Abstract
Many intriguing facets of lipoxygenase (LOX) catalysis are open to a detailed structural analysis. Polyunsaturated fatty acids with two to six double bonds are oxygenated precisely on a particular carbon, typically forming a single chiral fatty acid hydroperoxide product. Molecular oxygen is not bound or liganded during catalysis, yet it is directed precisely to one position and one stereo configuration on the reacting fatty acid. The transformations proceed upon exposure of substrate to enzyme in the presence of O2 (RH + O2 → ROOH), so it has proved challenging to capture the precise mode of substrate binding in the LOX active site. Beginning with crystal structures with bound inhibitors or surrogate substrates, and most recently arachidonic acid bound under anaerobic conditions, a picture is consolidating of catalysis in a U-shaped fatty acid binding channel in which individual LOX enzymes use distinct amino acids to control the head-to-tail orientation of the fatty acid and register of the selected pentadiene opposite the non-heme iron, suitably positioned for the initial stereoselective hydrogen abstraction and subsequent reaction with O2. Drawing on the crystal structures available currently, this review features the roles of the N-terminal β-barrel (C2-like, or PLAT domain) in substrate acquisition and sensitivity to cellular calcium, and the α-helical catalytic domain in fatty acid binding and reactions with O2 that produce hydroperoxide products with regio and stereospecificity. LOX structures combine to explain how similar enzymes with conserved catalytic machinery differ in product, but not substrate, specificities.
Keywords: lipoxygenase, polyunsaturated fatty acids, linoleic acid, arachidonic acid, hydroperoxide oxygenation, oxylipins
Introduction
Lipoxygenase (LOX) enzymes are widely expressed in animals, plants, and fungi, and although also ubiquitous in cyanobacteria, LOX enzymes are quite rarely found in other prokaryotes.1 The specificities of their reactions provide fatty acid hydroperoxides with differing functional roles. LOX reactions commonly initiate pathways of lipid mediator biosynthesis,2,3 they are used for the mobilization of lipids for energy production or for structural roles4,5 and the hydroperoxide products possibly ramp up cellular peroxide tone, potentially with an activating effect on other pathways.6,7 With respect to nomenclature, mammalian 5-lipoxygenase oxygenates carbon-5 on arachidonic acid, a plant 13-LOX oxygenates the 13-carbon of linoleic or α-linolenic acids, and so on. LOX catalysis is always centered around a five-carbon unit with a CH2 methylene between a pair of cis double bonds (CH=CH=CH2=CH=CH). Molecular oxygen is introduced at one end or other of the pentadiene (regiospecificity), and in either the R or S configuration (stereospecificity). The reaction of fatty acid with O2 is covalently complete (RH + O2 → ROOH) and, therefore, no reducing cofactor is required.
The specificity with which LOX enzymes (1) oxygenate polyunsaturated fatty acids raises intriguing questions for the structural biologist on how these reactions are initiated and controlled. The typical LOX product is a single, chiral, fatty acid hydroperoxide (Scheme 18). Remarkably there is a LOX enzyme specific for most of the available positions on the common substrates, linoleic and arachidonic acids. LOX enzymes are known that oxygenate linoleic acid at three of its four available positions (9R, 9S, or 13S),9 and there are LOX enzymes specific for 10 of the 12 available positions on arachidonic acid (5R, 5S, 8R, 8S, 9R, 11R, 11S, 12R, 12S, and 15S are all known in Nature.1,10,11 The conceptual basis for this specificity is quite well established (see later), yet the structural basis remains an open issue. With the availability of genomic sequences providing any of these enzymes open for cloning and expression, the wealth of diversity provides opportunities for the structural biologist to compare and contrast enzymes with a view to explaining the distinct catalytic specificities.
Scheme 1.
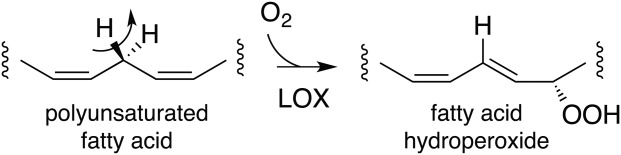
Scheme Lipoxygenase catalysis.
The first section of this review explains the chemistry of the LOX reaction and the conceptual basis of specificity, the next provides an overview of LOX enzyme structure. It then details our current understanding of structure and function.
The Conceptual Basis of Specificity
LOX enzymes initiate, control, and terminate a free radical reaction between the substrate fatty acid and molecular oxygen. The non-heme iron initiates the reaction by stereoselective H-abstraction on one face of the fatty acid and then oxygen reacts on the opposite face, an antarafacial relationship common to all LOX catalysis.12 A major element of stereo control is inherent in this antarafacial “rule” but with so many distinct hydroperoxide products emanating from LOX active sites of very similar overall structure, the basis of specificity has to include (i) the precise positioning of the fatty acid carbon chain in the active site, (ii) the head-to-tail orientation of the fatty acid, and (iii) the access of molecular oxygen to one end or other of the reacting pentadiene.
Point (i) involves a “frame-shift” as the fatty acid can slide to different depths into the active site and expose a selected pentadiene for reaction with the iron.13 For example, the difference between 12-S and 15-S oxygenation of arachidonic acid involves this shift in the register of substrate.14 Point (ii), switching of the head-to-tail orientation of the fatty acid in the active site, was first deduced in the early 1970s from the finding that plant 9S-LOX and 13S-LOX abstract the 11pro-R and 11pro-S hydrogen, respectively, from linoleic acid.15 As illustrated in Figure 1, the 9S and 13S positions are on opposite faces of linoleic acid, and therefore a logical deduction is that regio and stereo control is equivalent in all respects except that the linoleate substrate is turned around in a switched head-to-tail orientation in the two enzymes. This was a logical deduction at the time, although there was no information on the relatedness of the two enzymes. Now we know of the close structural homology within the LOX gene family and it is a compelling argument, and the only reasonable explanation for the many known LOX reactions with arachidonic acid. This is especially the case when we know that stereospecific 15S-HPETE synthesis can be followed by 5S or 8S oxygenation in the reversed orientation in the same LOX active site.8 Point (iii), the access of O2 to one end or other of the reacting pentadiene, is the ultimate determinant of R or S specificity. As illustrated in Figure 1, the R and S positions are at opposite ends on each face the pentadiene. Also shown in Figure 1, oxygenations occurring on the same face of the substrate (e.g., 9S/13R or 9R/13S) are initiated by the same stereospecific hydrogen abstraction.16 Mutagenesis studies reveal the importance of an Ala residue conserved in S-LOX and the equivalent Gly residue in R-LOX,17 yet how this comes into play is a structural issue waiting to be solved. Selective shielding of one end or the other of the pentadiene and directional channeling of O2 have both been proposed.18–22
Figure 1.
