates of this pathway. The precursors are initially translocated by the TOM complex, yet do not use the TIM23 complex, but bind to soluble TIM chaperones of the intermembrane space that are formed by small Tim proteins [Fig. 1, pathway (ii)].51 The small TIM chaperones guide the hydrophobic precursors to the carrier translocase of the inner membrane (TIM22 complex) that mediates membrane insertion of the proteins in a membrane potential (Δψ)-driven manner.52
It had been assumed that the mitochondrial intermembrane space is a reducing environment and thus proteins are in a reduced state. The identification of an oxidative protein import and folding system in the intermembrane space changed this view and led to the discovery of a large number of oxidized, disulfide-containing intermembrane space proteins.53–60 We termed the system the mitochondrial intermembrane space import and assembly (MIA) machinery. The MIA machinery accepts preproteins after their translocation through TOM, recognizes a cysteine-containing targeting signal and inserts disulfide bonds into the proteins [Fig. 1, pathway (iii)].61–66
The mitochondrial outer membrane contains two types of transmembrane proteins, proteins with α-helical transmembrane segments and β-barrel proteins. The precursors of β-barrel proteins are initially recognized and translocated by the TOM complex. The precursors are transferred to the intermembrane space side and bind to the small TIM chaperones [Fig. 1, pathway (iv)]. Insertion of the proteins into the outer membrane is mediated by the sorting and assembly machinery (SAM).67–70
The biogenesis of α-helical outer membrane proteins involves several distinct pathways and is only understood in part. The mitochondrial import (MIM) complex promotes insertion of some α-helical proteins into the outer membrane, in particular proteins with amino-terminal membrane anchor (termed signal-anchored proteins) as well as multispanning outer membrane proteins [Fig. 1, pathway (v)].71–75 Tom receptors can be involved in the recognition of multispanning proteins before their insertion by the MIM complex. As discussed below, a special signal-anchored protein uses the presequence pathway, followed by MIM-dependent outer membrane insertion.76,77 Various pathways have been discussed for proteins with carboxy-terminal membrane anchor (tail-anchored proteins), including a lipid-mediated insertion into the outer membrane,78,79 and an involvement of the SAM complex for some of the proteins.80,81
In this article, we will discuss that physical contacts between protein complexes of mitochondrial outer and inner membranes promote the translocation of precursor proteins not only via the presequence pathway, but also via the MIA and β-barrel pathways.
TOM-TIM Contact Sites and the Presequence Pathway
The first evidence for import of mitochondrial proteins via contact sites was obtained by the accumulation of translocation intermediates of cleavable preproteins across both mitochondrial membranes.82 The two-membrane spanning preproteins were present in a hydrophilic membrane environment, suggesting that they were accumulated in hydrophilic transport channels.83 The copurification of TOM and TIM23 complexes together with an accumulated preprotein directly demonstrated the existence of translocation contact sites.42,84–94 TOM and TIM23 complexes transiently interact in the absence of preproteins,95–97 yet their interaction is strongly stabilized by a preprotein in transit [Fig. 2(A)]. The channels formed by the TOM and TIM23 translocases are so narrow that folded proteins cannot pass98–102 and thus preproteins have to be translocated in an unfolded conformation.29,41,103–113
Figure 2.
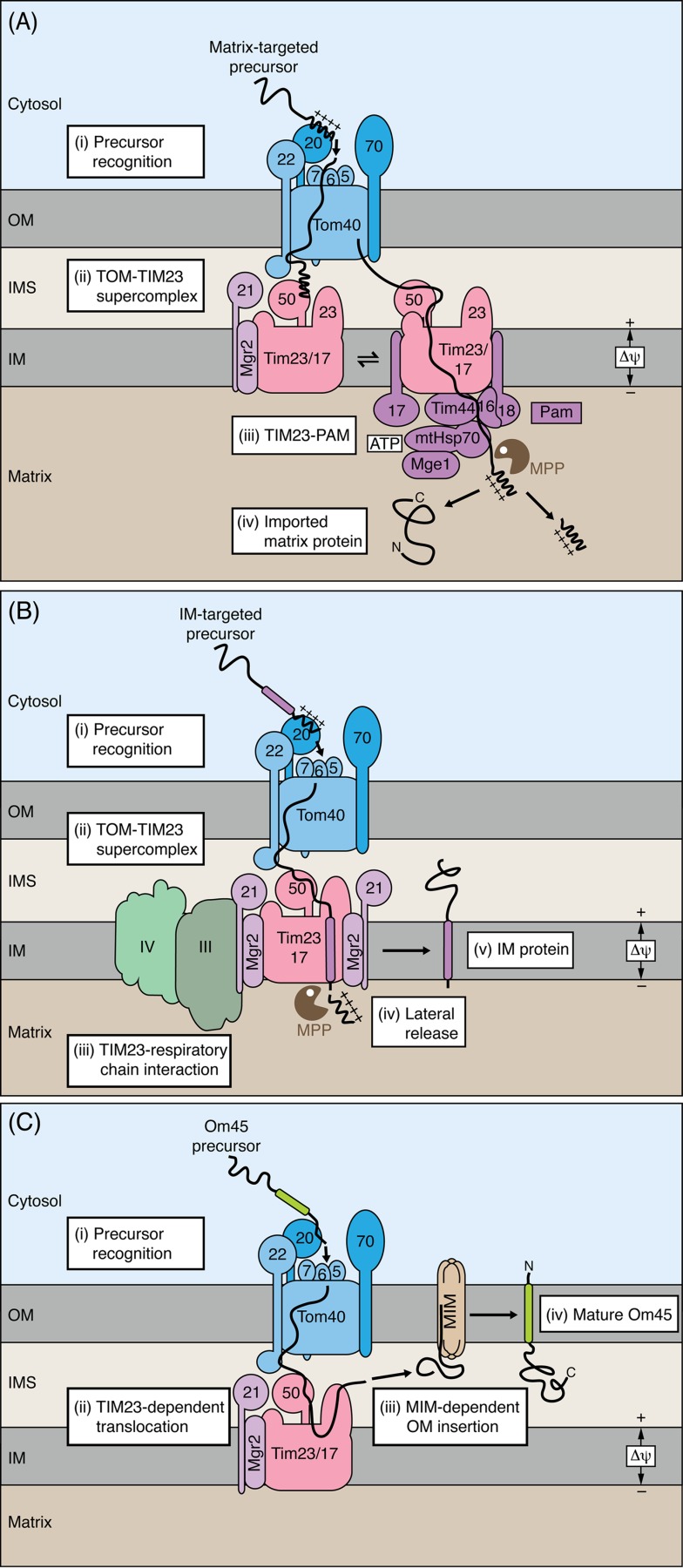
Translocation contact sites as core of the presequence pathway of mitochondria. (A) Matrix-targeted precursor proteins typically carry cleavable amino-terminal presequences. The presequences are recognized by receptors of the translocase of the outer membrane (TOM). Upon translocation through the Tom40 channel, the preproteins are transferred to the presequence translocase of the inner membrane (TIM23 complex). Several Tom and Tim proteins cooperate in the formation of dynamic TOM-TIM23 translocation contact sites. Two forms of the TIM23 complex are in dynamic exchange with each other. Both TIM23 forms are involved in the formation of TOM-TIM23 supercomplexes. In an early stage of translocation, Tim21 is associated with TIM23, whereas in later stages, the presequence translocase-associated motor (PAM) is associated with TIM23. The mitochondrial processing peptidase (MPP) removes the presequences. OM, outer membrane; IMS, intermembrane space; IM, inner membrane. (B) Cleavable preproteins inserted into the inner membrane (IM) carry a hydrophobic sorting signal behind the positively charged matrix targeting signal. Insertion of these preproteins into the inner membrane can be mediated by the Tim21-containing TIM23 complex without PAM. Mgr2 functions as lateral gatekeeper of the TIM23 complex and controls the proper sorting of preproteins into the inner membrane. The respiratory chain complexes III and IV interact with the Tim21-containing TIM23 complex and stimulate the Δψ-dependent preprotein insertion. (C) The outer membrane protein Om45 uses an unusual biogenesis pathway that involves TOM, TIM23 and the mitochondrial import (MIM) complex.
