Abstract
Cytochrome c555 from hyperthermophilic bacteria Aquifex aeolicus (AA cyt c555) is a hyperstable protein belonging to the cyt c protein family, which possesses a unique long 310-α-310 helix containing the heme-ligating Met61. Herein, we show that AA cyt c555 forms dimers by swapping the region containing the extra 310-α-310 helix and C-terminal α-helix. The asymmetric unit of the crystal of dimeric AA cyt c555 contained two dimer structures, where the structure of the hinge region (Val53–Lys57) was different among all four protomers. Dimeric AA cyt c555 dissociated to monomers at 92 ± 1°C according to DSC measurements, showing that the dimer was thermostable. According to CD measurements, the secondary structures of dimeric AA cyt c555 were maintained at pH 2.2–11.0. CN- and CO bound to dimeric AA cyt c555 in the ferric and ferrous states, respectively, owing to the flexibility of the hinge region close to Met61 in the dimer, whereas these ligands did not bind to the monomer under the same conditions. In addition, CN- and CO bound to the oxidized and reduced dimer at neutral pH and a wide range of pH (pH 2.2–11.0), respectively, in a wide range of temperature (25–85°C), owing to the thermostability and pH tolerance of the dimer. These results show that the ligand binding character of hyperstable AA cyt c555 changes upon dimerization by domain swapping.
Keywords: domain swapping, cytochrome c, thermostable, pH tolerance, ligand binding
Introduction
The function of a protein is related to its three-dimensional structure, and many attempts have been made to alter the structure, including mutagenesis and chemical modification.1–17 However, since the amino acid sequence defines the unique three-dimensional structure of a protein, conversion of the function of a protein without modifying its amino acid sequence is limited. An example of a native bifunctional heme protein is dehaloperoxidase, which has been shown to convert with its substrate radical from an O2-binding hemoglobin to a peroxidase.18,19
Proteins in the cytochrome c (cyt c) protein family have been studied extensively in terms of protein folding and stability.20–35 Met80 and His18 are coordinated to the heme iron in cyt c, creating a relatively high redox potential for electron transfer. The redox potential of M80A cyt c was lower than that of wild-type cyt c, due to the loss of the Met80-heme iron coordination bond,36 whereas M80A cyt c exhibited unusual O2 and CO binding properties.37 Cytochrome c555 from hyperthermophilic bacteria Aquifex aeolicus (AA cyt c555) is one of the most stable cyt c proteins, and its denaturation temperature is about 130°C.30 AA cyt c555 exhibits 20.7% homology in its amino acid sequence with that of horse cyt c. AA cyt c555 is globular as other cyt c proteins, but possesses an exceptionally long, extra 310-α-310 helix containing heme-ligating Met61.29 The high thermostability of AA cyt c555 has been ascribed to the possession of the extra 310-α-310 helix and tight packing of its hydrophobic residues inside the protein.29,33
Domain swapping is a protein oligomerization mechanism, which has been suggested as a general property of all proteins.38–48 It has been observed for many proteins, including proteins which may form amyloid fibrils.43,49–52 We have previously shown that horse cyt c forms oligomers by swapping its C-terminal α-helix intermolecularly, without changing its secondary structures significantly.53 Since Met80 dissociated from the heme iron in dimeric horse cyt c, CN− and H2O2 bound to the heme iron easier in the dimer compared with the monomer.54,55 These results indicated that the properties of cyt c may be modified by domain swapping without changing its amino acid sequence, although the dimer may dissociate to monomers at high temperatures or acidic pH.53 Cytochrome c552 from thermophilic bacteria Hydrogenobacter thermophilus (HT cyt c552) is also a thermostable protein belonging to the cyt c protein family. HT cyt c552 forms a domain-swapped dimer exchanging the region containing the N-terminal helix and heme between molecules, but the active site coordination structure of the dimer and the positions of the amino acids near the heme-coordinating Met were the same as those of the monomer.56 Dimeric HT cyt c552 exhibited a similar redox potential to that of the monomer, due to coordination of Met and His to the heme iron. In this study, we show that the ligand binding properties of a hyperstable electron transfer protein, AA cyt c555, changes by domain swapping.
Results
X-ray crystal structure of dimeric AA cyt c555
Dimeric AA cyt c555 exhibited a domain-swapped structure, according to X-ray crystallographic analysis at 1.6 Å resolution (PDB code: 3X15) (Fig. 1; Supporting Information Table SI). There were four independent protomers forming two dimers in the asymmetric unit of the dimeric AA cyt c555 crystal. The region containing the C-terminal α-helix and the extra 310-α-310 helix before the C-terminal helix were exchanged with the corresponding region of another protomer in both dimers. The structure of the hinge region (Val53–Lys57) was different among the protomers and the monomer. The distance between the heme irons in the dimer was different between the two dimers, 26.3 and 23.3 Å [Fig. 2(A)], and the orientation of a heme relative to the other heme in a protomer was different for 29.5° between the two protomers [Fig. 2(B)]. A loop-to-helix transition at the hinge region forming a long α-helix from Asp54 to Lys62 was also observed in one of the protomers (Fig. 2, green), and thus the distance between the hemes in the dimer containing this protomer was decreased (Fig. 2, magenta and green) from that in the other dimer (Fig. 2, red and blue). The structural polymorphism of the hinge region of dimeric AA cyt c555 indicates that the hinge region is flexible.