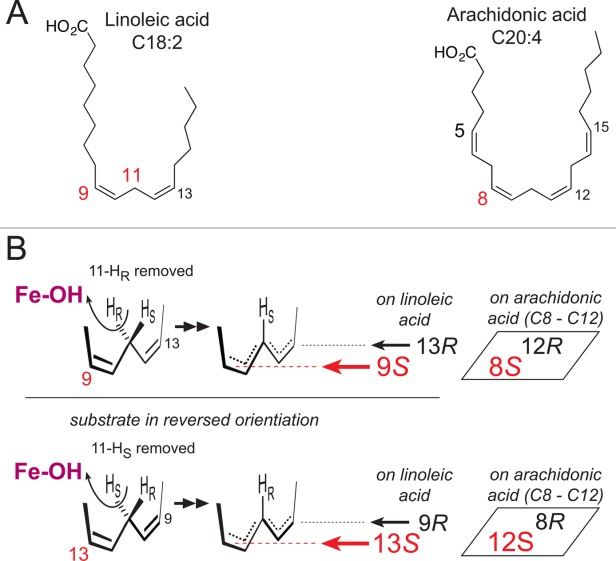
Figure The basis of LOX regio- and stereospecificity. (A)The structures of linoleic acid (18:2) and arachidonic acid (20:4). (B) A perspective view on LOX catalysis showing the reacting pentadiene. Top left: The catalytic iron (Fe-OH) abstracts the pro-R hydrogen from C11 and O2 is added antarafacially in the 9S position (in red, in the foreground).15 Lower left: with linoleic acid in a reversed head-to-tail orientation, the identical stereochemical relationships involve pro-S hydrogen abstraction at C11 and oxygenation in the 13S position (in red, in the foreground).15 Formation of the corresponding 9R or 13R configuration products entails antarafacial reaction of O2 at the opposite end of the pentadiene.16 In boxes on the right side: an example of the corresponding positional specificities of arachidonic acid, illustrating the positions around the 8–12 pentadiene on arachidonic acid.
Lipoxygenase Structure
In 1993, Boyington et al.23 described the first crystal structure of a LOX, the L1 isoform from Soybean. This structure established the molecular framework common to plant and animal enzymes: an amino terminal β-barrel, now known as a PLAT (Polycystin-1, Lipoxygenase, Alpha-Toxin) domain and a much larger α-helical domain that houses the catalytic iron. The plant enzymes are significantly larger than the animal enzymes (∼900 vs. ∼650 amino acids, respectively), and the smaller animal enzymes are simply pared down by the omission of several plant-specific loop regions. More recently, the structure of a LOX from the opportunistic pathogen P. aeruginosa has been reported.24 This enzyme is reduced further still: it lacks the amino terminal β-barrel domain. However, an α-helical insertion expands the catalytic domain and an additional amino terminal helix shields the surface covered by the PLAT domain of the eukaryotic enzymes. Despite these differences, a large helical core, along with the relative placements of most of the ∼17 helices that comprise it, is conserved (Fig. 2). At the heart of the core is the catalytic iron, positioned by invariant histidine side chains contributed by the two longest helices in the common core (∼40–50 amino acids in length, helices α7 and α14 (green, orange, respectively, Fig. 3), as well as the main chain carboxyl at the C-terminus provided by an invariant Ile. On the opposite side of the iron are helix α8 (lime-green) and the penultimate (red) helix. An unusual structural feature of helix α8, a unique insertion which gives it a distinct curvature,25 has been observed in all LOX structures to date (bright pink, Fig. 2). This helix, subsequently referred to as the “arched” helix, helps shield a U-shaped cavity, at the base of which lies the catalytic center. The remainder of the common core is an awkward bundling of helices, primarily in antiparallel orientations. The protein data bank codes for the four plant,23,25–27 seven animal,28–34 and single bacterial enzyme24 are given in TableI.
Figure 2.
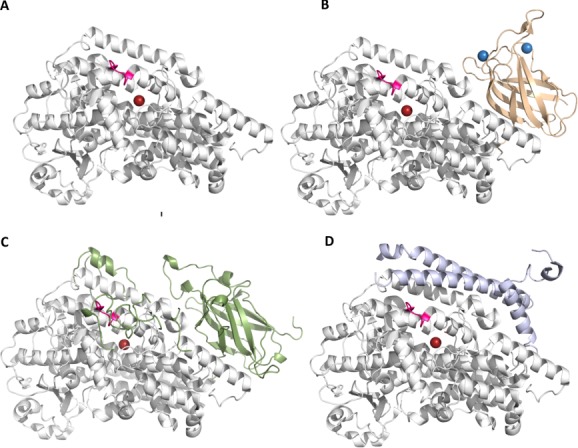
Figure The common core of LOXs is shared by plant, animal and bacterial enzymes. (A) The core domain is a large bundle of helices that houses the catalytic iron. A distinct insertion that contributes to a helix that forms an arch over the active site is colored in bright pink. (B) and (C) The animal (flesh) and plant (green) enzymes have amino terminal domain PLAT domains that may harbor Ca2+ binding sites. (D) The bacterial LOX lacks the PLAT domain, but has additional helices. An interactive view is available in the electronic version of the article.
Figure 3.
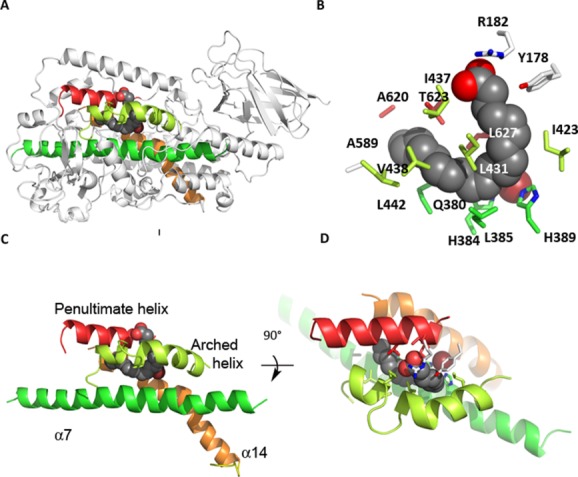
Figure Four helices form the heart of the common core structure (depicted in 8R-LOX). Amino acids from these helices contribute the major part of the side chains that form the iron binding site and substrate binding cavity. (A) The location of these four helices in the LOX overall structure. (B) The amino acids that form the arachidonic acid binding site in 8R, colored accordingly. (C) and (D) Details of the helical core with 90° rotation so that the cavity entrance can be visualized. An interactive view is available in the electronic version of the article.
Table I.
LOX Structures in the Protein Data Bank
| Lipoxygenase | PDB codes | Inhibitor/substrate structures | Resolution (Å) | References |
|---|---|---|---|---|
| Glycine max LOX1 | 2SBL/1YGE | — | 2.6/1.4 | 23,25 |
| Glycine max LOX3 | 1LNH | 1IK3, 1HU9, 1NO3, IN8Q, 1JNQ, 1ROV | 2.6 | 26,48 |
| Glycine max LOXB | 2IUJ | — | 2.4 | 27 |
| Glycine max LOXD | 2IUK | — | 2.4 | 27 |
| O. cuniculus 15-LOX-1 | /2P0M | 1LOX/2P0M | 2.4 | 28,29 |
| P. homomalla 8R-LOX | 2FNQ/3FG1 | 4QWT | 3.1/1.85/2.0 | 40, 41, 34 |
| H. sapiens 5-LOX | 3O8Y | — | 2.4 | 30 |
| G. fruticosa 11R-LOX | 3VF1 | — | 2.5 | 31 |
| S. scrofa 12-LOX (truncation) | 3RDE | 3RDE | 1.89 | 32 |
| H. sapiens 15-LOX-2 | 4NRE | 4NRE | 2.63 | 33 |
| H. sapiens 12-LOX (truncation) | 3D3L | — | 2.6 | — |
| P. aeruginosa | 4G32/4G33 | 4G32,4G33 | 1.75,2.03 | 24 |
The Carboxy Terminus Fills the Iron Coordination Sphere
The iron coordination sphere adopts a pseudo octahedral geometry.35 A remarkable feature of the coordination sphere is the utilization of the main-chain carboxy-terminus as an iron ligand (Fig. 4). In order for the main-chain carboxyl to fill the sphere the terminal Ile must insert into the helical bundle to position itself at the iron, deep in the four-helix bundle. The insertion is made possible by irregularities in the helices that form the core. While the inserted peptide fills what would otherwise be a gaping hole in the helical bundle, it does not fill it completely, and in the high resolution structure of P. homomalla 8R-LOX an intricate network of water molecules solvates the “buried” C-terminus, including the terminal Ile side chain (Fig. 4). A similar solvent channel is apparent in the 1.4 Å resolution structure of Soybean LOX1.25 The majority of the hydrogen bonds that restrain the C-terminus in 8R-LOX are main-chain to main-chain; however, a highly conserved Asn four amino acids upstream of the C-terminus participates in main-chain to main-chain, main-chain to side-chain, and side-chain to main-chain H-bonds. The numerous H-bonds mediated by this residue are consistent with its high degree of conservation. In contrast the Ile sidechain makes limited van der Waals contacts within the protein core and, although “buried,” is highly solvated, and it is not obvious why it is virtually invariant.