Several Tom and Tim proteins participate in the formation of TOM-TIM23 translocation contact sites: the intermembrane space domain of the receptor Tom22, Tom40, and Tom7 of the outer membrane translocase,88,96,114–116 as well as Tim50, Tim23, and Tim21 of the inner membrane translocase85,86,89,95,97,117,118 [Fig. 2(A)]. Tim50 plays a central role in the TOM-TIM23 reaction cycle. In the absence of preproteins, the intermembrane space domain of Tim50 keeps the translocation channel formed by Tim23 in a closed state.119 Presequence-carrying preproteins are initially recognized by the receptors Tom20 and Tom22 on the cytosolic side of the outer membrane.120–124 Upon passing through the Tom40 channel of the outer membrane, the presequences bind to the intermembrane space domain of Tom22.125–128 Tom7 may be involved at this import stage. Tim50 promotes the efficient binding of preproteins to Tom22 at the trans side of the TOM complex,88,95,117,118 underscoring the close cooperation between Tom and Tim proteins. Tim50 then functions as inner membrane receptor for preproteins and in cooperation with the intermembrane space domain of Tim23 transfers the preproteins to the Tim23 channel.85,95,100,117,129–134 Tim21 plays a regulatory role in preprotein transfer through the intermembrane space.88,132 The membrane potential (Δψ) across the inner membrane activates the Tim23 channel and exerts an electrophoretic effect on the positively charged presequences.100,135
The TIM23 complex functions in a dynamic manner and distinct TIM23 forms have been identified [Fig. 2(A)].88,91,92,136 In an early stage of translocation, the TIM23 core complex consisting of Tim50, Tim23, and Tim17 is associated with Tim21 and the recently identified subunit Mgr2.137,138 The motor subunit Pam17 plays a regulatory role. Pam17 replaces Tim21 at the translocase and initiates assembly of further motor subunits to the TIM23 complex.90,91,132,139,140 Tim44 functions as membrane docking site for the central motor component, the matrix heat shock protein 70 (mtHsp70).139,141–145 Two further membrane-bound cochaperones, the J-protein Pam18 (also termed Tim14) and its partner Pam16 (Tim16) regulate the ATP-driven activity of mtHsp70.42–44,87,146–153 The nucleotide release factor Mge1 promotes the release of ADP from mtHsp70 to initiate a new round of the mtHsp70 reaction cycle.154–157 The mtHsp70 motor drives the ATP-dependent import of preproteins into the matrix by a combination of pulling and trapping of the precursor polypeptide and thus promotes unfolding of carboxy-terminal precursor domains on the mitochondrial surface.40,41,93,158–160
During preprotein import, both forms of the TIM23 complex, TIM23-Tim21 and TIM23-PAM, cooperate with TOM to form TOM-TIM23-preprotein supercomplexes.92 TOM-TIM23-Tim21 is found in translocation contact sites at early import stages and TOM-TIM23-PAM preferentially at later stages. The translocation of presequence-carrying preproteins across the mitochondrial outer and inner membranes thus involves a dynamic interaction between TOM, different TIM23 states and the motor PAM and is driven by presequence binding, Δψ and ATP. The presequences sequentially interact with binding sites at multiple steps of their translocation pathway, including receptors on the cytosolic side, the channel protein Tom40, the intermembrane space domain of Tom22, Tim50, Tim23, and chaperones.40,122,123,128,130,132,161–166 During or after translocation into the matrix, the preproteins are cleaved by MPP to remove the positively charged presequences.
Imaging studies revealed that mitochondrial import sites are not equally distributed across the membranes, but cluster in microdomains,167,168 suggesting that several TOM-TIM23-preprotein supercomplexes are organized into larger translocation contact sites. Electron cryotomography shows that the contact sites are connected via protein–protein interactions, not via direct contact of the lipid phases of the membranes.168 The high resolution structures of various hydrophilic import components and domains have been determined and provided important information on the recognition of presequences, function of cochaperones and processing of preproteins.48,114,116,123,131,149,169–175 However, high resolution structures of the membrane domains (channels) of TOM and TIM complexes are lacking so far. This is a major reason that despite a wealth of knowledge on individual reaction steps, the exact mechanisms of preprotein translocation and the complete TOM-TIM23-PAM reaction cycle have not been elucidated yet.
Lateral Escape from the Presequence Pathway
TOM-TIM23 supercomplexes efficiently guide preproteins from the outer membrane to the inner membrane. However, they do not form completely sealed structures, but preproteins in transit are partially exposed to the intermembrane space.176
At the level of the inner membrane, the import of cleavable preproteins branches into the matrix route and the inner membrane sorting route.45,46 Hydrophilic preproteins are fully translocated into the matrix, whereas preproteins with a hydrophobic sorting signal behind the positively charged matrix targeting signal are typically arrested in the TIM23 complex and are laterally released into the inner membrane (stop transfer pathway).45 The TIM23-Tim21 complex functions as insertase that can insert preproteins into the inner membrane without the motor PAM136 [Fig. 2(B)] (whereas an association with PAM is strictly required for preprotein translocation into the matrix). The small hydrophobic TIM23 subunit Mgr2 acts as lateral gatekeeper of the translocase. Mgr2 is not required for the translocation of preproteins into the matrix, but plays a regulatory role by delaying the release of preproteins into the lipid phase of the inner membrane.94 Mgr2 recognizes the inner membrane-sorting signal and permits its lateral exit, whereas preproteins without a functional sorting signal are prevented from entering the inner membrane.
A direct connection between the TIM23 complex and the respiratory chain complexes III (cytochrome bc1 complex) and IV (cytochrome c oxidase) stimulates the insertion of preproteins into the inner membrane [Fig. 2(B)]. Tim21 provides the link between preprotein translocase and respiratory supercomplexes III-IV.137,177–179 The exact mechanism of how the TIM23-respiratory chain connection promotes preprotein import has not been identified. TIM23 complexes in the direct vicinity of proton-pumping respiratory complexes likely experience a higher proton-motive force and thus preprotein import is stimulated.180 The Tim21-respiratory chain connection may also play a role in the assembly of respiratory chain complexes. Human Tim21 was found to dynamically interact with the TIM23 complex and with assembly intermediates of the respiratory chain, termed MITRAC (mitochondrial translation regulation assembly intermediate of cytochrome oxidase).181 Human Tim21 thus helps to transfer preproteins from the presequence translocase to the respiratory chain. Additionally, a recent study showed that the mitochondrial ADP/ATP carrier associates with the TIM23 complex and may support preprotein import when the respiratory activity is low.182 When oxidative phosphorylation is inhibited, mitochondria do not export ATP, but can import glycolysis-generated ATP in exchange for ADP, thus generating a membrane potential and providing ATP to drive the TIM23-PAM machinery.
It should be added that not all preproteins with hydrophobic segments are arrested in the inner membrane. Some hydrophobic proteins or protein segments are transferred to the matrix via TIM23-PAM and are exported into the inner membrane by the OXA machinery that has been conserved from bacteria (YidC) to mitochondria. This pathway has been termed the conservative sorting pathway.183–188 A special case of conservative sorting represents the biogenesis of the Rieske iron-sulfur protein of respiratory complex III. After initial translocation of the precursor into the matrix and proteolytic processing, the folded iron-sulfur domain is exported to the intermembrane space by the AAA-ATPase Bcs1.189 Currently, many more inner membrane proteins sorted by a stop transfer mechanism are known than proteins using conservative sorting. Further work will be required to define the substrates of the conservative sorting pathway.
Recent studies surprisingly showed that TOM-TIM23 contact sites can even be used for protein translocation into the outer membrane. The protein Om45 is anchored in the outer membrane by an amino-terminal anchor sequence and exposes a large domain into the intermembrane space.76,77,190,191 The precursor of Om45 initially embarks on the presequence pathway using TOM and TIM23 complexes and the Δψ across the inner membrane [Fig. 2(C)]. However, the precursor is not translocated into the matrix but escapes from the presequence route and is inserted into the outer membrane.76,77 The outer membrane MIM complex, which has been known to insert various α-helical outer membrane proteins arriving from the cytosolic side, is involved in this final stage of Om45 biogenesis.76 MIM may mediate membrane insertion of Om45 from the intermembrane space side or promote the assembly of Om45 in the outer membrane. Thus, three translocases, TOM, TIM23 and MIM, cooperate to form a new intramitochondrial sorting pathway, revealing a high versatility of how contact sites are used for protein import into mitochondria.
In summary, the presequence translocase shows a remarkable flexibility. It dynamically interacts with TOM, PAM, ADP/ATP carrier, and respiratory chain complexes and thus TOM-TIM23 translocation contact sites undergo a multi-step reaction cycle and control different intramitochondrial sorting routes.
Mitochondrial Contact Site and Cristae Organizing System
The mitochondrial inner membrane displays a characteristic architecture with extended membrane invaginations, termed cristae. Two domains of the inner membrane can be distinguished: the inner boundary membrane that is closely opposed to the outer membrane and the cristae membranes [Fig. 3(A)].192 The two membrane domains differ in their protein distribution: preprotein translocases and components of the fusion machinery are predominantly localized in the inner boundary membrane, whereas the F1Fo-ATP synthase and respiratory chain complexes are mainly found in the cristae membranes.193–201 Both membrane domains are connected through tubular openings, termed crista junctions, which probably limit diffusion of molecules and help to create a microenvironment in the intracristal space enhancing the capacity of the oxidative phosphorylation system of mitochondria.34,198,202–205
Figure 3.