Figure 1.
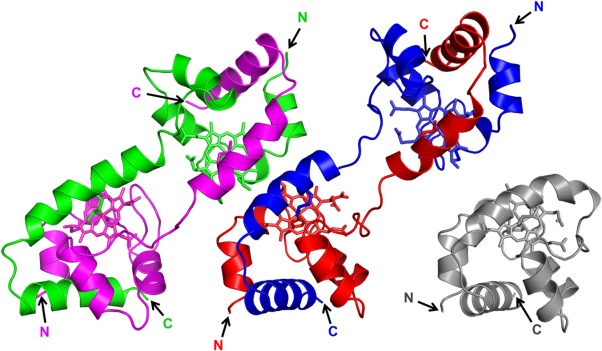
Crystal structures of dimeric AA cyt c555 (PDB code: 3X15). Two dimer structures (red-blue and magenta-green) were observed in the asymmetric unit. The red and blue regions and the magenta and green regions in the dimers belong to different protomers. The hemes are shown as stick models. Side chain atoms of heme-binding Cys12 and Cys15, and heme iron-coordinating His16 and Met61 are also shown as stick models. The N- and C-termini are labeled as N, C, respectively. Structure of monomeric AA cyt c555 (gray, PDB code: 2ZXY) is also shown.
Figure 2.
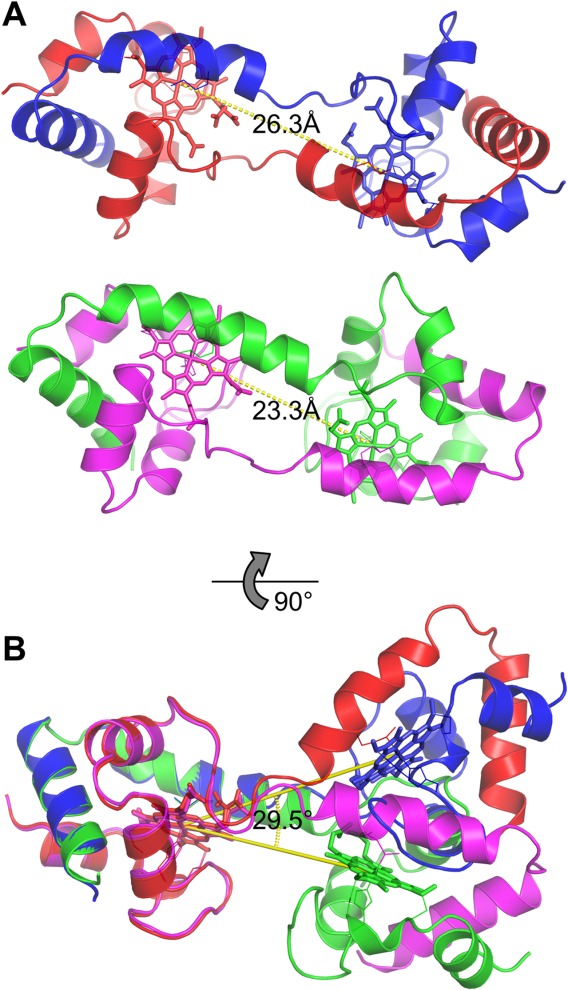
Heme–heme distance and difference in heme orientation between the two dimer structures in the asymmetric unit of dimeric AA cyt c555. A) The distance of two heme iron atoms in dimeric AA cyt c555. B) The difference in heme orientation between the two dimer structures. The hemes of the protomers depicted in red and magenta are superimposed.
The overall protein structure of dimeric AA cyt c555 corresponded well to that of monomeric AA cyt c555 (Supporting Information Fig. S1). The root-mean-square deviation (rmsd) values for the Cα atoms of the region containing the extra 310-α-310 helix and C-terminal helix (Glu58−His86, the position of Lys87 has not been determined in the crystal structure of the monomer) and those of the rest of the protein excluding the hinge region (Ala1−Ile52) between the monomer (PDB code: 2ZXY) and four protomers of the dimer were calculated to be 0.28−0.73 Å (Supporting Information Table SII). These results indicate that the structure of the region containing the extra 310-α-310 helix and C-terminal helix was similar between the monomer and the protomers of the dimer, as well as that of the rest of the protein excluding the hinge region. However, the rmsd value calculated for the region containing the extra 310-α-310 helix and C-terminal helix between the monomer and protomer D was relatively large (0.73 Å), due to the loop-to-helix transition of the hinge region in protomer D (Fig. 2, green).