Figure 4.
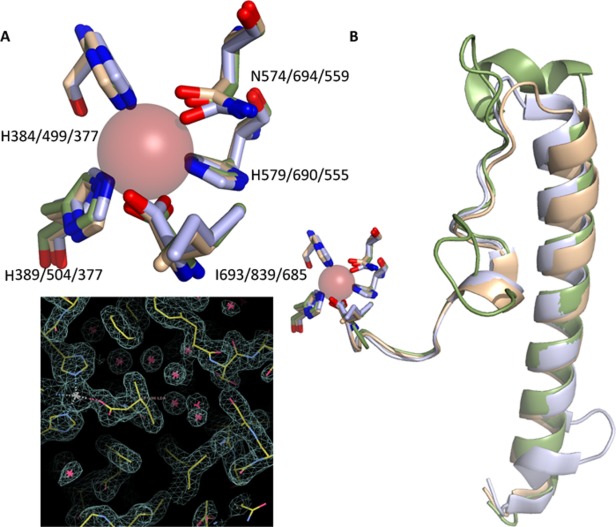
Figure The iron coordination sphere. (A) Superposition of the iron ligands from representative plant (1YGE) animal (4QWT) and bacterial (4G32) structures. (B) The C-terminal region of the enzymes; a long loop that follows the terminal α-helix makes its way to the catalytic iron so that the carboxyl can fill the coordination sphere. (C). A portion of the electron density map for 8R-LOX (3FG1), centered around the C-terminal Ile. Note the presence of a water-filled cavity that accommodates the terminal Ile.
Variations in the Common Core
While the orientations of most of the “common core” helices are maintained, a notable exception is helix α2 (Fig. 5). The positioning in plants is impacted by a plant specific insertion and it cannot occupy the same space as its counterparts in the animal and bacterial enzymes. Its placement occludes access to the catalytic machinery. A putative access portal that requires movement of the side chain in Thr-259 located on this helix, as well as Leu-541 from a neighboring helix, was inferred from a crystal structure of LOX1.36 More recently Bradshaw and Gaffney probed the dynamics of soybean LOX1 α2 by sampling along this helix with site-directed mutagenesis, spin-labeling and EPR. Their results, which allow them to observe mobility along the helix at a time scale relevant to catalysis and in the presence of substrate, put the “portal” just a few residues up from Thr-259 at a conserved π-helical segment of α2 at 261–267.37 Earlier work by the investigators led them to position the polar end of the fatty acid at the “entrance” end of the cavity.38 Their data suggest a model in which the hydrocarbon region of the substrate interacts with the π-helix segment to grease its way between the two helices (α2 and the arched helix) that shield the active site. The combination of dynamic EPR spin-label experiments and a crystal structure provides a compelling model for how substrate enters the encapsulated active site in plant LOXs.39
Figure 5.
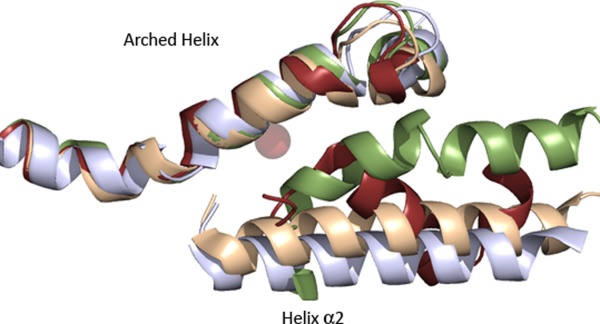
Figure The placement of α2 varies among LOXs. The “arched” helix and helix α2 in the plant (green, 1YGE) animal (flesh, 4QWT) and bacterial enzymes (light blue, 4G32) are superimposed. The “broken” α2 (brown, 3O8Y) of 5-LOX is included as well for reference. Note that while the arched helices from the diverse structure superpose, α2 varies in its placement.
In the animal enzymes three placements of α2, which fall into two classes, have been described. The more commonly observed α2 orientation is as a single helix that runs almost the length of the catalytic domain and packs against the penultimate helix of the common core. In this orientation it rims the active site cavity, but does not obscure access to the active site as its counterparts do in the plant enzymes. A variation of this orientation, in which α2 is displaced ∼10 Å from the commonly observed position, is found in the rabbit 15-LOX structure in the presence of an inhibitor. The displacement results in significant remodeling of the active site cavity. However, a similar displacement is not observed in other LOX structures reported with inhibitors that are substrate mimics32,33 or in 8R-LOX in the presence of arachidonic acid.34 Thus, a conformational change of such magnitude is not required for substrate entry into the active site for all isoforms.
Alternately, α2 has been observed as a “broken” helix that covers the active site cavity and contributes bulky, aromatic amino acids that plug an otherwise open U-shaped site. The “corking” aromatics are not unique to the closed enzymes, and no obvious sequence hallmarks, except perhaps the presence of a small side chain (Ala), which appears to make insertion possible, in the penultimate helix, can be inferred from the small number of structures reported. The two enzymes that display this alternate configuration (5S- and 11R- LOX, Ala at positions 606 and 612, respectively, TableII) share ∼35% identity and differ significantly in product specificity, and both structures were determined in the absence of inhibitor or substrate.30,31 However, other structures in the absence of inhibitor or substrate have open cavities (15-LOX-1,29 8R-LOX40,41), with an unbroken helical structure that anchors the “corking” aromatic amino acids far from the active site entrance. Interestingly, the 12-LOX inhibitor, which was necessary for crystallization of the catalytic domain of that enzyme, has an aromatic ring which occupies the same space as the “corking” side chain common to the human and coral enzymes,32 hence “corked” and “uncorked” LOXs can accommodate aromatic rings at the active site entrance (Fig. 6). This might suggest that the determining factor for “corking” is not an aromatic amino acid in helix α2, but whether α2 is “broken” such that its aromatic side chain(s) are in proximity to the cavity entrance.
Table II.