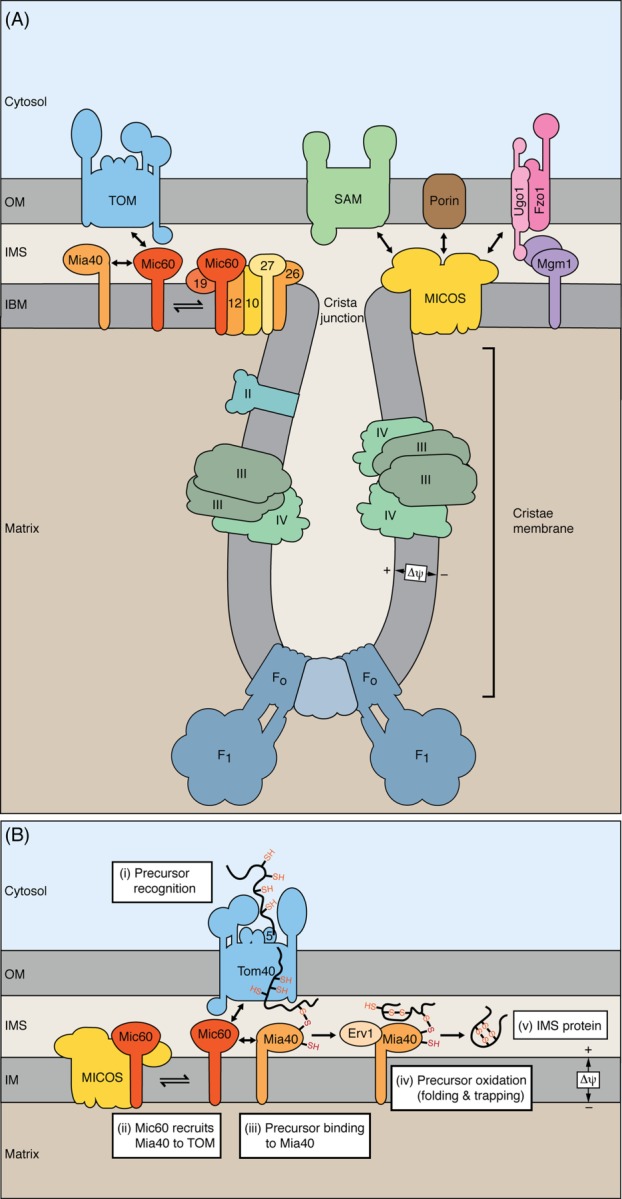
Mitochondrial contact site and cristae organizing system and protein import into intermembrane space. (A) The mitochondrial inner membrane consists of the inner boundary membrane (IBM), cristae membranes and the connecting crista junctions. Protein translocases are enriched in the IBM, whereas respiratory complexes and the F1Fo-ATP synthase are enriched in cristae membranes. The MICOS complex is enriched at crista junctions and exposes protein domains into the intermembrane space (IMS). MICOS is crucial for maintenance of the cristae structure and forms multiple contact sites with outer membrane (OM) protein complexes. Fzo1, Ugo1, and Mgm1, components of the mitochondrial membrane fusion machinery. (B) The precursors of intermembrane space proteins with cysteine motifs are translocated through the Tom40 channel and are recognized by the oxidoreductase Mia40 that functions as receptor in the intermembrane space. Mic60 interacts with TOM and Mia40 and thus positions Mia40 close to the Tom40 channel. In cooperation with the sulfhydryl oxidase Erv1, Mia40 oxidizes the imported proteins, leading to disulfide formation and folding of the proteins.
A large hetero-oligomeric protein complex of the inner mitochondrial membrane is enriched at crista junctions.206,207 The complex and its evolutionary conserved components originally received several different names,26,206–215 yet recently the nomenclature was unified and the complex is termed MICOS for mitochondrial contact site and cristae organizing system.216 MICOS is composed of six subunits that all expose domains to the intermembrane space [Fig. 3(A)]. The two core components, Mic10 and Mic60 (mitofilin/Fcj1), and three further subunits, Mic12, Mic26, and Mic27, are integral inner membrane proteins. The sixth subunit, Mic19, is a peripheral membrane protein. In addition, the inner membrane protein Aim24 is required for the integrity of MICOS, but is not a structural component of the mature complex.217 Cells lacking structural MICOS subunits show a strongly altered morphology of the inner membrane.26,206–208,213,218–220 Most crista junctions are lost and the cristae membranes form large internal membrane stacks that are detached from the inner boundary membrane. MICOS is thus required for maintaining the connections between inner boundary membrane and cristae membranes.
MICOS plays a dual role. In addition to maintaining crista junctions, multiple interactions of MICOS with protein complexes of the outer membrane were observed. MICOS was found to interact with TOM and SAM complexes, with the abundant outer membrane channel protein porin (VDAC) and with a component of the mitochondrial fusion machinery, the outer membrane protein Ugo1.26,207–209,221–223 MICOS is considered as an important element in the formation of outer–inner membrane contact sites.224 Super-resolution microscopy indicated that MICOS complexes are regularly distributed in the inner membrane, forming MICOS arrays.225 MICOS is thus involved in a network of interactions that are important for maintaining inner membrane architecture and formation of contact sites.
MICOS and Protein Import into Intermembrane Space and Outer Membrane
Cells lacking the largest MICOS subunit, Mic60, were found to import cysteine-rich intermembrane space proteins with reduced efficiency.26,226 Mutant mitochondria lacking other MICOS subunits imported these proteins like wild-type mitochondria. How does Mic60 influence import via the MIA pathway, yet none of the other MICOS subunits does? We propose that Mic60 can function in two modes, a MICOS-dependent one and a MICOS-independent one [Fig. 3(B)].
In the MICOS-dependent mode, Mic60 is of central importance for inner membrane architecture. In the MICOS-independent mode, Mic60 forms contact sites with the TOM complex and transiently interacts with Mia40, the core component of the oxidative protein import and folding machinery of the intermembrane space [Fig. 3(B)]. The oxidoreductase Mia40 functions as receptor for precursors upon their passage through the TOM channel.53–55,227,228 Since Mia40 interacts with the cysteine-containing targeting signals of the precursors as soon as they emerge from the TOM channel,229–232 a close spatial proximity of Mia40 to the TOM complex promotes efficient protein import via the MIA pathway.26 However, a direct interaction between Mia40 and TOM has not been observed, but Mic60 performs an adapter-like function. By interacting with both TOM and Mia40, Mic60 positions Mia40 close to the exit of the TOM channel. In cooperation with the sulfhydryl oxidase Erv1, Mia40 oxidizes the precursors, leading to formation of disulfides and folding of the mature proteins.56–58,233–236 Thus, a close proximity to the TOM complex is important for Mia40 to capture precursor substrates directly at the entry gate.
Mitochondria deficient in Mic60 are not only impaired in the import of intermembrane space proteins, but also in the import of β-barrel proteins of the outer membrane. β-barrel precursors engage both TOM and SAM complexes on their biogenesis pathway (Fig. 4, left half). The precursor proteins are initially translocated through the TOM channel to the intermembrane space, bind to small TIM chaperones that shield hydrophobic regions of the precursors, and are inserted into the outer membrane by the SAM complex. TOM and SAM complexes transiently interact to form a supercomplex that promotes an efficient transfer of β-barrel precursors.237 In addition to the Mic60-TOM interaction, MICOS also interacts with the SAM complex (Fig. 4),207,209,221–223 and thus both interactions may contribute to an efficient biogenesis of β-barrel precursors. However, by dissecting the translocation of β-barrel precursors into distinct stages, Bohnert et al.238 indicated that the early step of precursor translocation through the TOM complex is promoted by Mic60, whereas the subsequent precursor interaction with SAM is not influenced by MICOS. Thus, the Mic60-TOM interaction is critical for stimulating β-barrel import. The molecular mechanism of β-barrel biogenesis stimulation by Mic60 remains to be elucidated.
Figure 4.
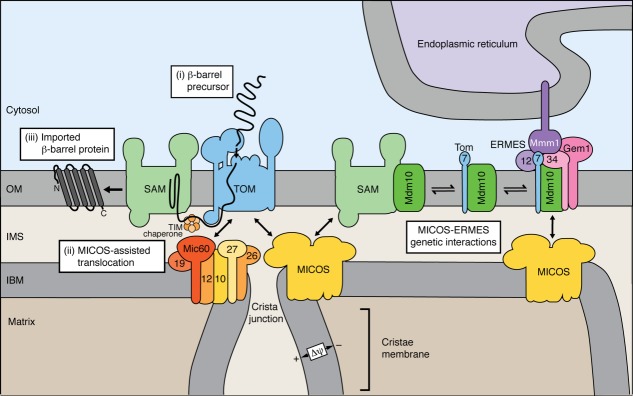
Biogenesis of mitochondrial outer membrane proteins and ER-mitochondria contact sites. Left half, β-barrel proteins of the outer membrane (OM) are imported into mitochondria via TOM, the small TIM chaperones of the intermembrane space (IMS) and the sorting and assembly machinery (SAM). MICOS interacts with TOM and SAM and stimulates β-barrel biogenesis. The transient interaction of Mic60 with TOM promotes translocation of β-barrel precursors to the intermembrane space. Right half, the SAM complex is in dynamic exchange with the ER-mitochondria encounter structure (ERMES) via the protein Mdm10. Tom7 binds to the SAM-free form of Mdm10 that can associate with ERMES. ERMES is composed of Mdm10, Mdm34, the adaptor protein Mdm12, the regulatory protein Gem1 and the ER localized protein Mmm1. MICOS and ERMES form genetic interactions.