Most of the interactions in the monomer were maintained in the dimer. Met61 also coordinated to the heme iron in dimeric AA cyt c555 as in the monomer, but it originated from the other protomer to which the heme belonged (Fig. 3). The Fe−His16 and Fe−Met61 distances in dimeric AA cyt c555 were 1.99 to 2.04 and 2.31 to 2.35 Å, respectively, which were similar to the corresponding distances in the monomer (Fe−His16, 2.0 Å; Fe−Met61, 2.3 Å) (Table I). The hydrogen-bonding network (<3.2 Å between heavy atoms) among Tyr34(Oη), Ile52 (N), Val53(N), and heme 17-propionate, is thought to stabilize monomeric AA cyt c555.29 This hydrogen-bonding network was maintained in the dimer (Supporting Information Fig. S2). The hydrophobic packing of Phe7, Leu27, Tyr34, Phe44, and Ile83, which also stabilizes the monomer was also maintained in the dimer,29,30,33 although Phe7, Leu27, Tyr34, and Phe44 originated from one protomer and Ile83 originated from the other.
Figure 3.
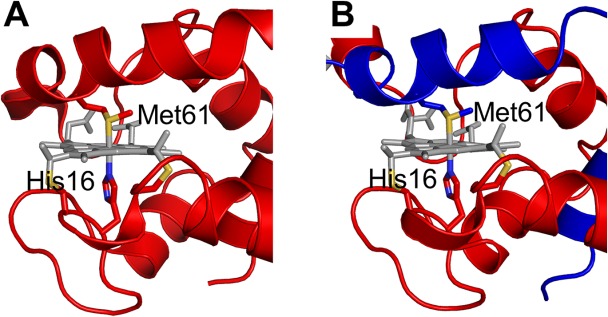
Active site structures of monomeric and dimeric AA cyt c555. A) Structure of monomeric AA cyt c555 (PDB code: 2ZXY). B) Structure of dimeric AA cyt c555 solved in this study (PDB code: 3X15). The red and blue regions in the dimer belong to different protomers. Side chain atoms of His16 and Met61 are shown as stick models. The hemes are shown as gray stick models. The sulfur atoms of the side chains of Cys12, Cys15, and Met81 are depicted in yellow, and the nitrogen atoms of the side chain of His16 are depicted in blue.
Table I.
Fe−His16 and Fe−Met61 Distances in Monomeric and Dimeric AA Cyt c555
| Fe−His16 (Å) | Fe−Met61 (Å) | |
|---|---|---|
| Monomera | 2.0 | 2.3 |
| Dimerb | ||
| Protomer A | 2.02 | 2.32 |
| Protomer B | 2.04 | 2.35 |
| Protomer C | 1.99 | 2.31 |
| Protomer D | 1.99 | 2.31 |
PDB code: 2ZXY.
Protomers A, B, C, and D of the dimers correspond to the protomers depicted in red, blue, magenta, and green, respectively, in the figures.
The hydrogen bond at Lys46(Nζ)/Asp73(Oδ1) in the monomer was broken in three of the four protomers in the dimers (Supporting Information Fig. S2), and the extra 310-α-310 helix and C-terminal helix interacted with the other protomer mainly by hydrophobic interactions in the dimer. Rearrangement of the hydrogen bonds around the hinge region was also observed in the dimer. A new hydrogen bond was formed within the protomers of the dimer at Ile60(O)/Glu64(Nε2), contributing to the stabilization of the extra 310-α-310 helix in all four protomers (Supporting Information Fig. S2). The hydrogen bonds at Ala51(N)/Glu58(Oε2), Asp54(N)/heme 13-propionate, and Asp54(O)/Lys57(N), contributing to the formation of the distorted 310 helix at Asp54–Lys57 of the extra 310-α-310 helix in the monomer, were broken, resulting in transition of the distorted 310 helix to a loop in three of the four protomers in the dimers (Supporting Information Fig. S2, red, blue, and magenta). These rearrangements around the hinge region caused relocation of the extra 310-α-310 helix and the heme, forming a space around the heme. As a result, a channel from the solvent to the heme moiety was created in the dimer (Supporting Information Fig. S3). The additional space around the heme moiety may allow exogenous ligands to access and bind easier to the heme iron in the dimer compared with the monomer, as indicated by the absorption spectra (see text below).
CD spectra of dimeric AA cyt c555
The far-UV circular dichroism (CD) spectrum of oxidized dimeric AA cyt c555 at pH 7.0, 25°C, was similar to that of the oxidized monomer under the same conditions (Fig. 4). These results show that the α-helical structures of dimeric AA cyt c555 were similar to those of the monomer under mild conditions. In the dimer, the Cotton effects at 190 to 220 nm became slightly more negative compared with those of the monomer, suggesting that the proportion of the random coil increased slightly in the dimer. The increase in the random coil structure may result from the increase in the fluctuation of the hinge region and/or its conversion from a distorted 310 helix to a loop by the dimerization.