The Amino Acids that Define the AA Binding Site in 8R-LOX and Their Counterparts in Other LOX Structures
| 8-R-LOX identity | 11-R-LOX 47% | 5 -LOX 38% | 15-LOX-2 33% | 15-LOX 30% | 12-LOX 30% | LOX1 28% | LOX3 28% | LOXB 27% | LOXD 27% | PA 33% |
|---|---|---|---|---|---|---|---|---|---|---|
| 4QWT | 3FG1 | 3O8Y | 4NRE | 2P0M | 3RDE | 1YGE | 1LNH | 2IUJ | 2IUK | 4G32 |
| Tyr-178 | Phe-185 | Phe-177 | Phe-184 | Phe-175 | Phe-175 | Ala-254 | Phe-272 | Phe-267 | Phe-277 | -- |
| Arg-182 | Gly-188 | Tyr-181 | Ala-188 | Leu-179 | Leu-179 | Gly-258 | Gly-276 | Gly-271 | Gly-281 | -- |
| Gln-380 | Gln-369 | Gln-363 | Glu-369 | Gln-357 | Glu-357 | Gln-495 | Gln-514 | Gln-509 | Gln-521 | Glu-373 |
| His-384 | 373 | 367 | 373 | 361 | 361 | 499 | 518 | 513 | 525 | 377 |
| Leu-385 | 374 | 368 | 374 | 362 | 362 | Trp-500 | Trp-519 | Trp-514 | Trp-526 | 378 |
| His-389 | 378 | 372 | 378 | 366 | 366 | 504 | 523 | 518 | 530 | 382 |
| Ile-423 | 412 | 406 | 412 | 400 | 400 | 538 | 557 | 552 | 564 | I416 |
| Gly-427 | Gly-416 | Ala-410 | Ala-416 | Ala-404 | Ala-404 | Ala-542 | Ala-561 | Ala-556 | Ala-568 | Ala-420 |
| Leu-431 | 420 | 414 | 420 | 408 | 408 | 546 | 565 | 560 | 572 | 424 |
| Ile-437 | Ala-426 | Leu-420 | Val-426 | Ile-414 | Ile-414 | Ile-552 | Val-571 | Ile-566 | Ile-578 | Phe-430 |
| Val-438 | Ala-427 | Phe-421 | Val-427 | Phe-415 | Phe-415 | Ile-553 | Ile-572 | Ile-567 | Ile-579 | Ile-430 |
| Leu-442 | Leu-431 | Asn-425 | Thr-431 | Met-419 | Leu-419 | Phe-557 | Phe-576 | Phe-571 | Phe-583 | Phe-435 |
| Ala-589 | Thr-575 | Pro-569 | Pro-572 | Cys-560 | Cys-560 | Thr-709 | Thr-728 | Thr-723 | Thr-735 | Ala-574 |
| Ala-620 | Met-606 | His-600 | Val-603 | Gln-590 | Gln-590 | Ser-747 | Asp-766 | Asp-761 | Asp-773 | Lys-605 |
| Thr-623 | Val-609 | Ala-603 | Ala-606 | Ile-593 | Ile-593 | Val-750 | Val-769 | Ile-764 | Ile-766 | Ile-608 |
| Ile-626 | Ala-612 | Ala-606 | Leu-609 | Gln-596 | Gln-596 | Ile-753 | Ile-772 | Ile-767 | Ile-779 | Leu-611 |
| Leu-627 | 613 | 607 | 610 | 597 | 597 | 754 | 773 | 768 | 780 | 612 |
Overall sequence identity with P. homomalla 8R-LOX is in indicated. The amino acids of the highly conserved active site core are in italics and shown in Bold font for reference structure.
Figure 6.
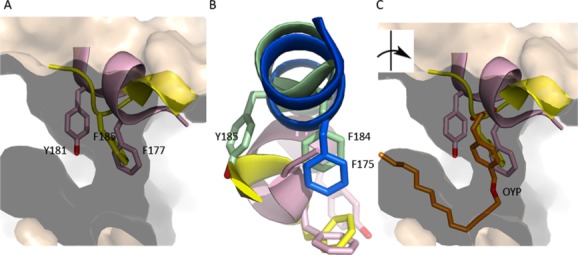
Figure The active site of animal LOXs can be “corked.” (A) The corking amino acids of 5-LOX (pink) and 11R-LOX (yellow) plug what would otherwise be an open U-shaped cavity (15-LOX-2, flesh). (B) The corking amino acids are conserved in “uncorked” LOXs (blue, 12-LOX, green, 15-LOX-2), however since α2 is not “broken” the side chains are distal and cannot seal the active site entrance. The image has been rotated relative to (A) for clarity. (C) In the inhibited structure of 12-LOX an aromatic ring of the inhibitor (gold, OYP) occupies the position of the “cork.”
The PLAT Domain and Ca2+ Dependent Membrane Binding
The amino terminal domain common to the plant and animal LOXs was originally known as a “C2-like” domain, given its resemblance to the Ca2+-dependent membrane binding domains of phospholipases. Moreover, like the phospholipase C2 domains, the PLAT domains were shown to confer membrane binding, most notably for the 5-LOX enzyme.42,43 Beyond the fact that this domain has been demonstrated to mediate membrane binding in experiments with PLAT-domain truncations, and the observation that amino terminal truncation mutants have impaired stability44 and reaction rates,45 a direct role for the PLAT domain in substrate acquisition has yet to be demonstrated. However, Eek et al. have suggested a plausible communication pathway connecting the PLAT Ca2+-binding site with helix α2 in the 11R-LOX enzyme via a cation-π interaction between an invariant Trp of the PLAT domain with K172 of that catalytic domain. A role for this cation-π interaction in linking the LOX domains is supported by its presence in both the plant and animal enzyme structures in the protein data bank, and by multiple sequence alignment.31
The PLAT domains display significantly less sequence conservation than the catalytic domains, notwithstanding a high degree of structural conservation. Early experiments with C2-like domain constructs demonstrated its importance in Ca2+-dependent membrane binding for 5-LOX.42,43 However, the amino acids that coordinate Ca2+ are not highly conserved among the LOX family. Moreover putative membrane insertion loops that emanate from the β–barrel framework differ among orthologs.
Crystal structures that provide insight into Ca2+ binding include that of the 8R-LOX,40 11-R-LOX31 and 15-LOX-2.33 While Ca2+-dependent membrane binding by 5-LOX has been studied extensively, mutations to the 5-LOX PLAT domain for crystallization make it a poor model for describing the Ca2+-binding sites. However 11R-LOX, which has an absolute requirement for membrane binding for catalytic activity even in cell-free assays,11 was crystallized in the absence of the cation and has intact membrane binding loops. Both 15-LOX-2 and the first 8R-LOX structure reported (2FNQ) have intact Ca2+-binding loops, and reveal density and coordination geometry consistent with Ca2+ binding. The Ca2+ ions appear to stabilize putative membrane-insertion loops that emerge from the edge of the amino terminal β-barrel. In Figure 7 the structures of 15-LOX-2 and 8R-LOX (∼33% sequence identity) with two and three Ca2+ bound, respectively, are superposed. Note the similarity in the placement of the loops. While there is some deviation in one loop, which could not be modeled in the 8R-structure, for the most part the loop regions of the barrel are superposable. In contrast, a LOX pair with greater sequence identity (47%) is superposed in panel B. In this case, one structure in the absence of Ca2+ and the other in the presence. In this pair, the loop structures are distinct. It seems reasonable to infer from this comparison that binding of Ca2+ may trigger a β-barrel loops conformational change that favors membrane binding.
Figure 7.
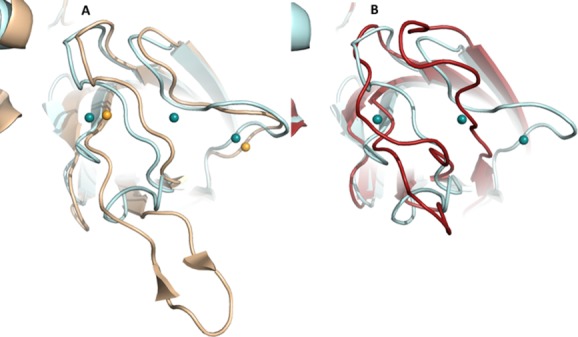
Figure Ca2+ binding appears to promote structural changes in the loops of the PLAT domain. (A) Superposition of cartoon renderings of 8R-LOX (light blue) and 15-LOX-2 structures (flesh), both in the presence of Ca2+. Three Ca2+ (spheres) sites are found in the former, and two in the latter. (B) Superposition of 8R-LOX in the presence of Ca2+ and 11-R-LOX in its absence. Despite a higher sequence identity in this pair of LOX, the loop conformations are divergent, presumably a reflection of differences in the Ca2+-bound- and Ca2+-free states.