Interestingly, for MIA substrates as well as for β-barrel precursors, only Mic60 is needed to stimulate their biogenesis, not any other MICOS subunit. Since the proper architecture of mitochondrial cristae depends on an intact MICOS complex, it is evident that the promotion of protein import by Mic60 does not depend on the morphology of the inner membrane. For example, in mitochondria lacking Mic10, most crista junctions are lost and large internal membrane stacks are formed like in mitochondria lacking Mic60.26,207,208,213 However, mic10Δ mitochondria possess fully active MIA and β-barrel pathways in contrast to mic60Δ mitochondria.26,238
We propose that the MICOS complex and in particular Mic60 represent a multifunctional molecular hub in the mitochondrial intermembrane space. Mic60 is crucial for maintaining mitochondrial inner membrane architecture and promotes biogenesis of proteins of the intermembrane space and outer membrane by forming contact sites with TOM. At present, two possibilities are conceivable to explain the distinct functional modes of Mic60 in wild-type mitochondria. (i) Mic60 molecules may be present in two distinct physical pools, a MICOS-integrated form and a form separated from MICOS, and thus perform MICOS-dependent and MICOS-independent functions (see model in Fig. 3). (ii) Alternatively, Mic60 may be part of MICOS in both functional modes (see model in Fig. 4). However, only the morphology function depends on the presence and activity of the other MICOS subunits. Mutant mitochondria show that Mic60 can interact with TOM and stimulate protein import also when other MICOS subunits are missing and MICOS is dissociated.26,238 Further studies will be required to define the molecular organization and functional mechanisms of Mic60.
Mitochondrial Organizing Network
We propose that translocation contact sites and MICOS are important parts of a large network of protein complexes that form an organizing center for mitochondrial biogenesis and activity. Mitochondrial outer and inner membranes are connected at sites that apparently involve numerous transient interactions. Probably none of the individual contact sites is sufficient to maintain the close connection of the inner boundary membrane with the outer membrane, but multiple contact sites are required.
The TOM complex is one of the important players in mitochondrial contact sites since it interacts with the TIM23 complex and the MICOS complex. Imaging studies show that TOM-TIM23 supercomplexes are clustered in the vicinity of crista junctions but not directly at the junctions,168 whereas MICOS complexes are enriched at crista junctions.206,207 Thus, the TOM-TIM23 and TOM-MICOS supercomplexes are in spatial vicinity, but form distinct contact sites that are part of a larger organizing network. It is open if these two membrane-spanning supercomplexes directly interact with each other. In addition, the TOM complex forms a transient supercomplex with the SAM complex in the outer membrane,237 and TOM is crucial for the import of carrier proteins, which are transferred to the small TIM chaperones but to our current knowledge do not use translocation contact sites.239,240 TOM complexes are thus engaged in several different pathways and interact with different partner complexes. Since TOM complexes are more abundant than the other mitochondrial protein translocases,84,241 they may exist in separate pools dedicated to different protein import pathways. Alternatively, a large dynamic pool of TOM complexes may be used to recruit TOM complexes to different partner complexes depending on the activity of the import pathways. Since each of the TOM-containing supercomplexes is of dynamic nature, we prefer the second scenario that TOM complexes engaged in formation of different supercomplexes are in exchange with each other.
The SAM complexes can interact with TOM as well as MICOS. In addition, SAM is in dynamic exchange with the endoplasmic reticulum (ER)-mitochondria encounter structure (ERMES), a protein complex that stably connects ER and the mitochondrial outer membrane.242–244 Though no evidence for a direct physical contact between SAM and ERMES has been reported so far, both complexes share a subunit. The mitochondrial distribution and morphology protein Mdm10 is a subunit of both complexes (Fig. 4, right half). In the SAM complex, Mdm10 promotes assembly of the TOM complex,81,245,246 whereas its exact function in the ERMES complex is unknown.247 Mdm10 can shuttle between SAM and ERMES. Interestingly, Tom7, a small subunit of the TOM complex, has a dual localization as well. Tom7 is not only found in the TOM complex, but also forms a complex with Mdm10. Since Tom7 binds to the SAM-free form of Mdm10, Tom7 promotes segregation of Mdm10 from the SAM complex and may favor the assembly of Mdm10 with the ERMES complex.248–250
Further ERMES subunits that are not in exchange with SAM were also found to be involved in the assembly of β-barrel proteins.251 This finding links outer membrane biogenesis to contact sites between mitochondria and ER. Such interorganellar contacts sites have gained a lot of interest in recent years since many vital cellular functions ranging from calcium homeostasis to phospholipid synthesis and mitochondrial fission have been associated with these tethering structures.242,244,252–258 In addition to yeast ERMES, further direct ER-mitochondria connections have been described, including the ER membrane protein complex (EMC) in yeast259 and mitofusin 2 in mammals.260,261 Two recent studies identified a molecular tether between vacuoles and mitochondria, termed vCLAMP, that may have complementary functions to ERMES in phospholipid metabolism.262,263
Future studies will have to unravel the mechanisms of molecular crosstalk between interorganellar contact sites as well as their structural and functional relationship with the mediators of mitochondrial biogenesis and membrane architecture, like protein translocases and MICOS. The importance of such wide-ranging connections is highlighted by the fact that MICOS not only interacts with TOM and SAM, but also shows strong genetic interactions with ERMES components (Fig. 4).208 Thus, multiple interactions involving TOM, SAM, MICOS, and ERMES point to a large ER-mitochondria organizing network, termed ERMIONE, which connects three membranes and functions as organizing center for mitochondrial architecture and biogenesis.224 The network likely involves further components such as mitochondrial fusion proteins, lipid synthesizing enzymes, the prohibitin ring complexes and the proteins Mdm31 and Mdm32 of the inner membrane that expose domains to the intermembrane space.211,217,256,264–274 ERMES, MICOS, and Mdm31/32 are also involved in the organization of mtDNA into nucleoids.220,247,265,275–277 Thus, ERMIONE likely spans from the ER lumen to the mitochondrial matrix and is also involved in the proper maintenance of mtDNA.
It will be a major task of future research to define the molecular functions of this mitochondrial organizing network. We propose that ERMIONE forms a central platform for coordinating mitochondrial biogenesis, architecture and activity, including import and assembly of proteins, biosynthesis and transfer of lipids, interorganellar and intraorganellar contact sites and mtDNA organization. Elucidation of the mechanisms that regulate these machineries will be an important topic. It had been assumed previously that mitochondrial protein import works in a constitutive, non-regulated manner. Recent studies, however, revealed that the TOM complex is extensively regulated by at least four cytosolic protein kinases that depending on the cellular growth conditions, metabolic state and cell cycle phases, stimulate or inhibit TOM biogenesis and activity and thus control the main mitochondrial entry gate.9,278–281 The efficiency of protein translocation into and across the inner membrane depends on the magnitude of the membrane potential and thus the protein import machinery can serve as a sensitive indicator for mitochondrial fitness and dysfunction. A reduced protein import efficiency has been found to trigger a mitochondrial stress response or the degradation of dysfunctional mitochondria by autophagy.282,283 Translocation contact sites are thus not only crucial for the import of proteins into mitochondria but also for the regulation of mitochondria and their integration into cellular metabolism and signaling.