Figure 4.
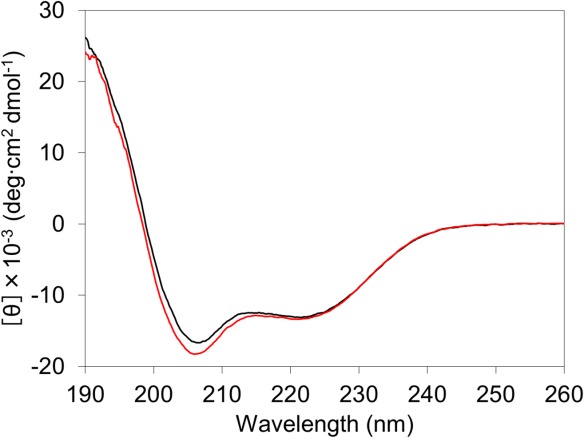
Far-UV circular dichroism spectra of oxidized monomeric (black) and dimeric (red) AA cyt c555. Measurement conditions: sample concentration (heme unit), 10 μM; solvent, 50 mM potassium phosphate buffer; pH, 7.0; temperature, 25°C.
A large amount of oxidized dimeric AA cyt c555 remained as dimers even after heating at 80 to 90°C for 10 min, or incubating at pH 2.2 or 11.0, 25°C for 10 min, according to size exclusion chromatography analysis (Supporting Information Fig. S4). The CD spectra of dimeric AA cyt c555 also exhibited negative ellipticity around 209 and 222 nm at pH 7.0, 85°C, and pH 2.2 to 11.0, 25°C (Supporting Information Fig. S5). These results show that dimeric AA cyt c555 exhibits thermostablity and pH tolerance, maintaining its secondary structures at high temperatures and a wide range of pH.
Thermodynamics of dimeric AA cyt c555
Differential scanning calorimetry (DSC) measurements of oxidized dimeric AA cyt c555 were performed to reveal its thermodynamic properties (Fig. 5). A negative peak was observed at 92 ± 1°C in the heat capacity (Cp) curve of the dimer, whereas no peak was observed at 90 to 95°C in the Cp curve of the monomer. Therefore, the peak temperature at 92°C corresponded to the dissociation temperature (Tm) of the dimer to monomers. The Tm value of dimeric AA cyt c555 was close to that of dimeric HT cyt c552 (92°C),56 and it was even higher than the denaturation temperature of the monomer of other cyt c proteins, such as horse cyt c, Pseudomonas aeruginosa cyt c551, and Shewanella cyt c5 (80–86°C).26,35,53 Mainly monomers were detected by the gel chromatography analysis when the AA cyt c555 dimer solution was heated up to 100°C [Supporting Information Fig. S4(A)], confirming the dissociation of the dimers by heating. The peak area in the DSC curve represents the enthalpy change (ΔH) for the dissociation of the dimer to monomers. ΔH exhibited a negative value, −14 ± 2 kcal/mol, for dimeric AA cyt c555, indicating that dimeric AA cyt c555 was enthalpically unstable compared with its monomer, as in the case of horse cyt c.53 The negative ΔH may be attributed to the rearrangement in the hydrogen bonds by the dimerization (Fig. S2), whereas a weaker coordination bond between Met61 and the heme iron in dimeric AA cyt c555 compared with its monomer may contribute to the negative ΔH value, since the intensity of the absorption band around 695 nm, which is related to the Met61–heme iron bond, decreased by the dimerization as in the case of horse cyt c (see text below).53
Figure 5.

Differential scanning calorimetry thermograms of oxidized monomeric (black) and dimeric (red) AA cyt c555. Measurement conditions: sample concentration (heme unit), 100 μM; buffer, 50 mM potassium phosphate buffer; pH, 7.0.
Ligand binding to dimeric cyt c555
The absorption spectra of dimeric AA cyt c555 were monitored to investigate its ligand binding properties. The Soret band of oxidized dimeric AA cyt c555 blue shifted from 412 nm in the monomer to 410 nm [Fig. 6(A)]. The intensity of the absorption band around 695 nm decreased in the dimer compared with that in the monomer [Fig. 6(A)].57 These results show that the Met61–heme iron bond was perturbed in oxidized dimeric AA cyt c555, as in the case of oxidized dimeric horse cyt c, although the bond was formed in the crystal structure of dimeric AA cyt c555. The Soret band of oxidized dimeric AA cyt c555 red shifted to 413 nm and the 695 nm absorption band disappeared by an addition of 1 mM potassium cyanide at pH 7.0, although no change was observed in the spectra by the addition of potassium cyanide to the oxidized monomer under the same conditions. A similar spectral change has been observed in CN- binding to oxidized dimeric horse cyt c.55 The absorption changes in the spectrum of oxidized dimeric AA cyt c555 by an addition of CN− were also observed at 85°C; this was not the case in the oxidized monomer [Supporting Information Fig. S6(A)].