Given the clear modular structures of LOXs, the question of whether the two domains can “flex” about a hinge region has been raised. Small angle x-ray scattering data are consistent with interdomain movement in 15-LOX-1.46 However, to date no crystal structure has revealed an alternate inter domain packing or placement.
The Substrate Binding Cavity
Over the years individual LOX structures have led to models for substrate recognition by this enzyme family. A “boot-shaped” model was derived from the inhibited 15-LOX structure reported in 199728 and suggested that Ile-418, Phe-353 and Ile-593 define cavity depth, and that Arg-403 tethered the substrate carboxylate.28,47 Although, the “boot shaped” model nicely explained the supporting mutagenesis data, in retrospect it was incomplete due to what was then interpreted as disordering of helix α2.28,29 Additionally, it was based on an inhibitor that lacks the structural flexibility of arachidonic acid and so might include conformational changes not required for the natural substrate to enter.
Subsequently, the structure was reported of soybean LOX-3 with the product 13-hydroperoxy-octadecadienoic acid (13-HPODE) in a peroxy-iron complex, the “purple lipoxygenase.”48 Purple LOX is a transient intermediate formed by reaction of excess hydroperoxide product with the enzyme, capturing the iron in the ferric state.49 Remarkably this induced little change in conformation of LOX-3 from the native ferrous state.26,48 Experimental and computational evidence indicates the purple complex is not involved in the normal catalytic cycle,50 and while the overall shape of the 13-HPODE is substrate-like, its orientation in the active site does not necessarily reflect substrate head-to-tail orientation during catalysis. Despite this caveat, the structure revealed that a boomerang-shaped polyunsaturated fatty acid could be positioned, with minimal protein conformational change, in the large enclosed cavity that houses the catalytic iron.48
Currently, the structures of a (1) bacterial LOX in complex with phospholipid,24 (2) the catalytic domain of 12-LOX in complex with inhibitor,32 (3) 15-LOX-2 with a substrate mimic and finally,33 (4) an anaerobic structure of the aborted complex of 8R-LOX with arachidonic acid34 combine to provide a robust structural context for understanding product specificity and reveal a U-shaped cavity with highly conserved Leu and Ile side chains to position the pentadiene for attack. Since the substrates for the plant enzyme have fewer double bonds, the cavities are more boomerang in shape, but as expected, the core of the active site is conserved throughout the extensive LOX family.
Invariant Amino Acids Position the Pentadiene for Attack
In the structure of the abortive complex of 8R-LOX with arachidonic acid, obtained from crystals grown in the absence of O2, the substrate is poised for attack in the U-shaped cavity.34 The arched helix, which lies above that catalytic iron and helps define the active site cavity, contributes one of several invariant leucines that line binding pocket. Leu-431 (invariant) and Ile-437 define the arch and make contacts with the arachidonic acid: Leu-431 with the central carbon of the pentadiene centered at C10 and Ile-437 at the top of the “U” between the carboxyl head (C1) and C17 at the tail. These amino acids side chains fill the U from the side opposite the catalytic iron, where just across from Leu-431 sits invariant Leu-627. Leu-627 and Leu-431 are equidistant from the substrate, and the two side chains appear to clamp the C10 pentadiene in place. The base of the U is positioned by invariant Leu-385 on one side, and the Fe and His-384 and −389 on the other. Leu-385 and the catalytic iron cradle the base of the “U” (Fig. 8).
Figure 8.
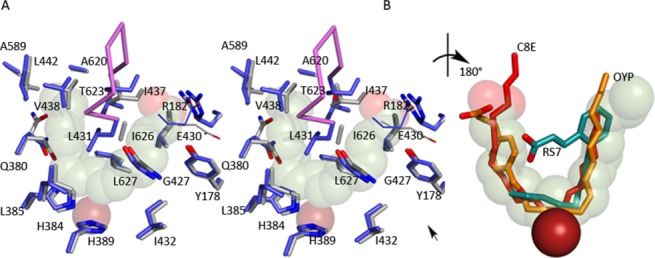
Figure (A) The substrate adopts a horseshoe shape in the U-shaped channel (stereo). The side chains of highly conserved amino acids line the base of the active site, along with Gly-427. Glu-430, part of an inter-helical charge cluster that includes the substrate carboxylate, is shown in line rendering. The Fe2+ (transparent rust sphere) is positioned behind the substrate. (B) Detail of the superposition of inhibitors (rotated ∼180° with respect to (A)) observed in 15-LOX-2 (red) 12-LOX (gold C, red O) and 15-LOX-1 (teal C, red O). The 15-LOX-2 and 12-LOX inhibitors conform to the arachidonic acid placement, while the 15-LOX-1 inhibitor overlaps partially. The Fe2+, solid rust sphere, is in front of the substrate in this view. An interactive view is available in the electronic version of the article.
Pocket depth, which allows the tail to slide in so that C10 is attacked, appears to be conferred by Ala-620, which with Ala-589 makes for a very a deep cavity. Leu-381 and Val-428 define the wall of the cavity at C15, and the polar and charged amino acids Gln-380 and Asp-424 (highly conserved) flank the cluster of histidines that hold the iron in place. In the absence of substrate, Tyr-181 and Arg-182 of helix α2 participate in an interhelical charge cluster with Glu-430 of the arched helix. In the substrate-bound structure, the fatty acid carboxylate expands this charge network with only a slight (∼1.3 Å) outward shift in the arched helix and the reorientation of the side chain of Glu-430 to allow Arg-182 to interact with the substrate carboxylate as well as Glu-430.
While conserved/invariant amino acids line the binding cavity, a counterpart for Arg-182, that is, a neutralizing amino acid from helix α2, is not conserved among LOXs. Yet this amino acid plays an important role in positioning the polyunsaturated fatty acid in a catalytically competent conformation in the active site in 8R-LOX, as the mutation R182A leads to profound substrate inhibition.34 Such an observation suggests that arachidonic does indeed enter the cavity in the absence of the neutralizing amino acid, but van der Waals contacts alone (the steric complementarity of the arachidonic acid and the U-shaped active site) are not sufficient to adequately fix the geometry of the pentadiene at the catalytic iron in 8R-LOX (nonproductive binding). The long hydrocarbon tail of the substrate makes extensive contacts with the amino acid side chains lining the site, but for 8R-LOX helix α2 must provide a polar/charged amino acid to interact with the substrate carboxyl group, perhaps to prevent it from penetrating deeper into the cavity, which extends beyond C20 of the substrate. An anchoring residue may not be necessary in all LOXs, especially those with shallower cavities that do not require as much “ease” built in to allow substrate entry and product exit.