References
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol. 2011;3:a007559. doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Rutter J. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 2013;27:2615–2627. doi: 10.1101/gad.229724.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria—a regulatory hub in metabolism, stress, and disease. Cell Metab. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokisch H, Scharfe C, Camp DG, II, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, Hixson KK, Mottaz HM, Zischka H, Ueffing M, Herman ZS, Davis RW, Meitinger T, Oefner PJ, Smith RD, Steinmetz LM. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2004;2:e160. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Fiumera HL, Dujardin G, Fox TD. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767–775. doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa M, Schatz G. Import of proteins into mitochondria: translatable mRNAs for imported mitochondrial proteins are present in free as well as mitochondria-bound cytoplasmic polysomes. J Biol Chem. 1982;257:13048–13055. [PubMed] [Google Scholar]
- Verner K. Co-translational protein import into mitochondria: an alternative view. Trends Biochem Sci. 1993;18:366–371. doi: 10.1016/0968-0004(93)90090-a. [DOI] [PubMed] [Google Scholar]
- Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie JA, Payne RM. Ribosomes specifically bind to mammalian mitochondria via protease-sensitive proteins on the outer membrane. J Biol Chem. 2004;279:9803–9810. doi: 10.1074/jbc.M307167200. [DOI] [PubMed] [Google Scholar]
- Yogev O, Karniely S, Pines O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J Biol Chem. 2007;282:29222–29229. doi: 10.1074/jbc.M704201200. [DOI] [PubMed] [Google Scholar]
- Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, Rapaport D, Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol Cell Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, Müller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, Hutu DP, Zerbes RM, Schulze-Specking A, Meyer HE, Martinou J-C, Rospert S, Rehling P, Meisinger C, Veenhuis M, Warscheid B, van der Klei IJ, Pfanner N, Chacinska A, van der Laan M. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenbrock CR. Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc Natl Acad Sci U S A. 1968;61:598–605. doi: 10.1073/pnas.61.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Guiard B, Wienhues U, Herzog V, Hartl FU, Neupert W. Translocation arrest by reversible folding of a precursor protein imported into mitochondria: a means to quantitate translocation contact sites. J Cell Biol. 1989;109:1421–1428. doi: 10.1083/jcb.109.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon L, Moll T, Vestweber D, Marshallsay B, Schatz G. Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J Cell Biol. 1989;109:2603–2616. doi: 10.1083/jcb.109.6.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Renken C, Martone ME, Young SJ, Ellisman M, Frey T. Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Frangakis AS, Typke D, Baumeister W. Cryo-electron tomography of Neurospora mitochondria. J Struct Biol. 2000;129:48–56. doi: 10.1006/jsbi.1999.4204. [DOI] [PubMed] [Google Scholar]
- Reichert AS, Neupert W. Contact sites between the outer and inner membrane of mitochondria—role in protein transport. Biochim Biophys Acta. 2002;1592:41–49. doi: 10.1016/s0167-4889(02)00263-x. [DOI] [PubMed] [Google Scholar]
- Mannella CA. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta. 2006;1763:542–548. doi: 10.1016/j.bbamcr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Neupert W, Rapaport D. Analysis and prediction of mitochondrial targeting signals. Methods Cell Biol. 2007;80:761–781. doi: 10.1016/S0091-679X(06)80035-X. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Hutu DP, Rehling P. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta. 2010;1803:732–739. doi: 10.1016/j.bbamcr.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W. The many faces of the mitochondrial TIM23 complex. Biochim Biophys Acta. 2010;1797:1045–1054. doi: 10.1016/j.bbabio.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Matouschek A, Azem A, Ratliff K, Glick BS, Schmid K, Schatz G. Active unfolding of precursor proteins during mitochondrial protein import. EMBO J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Voos W, Frazier AE, Lind M, Li Y, Geissler A, Dudek J, Müller H, Sickmann A, Meyer HE, Meisinger C, Guiard B, Rehling P, Pfanner N. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci U S A. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Brandt A, Cunningham K, Müller S, Hallberg RL, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Bömer U, Meijer M, Guiard B, Dietmeier K, Pfanner N, Rassow J. The sorting route of cytochrome b2 branches from the general mitochondrial import pathway at the preprotein translocase of the inner membrane. J Biol Chem. 1997;272:30439–30446. doi: 10.1074/jbc.272.48.30439. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9:615–625. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Mossmann D, Meisinger C, Vögtle FN. Processing of mitochondrial presequences. Biochim Biophys Acta. 2011;1819:1098–1106. doi: 10.1016/j.bbagrm.2011.11.007. [DOI] [PubMed] [Google Scholar]
- de Marcos-Lousa C, Sideris DP, Tokatlidis K. Translocation of mitochondrial inner-membrane proteins: conformation matters. Trends Biochem Sci. 2006;31:259–267. doi: 10.1016/j.tibs.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Koehler CM. The small Tim proteins and the twin Cx3C motif. Trends Biochem Sci. 2004;29:1–4. doi: 10.1016/j.tibs.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N. Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuan Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoé M, Ohwa Y, Ishikawa D, Ohshima C, Nishikawa S-I, Yamamoto H, Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- Terziyska N, Lutz T, Kozany C, Mokranjac D, Mesecke N, Neupert W, Herrmann JM, Hell K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579:179–184. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Rissler M, Wiedemann N, Pfannschmidt S, Gabriel K, Guiard B, Pfanner N, Chacinska A. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J Mol Biol. 2005;353:485–492. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J Mol Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Gabriel K, Milenkovic D, Chacinska A, Müller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J Mol Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics. 2012;11:1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim Biophys Acta. 2008;1783:601–609. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. The MIA system for protein import into the mitochondrial intermembrane space. Biochim Biophys Acta. 2008;1783:610–617. doi: 10.1016/j.bbamcr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Tienson HL. Redox regulation of protein folding in the mitochondrial intermembrane space. Biochim Biophys Acta. 2009;1793:139–145. doi: 10.1016/j.bbamcr.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris DP, Tokatlidis K. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13:1189–1204. doi: 10.1089/ars.2010.3157. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. Structural basis for the disulfide relay system in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13:1359–1373. doi: 10.1089/ars.2010.3099. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem. 2012;287:4426–4433. doi: 10.1074/jbc.R111.270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Neupert W, Rapaport D. Biogenesis of β-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Becker T, Vögtle FN, Stojanovski D, Meisinger C. Sorting and assembly of mitochondrial outer membrane proteins. Biochim Biophys Acta. 2008;1777:557–563. doi: 10.1016/j.bbabio.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta. 2010;1803:706–714. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Höhr AIC, Straub SP, Warscheid B, Becker T, Wiedemann N. Assembly of β-barrel proteins in the mitochondrial outer membrane. Biochim Biophys Acta. 2015;1853:74–88. doi: 10.1016/j.bbamcr.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem. 2008;283:120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- Popov-Celeketić J, Waizenegger T, Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol. 2008;376:671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likić VA, Gooley PR, Lithgow T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Krüger V, Lehmann W, Müller JM, Goroncy L, Zufall N, Lithgow T, Guiard B, Chacinska A, Wagner R, Meisinger C, Pfanner N. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J Cell Biol. 2011;194:387–395. doi: 10.1083/jcb.201102044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papic D, Krumpe K, Dukanovic J, Dimmer KS, Rapaport D. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J Cell Biol. 2011;194:397–405. doi: 10.1083/jcb.201102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz LS, Opaliński L, Schuler MH, Ellenrieder L, Ieva R, Böttinger L, Qiu J, van der Laan M, Wiedemann N, Guiard B, Pfanner N, Becker T. The presequence pathway is involved in protein sorting to the mitochondrial outer membrane. EMBO Rep. 2014;15:678–685. doi: 10.1002/embr.201338144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Tamura Y, Yoshihisa T, Endo T. A novel import route for an N-anchor mitochondrial outer membrane protein aided by the TIM23 complex. EMBO Rep. 2014;15:670–677. doi: 10.1002/embr.201338142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci. 2008;121:1990–1998. doi: 10.1242/jcs.024034. [DOI] [PubMed] [Google Scholar]
- Krumpe K, Frumkin I, Herzig Y, Rimon N, Özbalci C, Brügger B, Rapaport D, Schuldiner M. Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol Biol Cell. 2012;23:3927–3935. doi: 10.1091/mbc.E11-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Guiard B, Kozjak-Pavlovic V, Pfanner N, Meisinger C. Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J Cell Biol. 2007;179:881–893. doi: 10.1083/jcb.200706043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton N, Stroud DA, Milenkovic D, Guiard B, Pfanner N, Becker T. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J Mol Biol. 2010;396:540–549. doi: 10.1016/j.jmb.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Schleyer M, Neupert W. Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell. 1985;43:339–350. doi: 10.1016/0092-8674(85)90039-x. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Hartl FU, Guiard B, Neupert W. Mitochondrial precursor proteins are imported through a hydrophilic membrane environment. Eur J Biochem. 1987;169:289–293. doi: 10.1111/j.1432-1033.1987.tb13610.x. [DOI] [PubMed] [Google Scholar]