Figure 6.
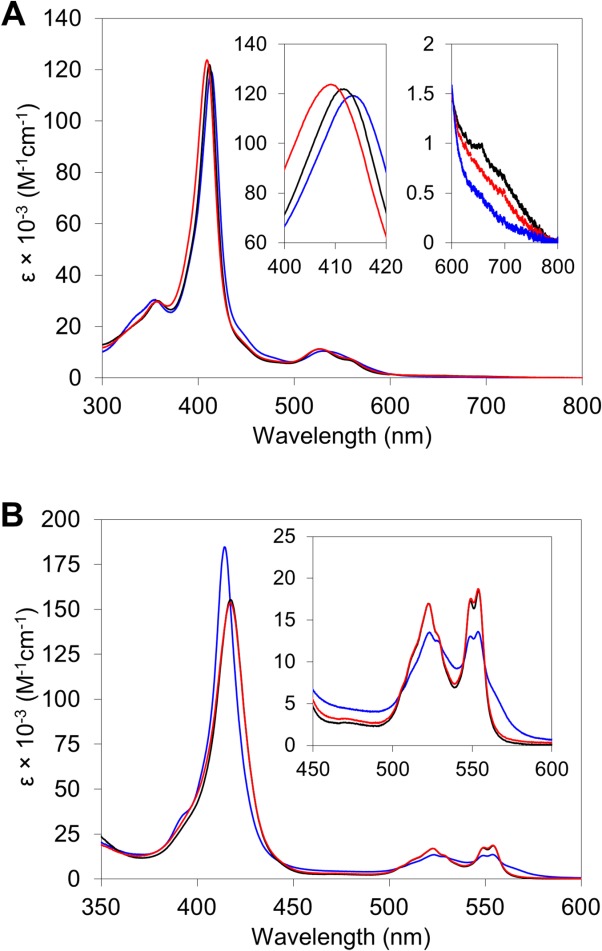
Absorption spectra of monomeric and dimeric AA cyt c555. A) Oxidized monomer (black) and dimer (red) in the absence of CN− and oxidized dimer in the presence of 1 mM CN− (blue). B) Reduced monomer (black) and dimer (red) under N2 atmosphere, and reduced dimer under CO atmosphere (blue). Measurement conditions: sample concentration (heme unit), 10 μM; solvent, 50 mM potassium phosphate buffer; pH, 7.0; temperature, 25°C.
Almost no difference was observed between the absorption spectra of dimeric AA cyt c555 and its monomer in the reduced state [Fig. 6(B)]. These results indicate that the Met–heme iron bond was formed in the dimer in the reduced state as in the monomer. However, the Soret band of reduced dimeric AA cyt c555 red shifted from 417 nm in the monomer to 414 nm with an increase in its intensity by exchanging the atmosphere from N2 to CO at pH 7.0, whereas no change was observed in that of the reduced monomer by exchanging the atmosphere [Fig. 6(A)]. These results indicate that CO bound to the reduced dimer but not to the reduced monomer at pH 7.0. Changes were also observed in the spectra of the dimer at 85°C, pH 7.0, and 25°C, pH 2.2 or 11.0 [Supporting Information Figs. S6(B) and S7]. However, almost no change was observed in reduced monomeric AA cyt c555 by introducing CO at 85°C, pH 7.0, or 25°C, pH 2.2–8.0, although CO bound to the monomer at pH higher than 9.0 (Supporting Information Fig. S8), owing to the dissociation of Met61 from the heme iron as reported for other cyts c.58,59 These results show that exchange of Met61 with exogenous ligands for the heme coordination was easier in dimeric AA cyt c555 compared with the monomer.
Discussion
Reports on proteins with domain-swapped structures have been increasing.39–53,56 We have reported that horse cyt c forms oligomers by domain swapping its C-terminal helix, whereas HT cyt c552 forms oligomers by swapping the domain containing the N-terminal helix and heme.53,56 In this study, AA cyt c555 formed oligomers by swapping the domain containing the extra 310-α-310 helix and C-terminal helix, where the swapping region of AA cyt c555 was similar to that of horse cyt c, except for the extra 310-α-310 helix (Fig. 1). The N-terminal region of monomeric AA cyt c555 interacted with the rest of the protein by a hydrogen-bonding network, along with hydrophobic interactions. Nine hydrogen bonds were formed at Cys15−Lys29 (within a loop and part of a helix), with a hydrogen-bonding network formed by Tyr34(Oη), Ile52(N), Val53(N), and heme 17-propionate in monomeric AA cyt c555 [Fig. 7(A)]. For the extra 310-α-310 helix and C-terminal helix, these helices interacted with each other through hydrophobic residues (Leu68, Leu71, Leu76, and Leu79) in the monomer, exhibiting a tight hydrophobic packing protein interior (Supporting Information Fig. S9). The extra 310-α-310 helix and the C-terminal helix each formed only one hydrogen bond, Ala51(N)/Glu58(Oε2) and Lys46(Nζ)/Asp73(Oδ1), respectively, with the rest of the protein in monomeric AA cyt c555 [Fig. 7(B)], and the extra 310-α-310 helix and C-terminal helix interacted with other regions of the protein also mainly by hydrophobic interactions in the monomer (Supporting Information Fig. S9). The hydrophobic interactions may have let the region containing the extra 310-α-310 helix and C-terminal helix exchange between molecules when forming the domain-swapped dimer.