Reconciling the U-Shaped Active Sites that Require Inverse Entry
To account for the product diversity among LOXs, a wealth of experimental data has indicated that LOXs differ with respect to which “end” of the substrate is innermost. One can directly extrapolate from the model of the 8R-enzyme with arachidonic acid to the 15-LOX-2 and 12-LOX structures solved with substrate mimics. Indeed, as one can see from the superposition of arachidonic acid, the 15-LOX-2 and 12-LOX inhibitors [Fig. 8(b)], the 8R-LOX is likely a robust, consensus model for substrate recognition in these enzymes. In contrast, in order for 5-LOX to produce the 5S- isomer of HPETE, the substrate carboxyl must be innermost in the cavity. Again, the 8R-model nicely explains this observation as well. Take as an example 15-LOX-2 and 5-LOX, two enzymes that differ in the direction of substrate head-to-tail orientations. As one can see in the schematic provided in Figure 9, given a cavity of equal depth, if arachidonic acid enters “tail first” into the cavity C15 is positioned at the site of oxygenation, while head-first entry places C5 at this position. Moreover, the product stereochemistries of 15S and 5S are a consequence of opposite faces of the substrate aligning at the catalytic center. This schematic predicts that 5-LOX and 15-LOX cavities are of similar depth. As one can see from panel B, in which the cavities of 15-LOX-2 and 5-LOX are depicted with a U-shaped 15-LOX-2 inhibitor positioned as a substrate mimic, this is indeed the case. Note the fork-like contour of the 5-LOX pocket appears to be a perfect complement for a carboxyl group. It is here where there is a His (600) in the 5-LOX site, but a Val (603) in the 15-LOX-2 site. However recall that the active site in 5-LOX is “corked.” Although the cavity depth is consistent of 5-LOX, one must assume that the cavity is “uncorked” in the active enzyme. Exactly what “uncorking” involves is not clear, although a structure of an “open” 5-LOX has been reported, the product of this enzyme is not 5S-HPETE.51 In this structure a dramatic opening of the active site that results in a loss of product specificity is revealed.
Figure 9.
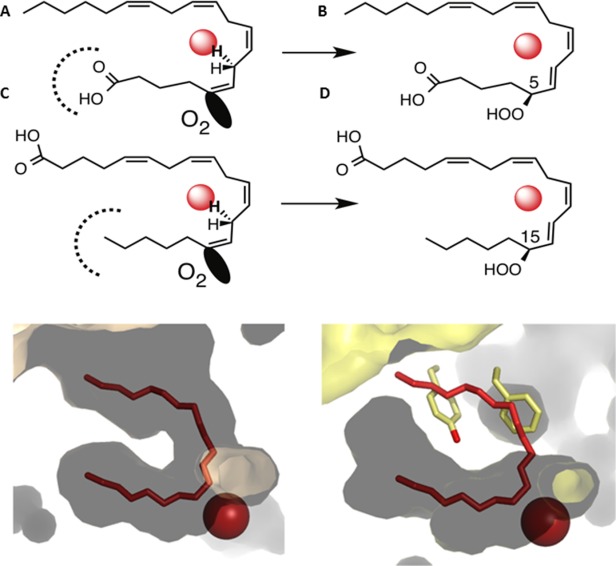
Figure Inverse entry of substrate is consistent with cavity depth in 15-LOX-2 and 5-LOX. (A) A schematic depicting how the products 15S-HPETE and 5S-HPETE can be explained by substrate entry in 15-LOX-2 as “tail-first” and that in 5-LOX as “head-first.” (B) The cavity depths in 15-LOX-2 (flesh) and 5-LOX (yellow) are consistent with this interpretation. The innermost part of the 5-LOX cavity should accommodate a carboxyl, rather than hydrocarbon.
In TableII the counterparts of the amino acids that line the 8R-LOX active site are given for all LOX structures in the Protein Data Bank. Note the invariant nature of many amino acids that envelop the pentadiene to be attacked; this level of conservation is certainly not surprising given that the enzymes share fundamental aspects of substrate recognition and catalysis. One hallmark of the plant enzymes is the presence of Trp where nonplant enzymes have a Leu. This Trp wedges right up against the pentadiene to be attacked and at the same time forms the proximal wall of a possible O2 access channel. This plant–specific sequence difference is coupled to a second substitution one turn farther along the helix: another invariant Leu in the animal enzymes is substituted by an Ala, presumably to compensate for the added bulk of the Trp. Interestingly, this second highly conserved amino acid has been implicated in an O2 access pathway as well in mammalian LOXs,21 as discussed in the following section.
Although the U-shaped model for the active site is consistent with the inhibited structures of 12-LOX and 15-LOX-2, it is not in agreement with the inhibited 15-LOX-1 structure. Porcine 12-LOX and rabbit 15-LOX-1 share ∼80% identity and exhibit the same overall U-shaped binding cavity described above in 8R-LOX. However, in apo-15-LOX-1 there is a conspicuous constriction of the internal end of the cavity made by Phe-415 and Leu-597; both amino acids are conserved in the porcine 12-LOX, and Leu-597 is invariant. Indeed, the amino acids lining the active site cavity are identical in the pair. Both enzymes have product stereochemistry consistent with tail-first entry, but the porcine 12-LOX cavity can allow the arachidonic acid to slide in deep enough to position the pentadiene centered at C10. Although the rabbit enzyme does produce the 12S isomer, the bulk of the product is 15S (1:9 ratio). Why do these highly homologous enzymes have different product profiles? This may due to the obvious constriction 15-LOX-1, which must be reconfigured to allow substrate entry and positioning of the C13 pentadiene. As mentioned above, the crystal structure of 15-LOX-1 with the inhibitor RS7 revealed a ∼10 Angstrom displacement of α2, and this structure has been used a template for models of 15-LOX-1 in complex with substrate. While it is clear that due to the constriction a conformational change must occur in the enzyme for substrate to fully enter the active site, whether arachidonic acid binding induces the RS7-induced conformational change in 15-LOX-1 is not clear. The highly homologous 12-LOX does not require displacement of α2 for its inhibitor, which adopts a U-like structure, to occupy the site. It is possible that the RS7 conformation is unique to the inhibitor, which is constrained in a conformation inconsistent with the U-shaped cavity.
An Oxygen Access Pathway
Common to LOX structures is a long tubular opening that emanates from the base of the U-shaped cavity. The tubular opening and active site iron lie on opposite sides of the substrate, as observed in the substrate bound 8R-LOX structure. Minor et al.25 first described this possible O2 channel, bounded by Trp-500 and Ile-553 in the 1.4 Å resolution structure of Soybean LOX L1 and experiments by Klinman and colleagues18,19 supported a role for Ile553 in controlling O2 access in that enzyme.
Subsequently, Coffa and Brash16,17 described a Gly/Ala “switch” that impacts product stereochemistry. 8R- and 11R- enzymes have a Gly at this residue, while the 12S- 15S- and 5S- enzymes all have Ala. One way to think about this “switch” is that it directs O2 to the carbon which is either +2 or −2 from the central carbon of the attacked pentadiene and thus the Gly/Ala switch nudges the channel toward the substrate carbon that will carry the peroxide group.
In a computational approach with the 15-LOX, Saam et al. identified the counterpart of 8R-LOX Leu-390(Leu-367 in the 15-LOX-1 enzyme) as critical in controlling O2 access. According to sequence analysis with Evolutionary Trace52 Leu-390 is invariant in animal LOXs and lies one helical turn from the animal LOX invariant active site Leu-385 in 8R-LOX. Leu-385 is the animal counterpart of Trp-500, which helps define the O2 channel in Soybean LOX1. Thus in plants a Trp-Ala pair define one wall of the O2 channel, and it animal LOX it is a Leu-Leu pair. The spatial relationship among these channel-defining amino acids is depicted in the structure of the abortive complex of 8R-LOX with substrate (Fig. 10). In 8R-LOX, the channel provides direct access to the enzyme surface. This is not the case for LOXs in general, and the full path that molecular oxygen must travel to reach the substrate is not obvious.