- Dekker PJT, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C. Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 2003;22:5370–5381. doi: 10.1093/emboj/cdg532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, Bilanchone V, Cumsky MG, Truscott KN, Pfanner N, Rehling P. Pam16 has an essential role in the mitochondrial protein import motor. Nat Struct Mol Biol. 2004;11:226–233. doi: 10.1038/nsmb735. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Popov-Celeketić D, Hell K, Neupert W. Role of Tim21 in mitochondrial translocation contact sites. J Biol Chem. 2005;280:23437–23440. doi: 10.1074/jbc.C500135200. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol Cell Biol. 2005;25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov-Celeketić D, Mapa K, Neupert W, Mokranjac D. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 2008;27:1469–1480. doi: 10.1038/emboj.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, van der Laan M, Mehnert CS, Guiard B, Mick DU, Hutu DP, Truscott KN, Wiedemann N, Meisinger C, Pfanner N, Rehling P. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol. 2010;30:307–318. doi: 10.1128/MCB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Rehling P. Remodelling of the active presequence translocase drives motor-dependent mitochondrial protein translocation. Nat Commun. 2014;5:4349. doi: 10.1038/ncomms5349. [DOI] [PubMed] [Google Scholar]
- Ieva R, Schrempp SG, Opaliński L, Wollweber F, Höß P, Heißwolf AK, Gebert M, Zhang Y, Guiard B, Rospert S, Becker T, Chacinska A, Pfanner N, van der Laan M. Mgr2 functions as lateral gatekeeper for preprotein sorting in the mitochondrial inner membrane. Mol Cell. 2014;56:641–652. doi: 10.1016/j.molcel.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Popov-Celeketić D, Mapa K, Gevorkyan-Airapetov L, Zohary K, Hell K, Azem A, Neupert W. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol Biol Cell. 2009;20:1400–1407. doi: 10.1091/mbc.E08-09-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci U S A. 2011;108:15179–15183. doi: 10.1073/pnas.1105921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Popov-Čeleketić D, Neupert W, Azem A, Mokranjac D. Cooperation of TOM and TIM23 complexes during translocation of proteins into mitochondria. J Mol Biol. doi: 10.1016/j.jmb.2014.07.015. in press doi: 10.1016/j.jmb.2014.07.015. PMID: 25083920 [Medline] [DOI] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner F, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Künkele KP, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat Struct Biol. 2001;8:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Bannwarth M, Meisinger C, Hill K, Model K, Krimmer T, Casadio R, Truscott KN, Schulz GE, Pfanner N, Wagner R. Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J Mol Biol. 2005;353:1011–1020. doi: 10.1016/j.jmb.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Eilers M, Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Pfaller R, Kleene R, Ito M, Tropschug M, Neupert W. Role of ATP in mitochondrial protein import: conformational alteration of a precursor protein can substitute for ATP requirement. J Biol Chem. 1988;263:4049–4051. [PubMed] [Google Scholar]
- Vestweber D, Schatz G. Point mutations destabilizing a precursor protein enhance its post-translational import into mitochondria. EMBO J. 1988;7:1147–1151. doi: 10.1002/j.1460-2075.1988.tb02924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienhues U, Becker K, Schleyer M, Guiard B, Tropschug M, Horwich AL, Pfanner N, Neupert W. Protein folding causes an arrest of preprotein translocation into mitochondria in vivo. J Cell Biol. 1991;115:1601–1609. doi: 10.1083/jcb.115.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, von Ahsen O, Müller H, Guiard B, Rassow J, Pfanner N. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J. 1996;15:2668–2677. [PMC free article] [PubMed] [Google Scholar]
- Bömer U, Maarse AC, Martin F, Geissler A, Merlin A, Schönfisch B, Meijer M, Pfanner N, Rassow J. Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J. 1998;17:4226–4237. doi: 10.1093/emboj/17.15.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume B, Klaus C, Ungermann C, Guiard B, Neupert W, Brunner M. Unfolding of preproteins upon import into mitochondria. EMBO J. 1998;17:6497–6507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Martin F, Guiard B, Pfanner N, Voos W. The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J. 2001;20:941–950. doi: 10.1093/emboj/20.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ratliff KS, Matouschek A. Protein unfolding by the mitochondrial membrane potential. Nat Struct Biol. 2002;9:301–307. doi: 10.1038/nsb772. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Choy J, Bustamante C, Matouschek A. Effect of protein structure on mitochondrial import. Proc Natl Acad Sci U S A. 2005;102:15435–15440. doi: 10.1073/pnas.0507324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci U S A. 2006;103:6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht R, Rehling P, Chacinska A, Brix J, Cadamuro SA, Volkmer R, Guiard B, Pfanner N, Zeth K. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 2006;7:1233–1238. doi: 10.1038/sj.embor.7400828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin J, Schulz C, Wrobel L, Bernhard O, Chacinska A, Jahn O, Schmidt B, Rehling P. Presequence recognition by the Tom40 channel contributes to precursor translocation into the mitochondrial matrix. Mol Cell Biol. 2014;34:3473–3485. doi: 10.1128/MCB.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj R, Jaremko L, Jaremko M, Becker S, Zweckstetter M. Molecular basis of the dynamic structure of the TIM23 complex in the mitochondrial intermembrane space. Structure. 2014;22:1501–1511. doi: 10.1016/j.str.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol. 2009;184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, Rehling P. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 2006;312:1523–1526. doi: 10.1126/science.1127628. [DOI] [PubMed] [Google Scholar]
- Mayer A, Nargang FE, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A, Kübrich M, Moczko M, Gärtner F, Mallet L, Bussereau F, Eckerskorn C, Lottspeich F, Dietmeier K, Jacquet M. The mitochondrial receptor complex: Mom22 is essential for cell viability and directly interacts with preproteins. Mol Cell Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Rimmer KA, Foo JH, Ng A, Petrie EJ, Shilling PJ, Perry AJ, Mertens HD, Lithgow T, Mulhern TD, Gooley PR. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J Mol Biol. 2011;405:804–818. doi: 10.1016/j.jmb.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Bolliger L, Junne T, Schatz G, Lithgow T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995;14:6318–6326. doi: 10.1002/j.1460-2075.1995.tb00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court DA, Nargang FE, Steiner H, Hodges RS, Neupert W, Lill R. Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol Cell Biol. 1996;16:4035–4042. doi: 10.1128/mcb.16.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol. 1997;17:6574–6584. doi: 10.1128/mcb.17.11.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the 'acid chain' hypothesis. EMBO J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J Cell Biol. 2011;195:643–656. doi: 10.1083/jcb.201105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom M, Dayan D, Demishtein-Zohary K, Mokranjac D, Neupert W, Azem A. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J Biol Chem. 2011;286:43809–43815. doi: 10.1074/jbc.M111.261040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Gebert M, Höpker J, Yan M, Li J, Wiedemann N, van der Laan M, Pfanner N, Sha B. Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J Mol Biol. 2011;411:513–519. doi: 10.1016/j.jmb.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko O, Melin J, Schulz C, Kilisch M, Hutu DP, Rehling P. Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 2013;32:886–898. doi: 10.1038/emboj.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman B, Kawano S, Yunoki-Esaki K, Anzai T, Endo T. NMR analyses on the interactions of the yeast Tim50 C-terminal region with the presequence and Tim50 core domain. FEBS Lett. 2014;588:678–684. doi: 10.1016/j.febslet.2013.12.037. [DOI] [PubMed] [Google Scholar]
- Bauer MF, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import: ΔΨ drives the movement of presequences. J Biol Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- van der Laan M, Meinecke M, Dudek J, Hutu DP, Lind M, Perschil I, Guiard B, Wagner R, Pfanner N, Rehling P. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol. 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- Gebert M, Schrempp SG, Mehnert CS, Heißwolf AK, Oeljeklaus S, Ieva R, Bohnert M, von der Malsburg K, Wiese S, Kleinschroth T, Hunte C, Meyer HE, Haferkamp I, Guiard B, Warscheid B, Pfanner N, van der Laan M. Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J Cell Biol. 2012;197:595–604. doi: 10.1083/jcb.201110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Heißwolf AK, Gebert M, Vögtle FN, Wollweber F, Mehnert CS, Oeljeklaus S, Warscheid B, Meisinger C, van der Laan M, Pfanner N. Mitochondrial inner membrane protease promotes assembly of presequence translocase by removing a carboxy-terminal targeting sequence. Nat Commun. 2013;4:2853. doi: 10.1038/ncomms3853. [DOI] [PubMed] [Google Scholar]
- Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, van der Laan M. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol Biol Cell. 2008;19:2642–2629. doi: 10.1091/mbc.E07-12-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller D. Pam17 and Tim44 act sequentially in protein import into the mitochondrial matrix. Int J Biochem Cell Biol. 2009;41:2343–2349. doi: 10.1016/j.biocel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Maarse AC, Krainer E, Kübrich M, Müller H, Meijer M, Craig EA, Pfanner N. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- Kronidou NG, Oppliger W, Bolliger L, Hannavy K, Glick BS, Schatz G, Horst M. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc Natl Acad Sci U S A. 1994;91:12818–12822. doi: 10.1073/pnas.91.26.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Guiard B, Neupert W, Cyr DM. The ΔΨ- and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO J. 1996;15:735–744. [PMC free article] [PubMed] [Google Scholar]