Figure 7.
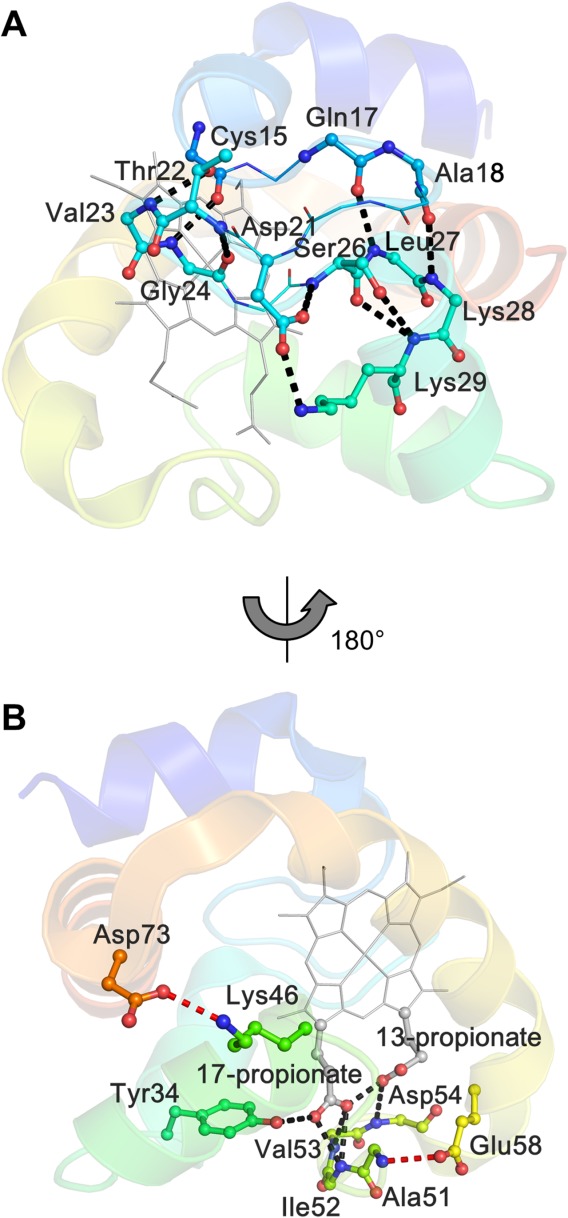
Hydrogen bonds in monomeric AA cyt c555. A) Hydrogen bonds formed at Cys15−Lys29. B) Hydrogen-bonding network around the heme propionates (black), together with the hydrogen bonds formed by the extra 310-α-310 helix and the C-terminal helix, respectively, with the rest of the protein (red). The protein is depicted with sequential colors from blue for the N terminal to red for the C terminal, and the heme is depicted in gray. The main chain or side chain of the residues forming the hydrogen bonds are shown as ball and stick models, and its nitrogen and oxygen atoms are depicted in blue and red, respectively.
The structure and ligand binding properties of the domain-swapped dimer differed among cyt c family proteins; HT cyt c552, horse cyt c, and AA cyt c555. The structure around the heme-ligating Met was not affected much and Met remained coordinated to the heme iron by the dimerization of HT cyt c552, where the hinge region was located away from the heme-ligating Met.56 When the hinge region was located close to the heme-ligating Met such as in horse cyt c, the structure around the heme-ligating Met was perturbed and Met dissociated from the heme iron by the dimerization,53 causing increase compared with the monomer in the binding properties of external ligands, such as CN− and H2O2.54,55 For the case of AA cyt c555, the hinge region was located slightly more away from the heme-ligating Met compared with that in horse cyt c owing to the extra 310-α-310 helix, which caused no dissociation of Met from the heme iron in the dimer crystal but increase in the ligand binding properties (Figs. 1 and 6). These results show that dissociation of the heme-ligating Met from the heme iron and increase in the ligand binding properties by domain swapping may depend on the distance between the heme-ligating Met and the hinge region.
The horse cyt c dimer was not stable against heat and acidic conditions, since the monomer unfolded at high temperature or acidic conditions,20,58 whereas AA cyt c555 is one of the most stable cyt c proteins.29,30 Dimeric AA cyt c555 exhibited high thermostability, where the dissociation temperature was 92 ± 1°C (Fig. 5). The secondary structures of dimeric AA cyt c555 were also pH tolerant [Supporting Information Fig. S5(B)], and the dimer did not dissociate to monomers at low or high pH [Supporting Information Fig. S4(B)]. The high thermostability and pH tolerance of dimeric AA cyt c555 are presumably due to maintenance of the interactions contributing to the hyperstability of the monomer, i.e., hydrogen-bonding network around the heme 17-propionate and the hydrophobic packing of the protein.