Figure 10.
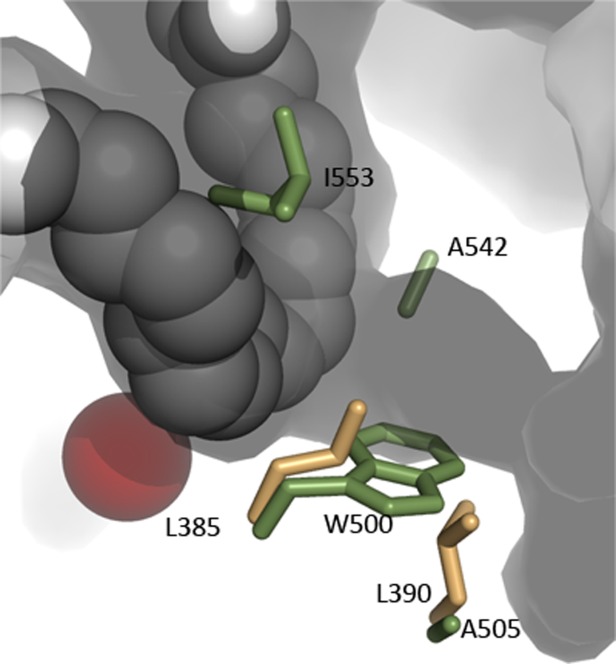
Figure A possible O2 access channel intersects with the U-shaped cavity on the side opposite the catalytic iron in 8R-LOX. Amino acids which have been shown to control O2 access in other LOXs are in included. Soybean LOX1 side chains (green) are Ile-553, Ala-542 (the Gly/Ala switch), Trp-500 and Ala-505. Leu-385 and Leu-390 are the animal counterparts of the Trp-Ala pair in plants.
Conclusions
As is commonly observed in protein families, minor modifications on a conserved structural framework can correlate with essential functional differences among family members. LOX structures have revealed a highly conserved Fe-coordination sphere positioned in a helical core. A hydrophobic U-shaped channel that can accommodate the polyunsaturated fatty acid substrate lies adjacent to the active site iron, with the metal ion positioned roughly at the base of the U. The invariant Leucines that line the base of the U appear to clamp a substrate pentadiene in place for hydrogen abstraction. Also at the base of the U, but on the opposite side of the substrate, is a putative O2 access cavity. Amino acids peripheral to this invariant core determine which pentadiene is aligned at the active site in the individual LOXs, as well head-to-tail positioning of the substrate. These features confer elegantly simple variations on a theme: a highly conserved core active site with alternate orientations for their common substrates, so that each enzyme generates a distinct product.
References
- Ivanov I, Heydeck D, Hofheinz K, Roffeis J, O'Donnell VB, Kuhn H, Walther M. Molecular enzymology of lipoxygenases. Arch Biochem Biophys. 2010;503:161–174. doi: 10.1016/j.abb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Samuelsson B, Dahlen SE, Lindgren JA, Rouzer CA, Serhan CN. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- Andreou A, Feussner I. Lipoxygenases—structure and reaction mechanism. Phytochemistry. 2009;70:1504–1510. doi: 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yin H, Boeglin WE, Elias PM, Crumrine D, Beier DR, Brash AR. Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: a proposed role in releasing omega-hydroxyceramide for construction of the corneocyte lipid envelope. J Biol Chem. 2011;286:24046–24056. doi: 10.1074/jbc.M111.251496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Herrfurth C, Brodhun F, Feussner I. Degradation of lipoxygenase-derived oxylipins by glyoxysomes from sunflower and cucumber cotyledons. BMC Plant Biol. 2013;13:177. doi: 10.1186/1471-2229-13-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaud O, Aronoff DM, Richardson JH, Marnett LJ, Oates JA. Determinants of the cellular specificity of acetaminophen as an inhibitor of prostaglandin H(2) synthases. Proc Natl Acad Sci USA. 2002;99:7130–7135. doi: 10.1073/pnas.102588199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronoff DM, Boutaud O, Marnett LJ, Oates JA. Inhibition of prostaglandin H2 synthases by salicylate is dependent on the oxidative state of the enzymes. J Pharmacol Exp Ther. 2003;304:589–595. doi: 10.1124/jpet.102.042853. [DOI] [PubMed] [Google Scholar]
- Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem. 1999;274:23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- Andreou A, Brodhun F, Feussner I. Biosynthesis of oxylipins in non-mammals. Prog Lipid Res. 2009;48:148–170. doi: 10.1016/j.plipres.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Hada T, Swift LL, Brash AR. Discovery of 5R-lipoxygenase activity in oocytes of the surf clam, Spisula solidissima. Biochim Biophys Acta. 1997;1346:109–119. doi: 10.1016/s0005-2760(96)00179-8. [DOI] [PubMed] [Google Scholar]
- Mortimer M, Jarving R, Brash AR, Samel N, Jarving I. Identification and characterization of an arachidonate 11R-lipoxygenase. Arch Biochem Biophys. 2006;445:147–155. doi: 10.1016/j.abb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Brash AR, Schneider C, Hamberg M. Applications of stereospecifically-labeled fatty acids in oxygenase and desaturase biochemistry. Lipids. 2012;47:101–116. doi: 10.1007/s11745-011-3612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H, Sprecher H, Brash AR. On singular or dual positional specificity of lipoxygenases. The number of chiral products varies with alignment of methylene groups at the active site of the enzyme. J Biol Chem. 1990;265:16300–16305. [PubMed] [Google Scholar]
- Sloane DL, Leung R, Craik CS, Sigal E. A primary determinant for lipoxygenase positional specificity. Nature. 1991;354:149–152. doi: 10.1038/354149a0. [DOI] [PubMed] [Google Scholar]
- Egmond MR, Vliegenthart JF, Boldingh J. Stereospecificity of the hydrogen abstraction at carbon atom n-8 in the oxygenation of linoleic acid by lipoxygenases from corn germs and soya beans. Biochem Biophys Res Commun. 1972;48:1055–1060. doi: 10.1016/0006-291x(72)90815-7. [DOI] [PubMed] [Google Scholar]
- Coffa G, Imber AN, Maguire BC, Laxmikanthan G, Schneider C, Gaffney BJ, Brash AR. On the relationships of substrate orientation, hydrogen abstraction, and product stereochemistry in single and double dioxygenations by soybean lipoxygenase-1 and its Ala542Gly mutant. J Biol Chem. 2005;280:38756–38766. doi: 10.1074/jbc.M504870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa G, Brash AR. A single active site residue directs oxygenation stereospecificity in lipoxygenases: Stereocontrol is linked to the position of oxygenation. 2004. Proc Natl Acad Sci USA [VOL:PAGE #S] [DOI] [PMC free article] [PubMed]
- Knapp MJ, Seebeck FP, Klinman JP. Steric control of oxygenation regiochemistry in soybean lipoxygenase-1. J Am Chem Soc. 2001;123:2931–2932. doi: 10.1021/ja003855k. [DOI] [PubMed] [Google Scholar]
- Knapp MJ, Klinman JP. Kinetic studies of oxygen reactivity in soybean lipoxygenase-1. Biochemistry. 2003;42:11466–11475. doi: 10.1021/bi0300884. [DOI] [PubMed] [Google Scholar]
- Brash AR, Yu Z, Boeglin WE, Schneider C. The hepoxilin connection in the epidermis. Febs J. 2007;274:3494–3502. doi: 10.1111/j.1742-4658.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- Saam J, Ivanov I, Walther M, Holzhutter HG, Kuhn H. Molecular dioxygen enters the active site of 12/15-lipoxygenase via dynamic oxygen access channels. Proc Natl Acad Sci USA. 2007;104:13319–13324. doi: 10.1073/pnas.0702401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Pratt DA, Porter NA, Brash AR. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyington JC, Gaffney BJ, Amzel LM. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- Garreta A, Val-Moraes SP, Garcia-Fernandez Q, Busquets M, Juan C, Oliver A, Ortiz A, Gaffney BJ, Fita I, Manresa A, Carpena X. Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa. Faseb J. 2013;27:4811–4821. doi: 10.1096/fj.13-235952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Steczko J, Stec B, Otwinowski Z, Bolin JT, Walter R, Axelrod B. Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- Skrzypczak-Jankun E, Amzel LM, Kroa BA, Funk MO. Structure of soybean lipoxygenase L3 and a comparison with its L1 isoenzyme. Proteins. 1997;29:15–31. [PubMed] [Google Scholar]