- Schilke BA, Hayashi M, Craig EA. Genetic analysis of complex interactions among components of the mitochondrial import motor and translocon in Saccharomyces cerevisiae. Genetics. 2012;190:1341–1353. doi: 10.1534/genetics.112.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat Struct Mol Biol. 2004;11:234–241. doi: 10.1038/nsmb734. [DOI] [PubMed] [Google Scholar]
- Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W. The presequence translocase-associated protein import motor of mitochondria: Pam16 functions in an antagonistic manner to Pam18. J Biol Chem. 2004;279:38047–38054. doi: 10.1074/jbc.M404319200. [DOI] [PubMed] [Google Scholar]
- D'Silva PR, Schilke B, Walter W, Craig EA. Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc Natl Acad Sci U S A. 2005;102:12419–12424. doi: 10.1073/pnas.0505969102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 2006;25:4675–4685. doi: 10.1038/sj.emboj.7601334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Berg A, Adam A, Neupert W, Hell K. Association of the Tim14-Tim16 subcomplex with the TIM23 translocase is crucial for function of the mitochondrial protein import motor. J Biol Chem. 2007;282:18037–18045. doi: 10.1074/jbc.M701895200. [DOI] [PubMed] [Google Scholar]
- D'Silva PR, Schilke B, Hayashi M, Craig EA. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol Biol Cell. 2008;19:424–432. doi: 10.1091/mbc.E07-08-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Joshi N, Chittoor B, Samji P, D'Silva P. Role of Magmas in protein transport and human mitochondria biogenesis. Hum Mol Genet. 2010;19:1248–1262. doi: 10.1093/hmg/ddq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais JE, Schilke B, Craig EA. Reevaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor. Mol Biol Cell. 2011;22:4740–4749. doi: 10.1091/mbc.E11-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Gambill BD, Craig EA. A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc Natl Acad Sci U S A. 1994;91:6481–6485. doi: 10.1073/pnas.91.14.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Gambill BD, Laloraya S, Ang D, Craig EA, Pfanner N. Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol Cell Biol. 1994;14:6627–6634. doi: 10.1128/mcb.14.10.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Prip-Buus C, Neupert W, Schwarz E. The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 1995;14:3452–3460. doi: 10.1002/j.1460-2075.1995.tb07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider HC, Westermann B, Neupert W, Brunner M. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 1996;15:5796–5803. [PMC free article] [PubMed] [Google Scholar]
- Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Brinker A, Paschen SA, Moarefi I, Hayer-Hartl M, Neupert W, Brunner M. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 2002;21:3659–3671. doi: 10.1093/emboj/cdf358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SY, Schilke BA, Hayashi M, Craig EA. Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor. J Biol Chem. 2014;289:28689–28696. doi: 10.1074/jbc.M114.588152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Rapaport D, Neupert W, Lill R. Mitochondrial protein import: Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- Esaki M, Kanamori T, Nishikawa S, Shin I, Schultz PG, Endo T. Tom40 protein import channel binds to non-native proteins and prevents their aggregation. Nat Struct Biol. 2003;10:988–994. doi: 10.1038/nsb1008. [DOI] [PubMed] [Google Scholar]
- Alder NN, Jensen RE, Johnson AE. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Malhotra K, Sathappa M, Landin JS, Johnson AE, Alder NN. Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat Struct Mol Biol. 2013;20:965–972. doi: 10.1038/nsmb.2613. [DOI] [PubMed] [Google Scholar]
- Wurm CA, Neumann D, Lauterbach MA, Harke B, Egner A, Hell SW, Jakobs S. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc Natl Acad Sci U S A. 2011;108:13546–13551. doi: 10.1073/pnas.1107553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold VA, Ieva R, Walter A, Pfanner N, van der Laan M, Kühlbrandt W. Visualizing active membrane protein complexes by electron cryotomography. Nat Commun. 2014;5:4129. doi: 10.1038/ncomms5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9–10 reveals a six-bladed α-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- Josyula R, Jin Z, Fu Z, Sha B. Crystal structure of yeast mitochondrial peripheral membrane protein Tim44p C-terminal domain. J Mol Biol. 2006;359:798–804. doi: 10.1016/j.jmb.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Igura M, Obita T, Ose T, Kojima R, Maenaka K, Endo T, Kohda D. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 2007;26:4777–4787. doi: 10.1038/sj.emboj.7601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T, Ohshima C, Maeda M, Endo T. Structural basis of functional cooperation of Tim15/Zim17 with yeast mitochondrial Hsp70. EMBO Rep. 2007;8:664–670. doi: 10.1038/sj.embor.7400990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qian X, Hu J, Sha B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J Biol Chem. 2009;284:23852–23859. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta. 2011;1808:955–970. doi: 10.1016/j.bbamem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Rassow J, Pfanner N. Mitochondrial preproteins en route from the outer membrane to the inner membrane are exposed to the intermembrane space. FEBS Lett. 1991;293:85–88. doi: 10.1016/0014-5793(91)81157-4. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol. 2007;179:1115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddar S, Dienhart MK, Stuart RA. The F1Fo-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J Biol Chem. 2008;283:6677–6686. doi: 10.1074/jbc.M708440200. [DOI] [PubMed] [Google Scholar]
- Rieger B, Junge W, Busch KB. Lateral pH gradient between OXPHOS complex IV and F0F1 ATP-synthase in folded mitochondrial membranes. Nat Commun. 2014;5:3103. doi: 10.1038/ncomms4103. [DOI] [PubMed] [Google Scholar]
- Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, Rehling P. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell. 2012;151:1528–1541. doi: 10.1016/j.cell.2012.11.053. [DOI] [PubMed] [Google Scholar]
- Mehnert CS, Rampelt H, Gebert M, Oeljeklaus S, Schrempp SG, Kochbeck L, Guiard B, Warscheid B, van der Laan M. The mitochondrial ADP/ATP carrier associates with the inner membrane presequence translocase in a stoichiometric manner. J Biol Chem. 2014;289:27352–27362. doi: 10.1074/jbc.M114.556498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Neupert W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science. 1990;247:930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Neupert W, Herrmann JM. Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria. J Cell Biol. 2005;170:881–888. doi: 10.1083/jcb.200505126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Mihara K. Topogenesis of mammalian Oxa1, a component of the mitochondrial inner membrane protein export machinery. J Biol Chem. 2009;284:14819–14827. doi: 10.1074/jbc.M809520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M, Rehling P, Guiard B, Herrmann JM, Pfanner N, van der Laan M. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- Park K, Botelho SC, Hong J, Österberg M, Kim H. Dissecting stop transfer versus conservative sorting pathways for mitochondrial inner membrane proteins in vivo. J Biol Chem. 2013;288:1521–1532. doi: 10.1074/jbc.M112.409748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener N, Ackermann M, Funes S, Neupert W. A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol Cell. 2011;44:191–202. doi: 10.1016/j.molcel.2011.07.036. [DOI] [PubMed] [Google Scholar]
- Burri L, Vascotto K, Gentle IE, Chan NC, Beilharz T, Stapleton DI, Ramage L, Lithgow T. Integral membrane proteins in the mitochondrial outer membrane of Saccharomyces cerevisiae. FEBS J. 2006;273:1507–1515. doi: 10.1111/j.1742-4658.2006.05171.x. [DOI] [PubMed] [Google Scholar]
- Lauffer S, Mäbert K, Czupalla C, Pursche T, Hoflack B, Rödel G, Krause-Buchholz U. Saccharomyces cerevisiae porin pore forms complexes with mitochondrial outer membrane proteins Om14p and Om45p. J Biol Chem. 2012;287:17447–17458. doi: 10.1074/jbc.M111.328328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation—linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Werner S, Neupert W. Functional and biogenetical heterogeneity of the inner membrane of rat-liver mitochondria. Eur J Biochem. 1972;25:379–396. doi: 10.1111/j.1432-1033.1972.tb01707.x. [DOI] [PubMed] [Google Scholar]
- Gilkerson RW, Selker JM, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546:355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- Vogel F, Bornhövd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm CA, Jakobs S. Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 2006;580:5628–5634. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Oostergetel GT, Lewejohann D, Braun HP, Boekema EJ. Row-like organization of ATP synthase in intact mitochondria determined by cryo-electron tomography. Biochim Biophys Acta. 2010;1797:272–277. doi: 10.1016/j.bbabio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci U S A. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt S, Wenzel D, Hildenbeutel M, Wurm CA, Herrmann JM, Jakobs S. The inner-mitochondrial distribution of Oxa1 depends on the growth conditions and on the availability of substrates. Mol Biol Cell. 2012;23:2292–2301. doi: 10.1091/mbc.E11-06-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B, Walter A, Horst A, Osiewacz HD, Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE. An electron microscope study of the mitochondrial structure. J Histochem Cytochem. 1953;1:188–211. doi: 10.1177/1.4.188. [DOI] [PubMed] [Google Scholar]
- Frey TG, Mannella CA. The internal structure of mitochondria. Trends Biochem Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- Frey TG, Renken CW, Perkins GA. Insight into mitochondrial structure and function from electron tomography. Biochim Biophys Acta. 2002;1555:196–203. doi: 10.1016/s0005-2728(02)00278-5. [DOI] [PubMed] [Google Scholar]