The Met ligand dissociated from the heme iron in the horse cyt c dimer, whereas it was coordinated to the heme iron in the HT cyt c552 dimer. CN- and peroxides have been shown to react with dimeric horse cyt c, owing to dissociation of Met80 from the heme iron.54,55 The region containing the extra 310-α-310 helix and C-terminal helix was swapped in dimeric AA cyt c555, and Met61 in the extra 310-α-310 helix was coordinated to the heme iron in its crystal structure. However, we found that CN- and CO bind to dimeric AA cyt c555 in its oxidized and reduced states, respectively (Fig. 6). The hinge region of AA cyt c555 (Val53–Lys57), which was located relatively close to Met61, was flexible and created a space around the heme and Met61 in the dimer (Fig. 1). The additional space may have allowed access of CN− and CO to the heme iron and a conformational change for ligand exchange from Met61 to exogenous ligands, whereas external ligands do not bind to the monomer due to its well-packed heme site.
Conclusion
Hyperstable AA cyt c555 belonging to the cyt c protein family formed a domain-swapped dimer, where the region containing the extra 310-α-310 helix and C-terminal helix swapped between protomers. The domain-swapped AA cyt c555 dimer exhibited structural polymorphism, showing flexibility at the Val53–Lys57 hinge region. Although Met61 was coordinated to the heme iron in oxidized dimeric AA cyt c555, CN− and CO bound to the oxidized and reduced dimer, respectively, at neutral pH, whereas these ligands did not bind to the monomer under the same conditions. Since dimeric AA cyt c555 exhibited high thermostability and pH tolerance, owing to the extra helix and hydrophobic interactions, it maintained its ligand binding properties at high temperatures and a wide range of pH. These results show that oligomerization by domain swapping may alter the properties of a protein, and thus domain swapping may be used for protein engineering.
Materials and Methods
Preparation of dimeric AA Cyt c555
Expression and purification of AA cyt c555 was performed as reported previously.29,30 Ethanol was added to the purified oxidized AA cyt c555 solution to a final concentration of 95 % (v/v) at room temperature. After centrifugation of the solution, the precipitate was lyophilized to remove residual ethanol. The lyophilized precipitate was dissolved in 50 mM potassium phosphate buffer, pH 7.0, at 90°C. Dimeric AA cyt c555 in the solution was separated from the monomer and higher-order oligomers with a HiLoad 26/600 Superdex 75 pg gel filtration column (GE Healthcare) using a fast protein liquid chromatography (FPLC) system (Biologic DuoFlow 10, Bio-rad, CA) with 50 mM potassium phosphate buffer, pH 7.0, at 4°C. The concentration of the protein was calculated from the absorbance of the Soret band using the coefficients of oxidized monomeric and dimeric AA cyt c555 obtained by the pyridine hemochrome method (monomer, 122,000 M−1 cm−1 at 412 nm; dimer 123,000 M−1 cm−1 at 410 nm).60
X-ray crystallographic analysis
Crystallization of dimeric AA cyt c555 was performed at 4°C using the sitting drop vapor diffusion method with crystal plates (CrystalClear D Strips, Douglas Instruments, Hampton Research, CA). Oxidized dimeric AA cyt c555 was dissolved in 10 mM Tris-HCl buffer, pH 8.0, at a protein concentration of 2.25 mM (heme unit). Droplets prepared by mixing 1 μL of the protein solution with 1 μL reservoir solution were equilibrated. The best reservoir solution was found to be 0.1 mM sodium cacodylate trihydrate, pH 6.5, containing 0.2 mM ammonium sulfate and 30% (w/v) polyethylene glycol 8000.
The diffraction data were collected at the BL38B1 beamline at SPring-8, Japan, using a Quantum315 detector (ADSC). The crystal was mounted on a cryo-loop and flash-frozen at 100 K in a nitrogen cryo system. The crystal-to-detector distance was 200 mm, and the wavelength was 1.0 Å. The oscillation angle was 1°, and the exposure time was 10 s per frame. The total number of frames was 180. The diffraction data were processed using the program, HKL2000.61 The preliminary structure was obtained by a molecular replacement method (MOLREP)62 using the atomic coordinates of the structure of monomeric AA cyt c555 (PDB code: 2ZXY) as a starting model. The structure refinement was performed using the program, REFMAC.63 The molecular model was manually corrected, and water molecules were picked up in the electron density map using the program, COOT. The data collection and refinement statistics are summarized in Table SI (Supporting Information). The three-dimensional structures of monomeric and dimeric AA cyt c555 were compared using the molecular graphics program, PyMOL.64
Size exclusion chromatography analysis
The solution containing monomeric or dimeric AA cyt c555 was incubated for 10 min in 50 mM potassium phosphate buffer, pH 7.0, at 70 to 100°C, 50 mM glycine-HCl buffer, pH 2.2, at 25°C, or 50 mM glycine-NaOH buffer, pH 11.0, at 25°C. The obtained AA cyt c555 solution was analyzed with a Superdex 75 10/300 GL gel filtration column (GE Healthcare) using the FPLC system (Biologic DuoFlow 10, Bio-rad) (flow rate, 0.5 mL/min; monitoring wavelength, 410 nm; solvent, 50 mM potassium phosphate buffer, pH 7.0; temperature, 4°C).