- Youn B, Sellhorn GE, Mirchel RJ, Gaffney BJ, Grimes HD, Kang C. Crystal structures of vegetative soybean lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms LOX-1 and LOX-3. Proteins. 2006;65:1008–1020. doi: 10.1002/prot.21182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity [published erratum appears in Nat Struct Biol 1998 Mar;5(3):242] Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- Choi J, Chon JK, Kim S, Shin W. Conformational flexibility in mammalian 15S-lipoxygenase: reinterpretation of the crystallographic data. Proteins. 2008;70:1023–1032. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, Newcomer ME. The structure of human 5-lipoxygenase. Science. 2011;331:217–219. doi: 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eek P, Jarving R, Jarving I, Gilbert NC, Newcomer ME, Samel N. Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J Biol Chem. 2012;287:22377–22386. doi: 10.1074/jbc.M112.343285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Mueser TC, Marnett LJ, Funk MO., Jr Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis. Structure. 2012;20:1490–1497. doi: 10.1016/j.str.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe MJ, Neau DB, Mitchell CE, Bartlett SG, Newcomer ME. The structure of human 15-lipoxygenase-2 with a substrate mimic. J Biol Chem. 2014;289:8562–8569. doi: 10.1074/jbc.M113.543777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neau DB, Bender G, Boeglin WE, Bartlett SG, Brash AR. Newcomer ME (in press) Crystal Structure of a Lipoxygenase in Complex with Substrate: the arachidonic acid binding site of 8R-lipoxygenase. J Biol Chem. 289:31905–31913. doi: 10.1074/jbc.M114.599662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney BJ. Lipoxygenases: structural principles and spectroscopy. Annu Rev Biophys Biomol Struct. 1996;25:431–459. doi: 10.1146/annurev.bb.25.060196.002243. [DOI] [PubMed] [Google Scholar]
- Minor WSJ, Stec B, Otwinowski Z, Bolin JT, Walter R, Axelrod B. Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- Bradshaw MD, Gaffney BJ. Fluctuations of an exposed pi-helix involved in lipoxygenase substrate recognition. Biochemistry. 2014;53:5102–5110. doi: 10.1021/bi500768c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney BJ, Bradshaw MD, Frausto SD, Wu F, Freed JH, Borbat P. Locating a lipid at the portal to the lipoxygenase active site. Biophys J. 2012;103:2134–2144. doi: 10.1016/j.bpj.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney BJ. Connecting lipoxygenase function to structure by electron paramagnetic resonance. Acc Chem Res. 2014;47:3588–3595. doi: 10.1021/ar500290r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham ML, Brash AR, Newcomer ME. Insights from the X-ray crystal structure of coral 8R-lipoxygenase: calcium activation via a C2-like domain and a structural basis of product chirality. J Biol Chem. 2005;280:39545–39552. doi: 10.1074/jbc.M506675200. [DOI] [PubMed] [Google Scholar]
- Neau DB, Gilbert NC, Bartlett SG, Boeglin W, Brash AR, Newcomer ME. The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity. Biochemistry. 2009;48:7906–7915. doi: 10.1021/bi900084m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarberg T, Provost P, Persson B, Radmark O. The N-terminal domain of 5-lipoxygenase binds calcium and mediates calcium stimulation of enzyme activity. J Biol Chem. 2000;275:38787–38793. doi: 10.1074/jbc.M006136200. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Das S, Funk CD, Murray D, Cho W. Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem. 2002;277:13167–13174. doi: 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- Ivanov I, Di Venere A, Horn T, Scheerer P, Nicolai E, Stehling S, Richter C, Skrzypczak-Jankun E, Mei G, Maccarrone M, Kuhn H. Tight association of N-terminal and catalytic subunits of rabbit 12/15-lipoxygenase is important for protein stability and catalytic activity. Biochim Biophys Acta. 2011;1811:1001–1010. doi: 10.1016/j.bbalip.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Walther M, Hofheinz K, Vogel R, Roffeis J, Kuhn H. The N-terminal beta-barrel domain of mammalian lipoxygenases including mouse 5-lipoxygenase is not essential for catalytic activity and membrane binding but exhibits regulatory functions. Arch Biochem Biophys. 2011;516:1–9. doi: 10.1016/j.abb.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Hammel M, Walther M, Prassl R, Kuhn H. Structural flexibility of the N-terminal beta-barrel domain of 15-lipoxygenase-1 probed by small angle X-ray scattering. Functional consequences for activity regulation and membrane binding. J Mol Biol. 2004;343:917–929. doi: 10.1016/j.jmb.2004.08.076. [DOI] [PubMed] [Google Scholar]
- Borngraber S, Browner M, Gillmor S, Gerth C, Anton M, Fletterick R, Kuhn H. Shape and specificity in mammalian 15-lipoxygenase active site. The functional interplay of sequence determinants for the reaction specificity. J Biol Chem. 1999;274:37345–37350. doi: 10.1074/jbc.274.52.37345. [DOI] [PubMed] [Google Scholar]
- Skrzypczak-Jankun E, Bross RA, Carroll RT, Dunham WR, Funk MO., Jr Three-dimensional structure of a purple lipoxygenase. J Am Chem Soc. 2001;123:10814–10820. doi: 10.1021/ja011759t. [DOI] [PubMed] [Google Scholar]
- de Groot JJ, Garssen GJ, Veldink GA, Vliegenthart JF, Boldingh J. On the interaction of soybean lipoxygenase-1 and 13-L-hydroperoxylinoleic acid, involving yellow and purple coloured enzyme species. FEBS Lett. 1975;56:50–54. doi: 10.1016/0014-5793(75)80109-8. [DOI] [PubMed] [Google Scholar]
- Bushnell EA, Jamil R, Gauld JW. Gaining insight into the chemistry of lipoxygenases: a computational investigation into the catalytic mechanism of (8R)-lipoxygenase. J Biol Inorg Chem. 2013;18:343–355. doi: 10.1007/s00775-013-0978-4. [DOI] [PubMed] [Google Scholar]
- Gilbert NC, Rui Z, Neau DB, Waight MT, Bartlett SG, Boeglin WE, Brash AR, Newcomer ME. Conversion of human 5-lipoxygenase to a 15-lipoxygenase by a point mutation to mimic phosphorylation at Serine-663. FASEB J. 2012;26:3222–3229. doi: 10.1096/fj.12-205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O, Sowa ME. Evolutionary predictions of binding surfaces and interactions. Curr Opin Struct Biol. 2002;12:21–27. doi: 10.1016/s0959-440x(02)00284-1. [DOI] [PubMed] [Google Scholar]