- Mannella CA. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim Biophys Acta. 2006;1762:140–147. doi: 10.1016/j.bbadis.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Körner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Körner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, Giaever G, Costanzo M, Troyanskaya OG, Caudy AA. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head BP, Zulaika M, Ryazantsev S, van der Bliek AM. A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Mol Biol Cell. 2011;22:831–841. doi: 10.1091/mbc.E10-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaja AK, Jans DC, Nikolov M, Vukotic M, Lytovchenko O, Ludewig F, Schliebs W, Riedel D, Urlaub H, Jakobs S, Deckers M. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Shi J, He Q, Lui K, Liu Y, Huang Y, Sheikh MS. CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem. 2012;287:7411–7426. doi: 10.1074/jbc.M111.277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber TA, Koob S, Heide H, Wittig I, Head B, van der Bliek A, Brandt U, Mittelborn M, Reichert AS. APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS One. 2013;8:e63683. doi: 10.1371/journal.pone.0063683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, Jakobs S, Ji J, Kozjak-Pavlovic V, Meisinger C, Odgren PR, Park SK, Rehling P, Reichert AS, Sheikh MS, Taylor SS, Tsuchida N, van der Bliek AM, van der Klei IJ, Weissman JS, Westermann B, Zha J, Neupert W, Nunnari J. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J Cell Biol. 2014;204:1083–1086. doi: 10.1083/jcb.201401006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner ME, Unger AK, Izawa T, Walther DM, Ozbalci C, Geimer S, Reggiori F, Brügger B, Mann M, Westermann B, Neupert W. Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife. 2014;3:e01684. doi: 10.7554/eLife.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JY, Lee TH, Kim JH, Yoo BH, Bahk YY, Koo HS, Han SS. Caenorhabditis elegans mitofilin homologs control the morphology of mitochondrial cristae and influence reproduction and physiology. J Cell Physiol. 2010;224:748–756. doi: 10.1002/jcp.22177. [DOI] [PubMed] [Google Scholar]
- Itoh K, Tamura Y, Iijima M, Sesaki H. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Mol Biol Cell. 2013;24:1842–1851. doi: 10.1091/mbc.E13-03-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C, Ross K, Straub S, Thiede B, Götz M, Goosmann C, Krischke M, Mueller MJ, Krohne G, Rudel T, Kozjak-Pavlovic V. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C, Barrera M, Dukanovic J, Eydt K, Harner M, Rabl R, Vogel F, Rapaport D, Neupert W, Reichert AS. The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol Biol Cell. 2012;23:2143–2155. doi: 10.1091/mbc.E11-10-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbes RM, Bohnert M, Stroud DA, von der Malsburg K, Kram A, Oeljeklaus S, Warscheid B, Becker T, Wiedemann N, Veenhuis M, van der Klei IJ, Pfanner N, van der Laan M. Role of MINOS in mitochondrial membrane architecture: cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J Mol Biol. 2012;422:183–191. doi: 10.1016/j.jmb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Bohnert M, Wiedemann N, Pfanner N. Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 2012;22:185–192. doi: 10.1016/j.tcb.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Jans DC, Wurm CA, Riedel D, Wenzel D, Stagge F, Deckers M, Rehling P, Jakobs S. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc Natl Acad Sci U S A. 2013;110:8936–8941. doi: 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varabyova A, Topf U, Kwiatkowska P, Wrobel L, Kaus-Drobek M, Chacinska A. Mia40 and MINOS act in parallel with Ccs1 in the biogenesis of mitochondrial Sod1. FEBS J. 2013;280:4943–4959. doi: 10.1111/febs.12409. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Cenacchi L, Ciofi-Baffoni S, Felli IC, Gallo A, Gonnelli L, Luchinat E, Sideris D, Tokatlidis K. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc Natl Acad Sci U S A. 2010;107:20190–20195. doi: 10.1073/pnas.1010095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicka A, Bragoszewski P, Chroscicki P, Wenz LS, Schulz C, Rehling P, Chacinska A. A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol Biol Cell. 2014;25:3999–4009. doi: 10.1091/mbc.E14-06-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Gabriel K, Guiard B, Schulze-Specking A, Pfanner N, Chacinska A. Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J Biol Chem. 2007;282:22472–22480. doi: 10.1074/jbc.M703294200. [DOI] [PubMed] [Google Scholar]
- Sideris DP, Tokatlidis K. Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol Microbiol. 2007;65:1360–1373. doi: 10.1111/j.1365-2958.2007.05880.x. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Ramming T, Müller JM, Wenz LS, Gebert N, Schulze-Specking A, Stojanovski D, Rospert S, Chacinska A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris DP, Petrakis N, Katrakili N, Mikropoulou D, Gallo A, Ciofi-Baffoni S, Banci L, Bertini I, Tokatlidis K. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol. 2009;187:1007–1022. doi: 10.1083/jcb.200905134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. Precursor oxidation by Mia40 and Erv1 promotes vectorial transport of proteins into the mitochondrial intermembrane space. Mol Biol Cell. 2008;19:226–236. doi: 10.1091/mbc.E07-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tienson HL, Dabir DV, Neal SE, Loo R, Hasson SA, Boontheung P, Kim SK, Loo JA, Koehler CM. Reconstitution of the Mia40-Erv1 oxidative folding pathway for the small Tim proteins. Mol Biol Cell. 2009;20:3481–3490. doi: 10.1091/mbc.E08-10-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol Cell. 2010;37:516–528. doi: 10.1016/j.molcel.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Böttinger L, Gornicka A, Czerwik T, Bragoszewski P, Loniewska-Lwowska A, Schulze-Specking A, Truscott KN, Guiard B, Milenkovic D, Chacinska A. In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol Biol Cell. 2012;23:3957–3969. doi: 10.1091/mbc.E12-05-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Wenz LS, Zerbes RM, Oeljeklaus S, Bohnert M, Stroud DA, Wirth C, Ellenrieder L, Thornton N, Kutik S, Wiese S, Schulze-Specking A, Zufall N, Chacinska A, Guiard B, Hunte C, Warscheid B, van der Laan M, Pfanner N, Wiedemann N, Becker T. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- Bohnert M, Wenz LS, Zerbes RM, Horvath SE, Stroud DA, von der Malsburg K, Müller JM, Oeljeklaus S, Perschil I, Warscheid B, Chacinska A, Veenhuis M, van der Klei IJ, Daum G, Wiedemann N, Becker T, Pfanner N, van der Laan M. Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol Biol Cell. 2012;23:3948–3956. doi: 10.1091/mbc.E12-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003;299:1747–1751. doi: 10.1126/science.1080945. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Becker K, Bauer MF, Walther E, Neupert W, Brunner M. Functional cooperation and stoichiometry of protein translocases of the outer and inner membranes of mitochondria. J Biol Chem. 1997;272:29963–29966. doi: 10.1074/jbc.272.47.29963. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Oeljeklaus S, Wiese S, Bohnert M, Lewandrowski U, Sickmann A, Guiard B, van der Laan M, Warscheid B, Wiedemann N. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413:743–750. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci U S A. 2011;108:14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Rissler M, Chacinska A, Sanjuán Szklarz LK, Milenkovic D, Kozjak V, Schönfisch B, Lohaus C, Meyer HE, Yaffe MP, Guiard B, Wiedemann N, Pfanner N. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 2010;11:187–193. doi: 10.1038/embor.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Wiedemann N, Rissler M, Strub A, Milenkovic D, Schönfisch B, Müller H, Kozjak V, Pfanner N. Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem. 2010;285:41222–41231. doi: 10.1074/jbc.M110.163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Thornton N, Stroud D, Meisinger C, Wiedemann N, Pfanner N. Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J Mol Biol. 2011;405:113–124. doi: 10.1016/j.jmb.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfannschmidt S, Rissler M, Milenkovic D, Becker T, Stojanovski D, Youngman MJ, Jensen RE, Chacinska A, Guiard B, Pfanner N, Wiedemann N. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Várnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G, Várnai P, Golenár T, Roy S, Purkins G, Schneider TG, Balla T, Hajnóczky G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26:1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, Loewen CJ, Prinz WA. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 2014;12:e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, Inatome R, Yanagi S. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem. 2004;279:28298–28303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Jakobs S, Vogel F, Altmann K, Westermann B. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J Cell Biol. 2005;168:103–115. doi: 10.1083/jcb.200410030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brügger B, Westermann B, Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338:815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Onguka O, Hobbs AE, Jensen RE, Iijima M, Claypool SM, Sesaki H. Role for two conserved intermembrane space proteins, Ups1p and Ups2p, in intra-mitochondrial phospholipid trafficking. J Biol Chem. 2012;287:15205–15218. doi: 10.1074/jbc.M111.338665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Thompson MN, Fei N, Hüttemann M, Greenberg ML. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J Biol Chem. 2012;287:17589–17597. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Korwitz A, Haag M, Tatsuta T, Dargazanli S, Baker M, Decker T, Lamkemeyer T, Rugarli EI, Langer T. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metab. 2014;20:158–171. doi: 10.1016/j.cmet.2014.04.016. [DOI] [PubMed] [Google Scholar]
- Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman MJ, Hobbs AE, Burgess SM, Srinivasan M, Jensen RE. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol. 2004;164:677–688. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Rao S, Schmidt O, Harbauer AB, Schönfisch B, Guiard B, Pfanner N, Meisinger C. Biogenesis of the preprotein translocase of the outer mitochondrial membrane: protein kinase A phosphorylates the precursor of Tom40 and impairs its import. Mol Biol Cell. 2012;23:1618–1627. doi: 10.1091/mbc.E11-11-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalińska M, Guiard B, Pfanner N, Meisinger C. Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metab. 2013;18:578–587. doi: 10.1016/j.cmet.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Opalińska M, Gerbeth C, Herman JS, Rao S, Schönfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science. 2014;346:1109–1113. doi: 10.1126/science.1261253. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]