Optical absorption and CD measurements
Optical absorption spectra of monomeric and dimeric AA cyt c555 (heme unit, 10 μM) in the oxidized and reduced states in 50 mM potassium phosphate buffer, pH 7.0, were obtained with a UV-2450 spectrophotometer (Shimadzu, Japan) using a 1-cm-pathlength quartz cell at 25°C. Binding of CN− to oxidized monomeric and dimeric AA cyt c555 was investigated by an addition of 1 mM potassium cyanide in 50 mm potassium phosphate buffer, pH 7.0, at 25°C. Reduced monomeric and dimeric AA cyt c555 were obtained by an addition of 2 mM sodium dithionite to the solution containing the corresponding oxidized protein under N2 atmosphere. Binding of CO to reduced monomeric and dimeric AA cyt c555 were investigated by incubation of the reduced sample at 25°C under CO atmosphere at pH 2.2 to 11.0: 50 mM glycine-HCl, pH 2.2; 50 mM sodium acetate, pH 3.0 to 6.0; 50 mM potassium phosphate, pH 7.0 and 8.0; 50 mM glycine-NaOH, pH 9.0 to 11.0.
CD spectra of monomeric and dimeric AA cyt c555 (heme unit, 10 μM) in the oxidized state were obtained with a JASCO J-720 spectrometer (JASCO, Japan) using a 0.1-cm-pathlength quartz cell in 50 mM potassium phosphate buffer, pH 7.0, at 25 to 85°C, and in the same buffers described above at pH 2.2 to 11.0, 25°C.
DSC measurements
DSC thermograms of monomeric and dimeric AA cyt c555 (heme unit, 100 μM) in the oxidized state were measured using VP-DSC (GE Healthcare) at a scan rate of 1°C/min in 50 mM potassium phosphate buffer, pH 7.0. Buffer baselines were recorded at the same scan rate, and subtracted from the sample thermograms to obtain the Cp curves. The Tm of thermal dissociation of the dimer to monomers and ΔH were obtained by fitting the data using the program, ORIGIN 8 (OriginLab, MA).
Acknowledgments
The authors thank Mr. Leigh McDowell, Nara Institute of Science and Technology, for his advice on manuscript preparation. The authors also thank the staff at beamline BL38B1 Spring-8, Japan (Proposal No. 2013A1851). The authors are also grateful to Mr. Masao Hada, Nara Institute of Science and Technology, for preliminary measurements, and Prof. Yoshihiro Sambongi, Hiroshima University, for a kind gift of pYU1 and pEC86 plasmids carrying the genes for AA cyt c555 and cytochrome c maturation proteins, respectively.
Glossary
Abbreviations:
- AA
Aquifex aeolicus
- CD
circular dichroism
- Cp
heat capacity
- cyt c
cytochrome c
- cyt c552
cytochrome c552
- cyt c555
cytochrome c555
- DSC
differential scanning calorimetry
- FPLC
fast protein liquid chromatography
- HT
Hydrogenobacter thermophilus
- rmsd
root-mean-square deviation
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tm
transition temperature.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- Tomlinson EJ, Ferguson SJ. Conversion of a c type cytochrome to a b type that spontaneously forms in vitro from apo protein and heme: implications for c type cytochrome biogenesis and folding. Proc Natl Acad Sci USA. 2000;97:5156–5160. doi: 10.1073/pnas.090089397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AJ, Bader R, Christodoulou J, MacPhee CE, Dobson CM, Barker PD. Cytochrome display on amyloid fibrils. J Am Chem Soc. 2006;128:2162–2163. doi: 10.1021/ja0565673. [DOI] [PubMed] [Google Scholar]
- Koder RL, Anderson JLR, Solomon LA, Reddy KS, Moser CC, Dutton PL. Design and engineering of an O2 transport protein. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Yeung N, Sieracki N, Marshall NM. Design of functional metalloproteins. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler V, Wilson YM, Lo C, Sardo A, Ward TR. Protein-based hybrid catalysts—design and evolution. Curr Opin Biotechnol. 2010;21:744–752. doi: 10.1016/j.copbio.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Nanda V, Koder RL. Designing artificial enzymes by intuition and computation. Nat Chem. 2010;2:15–24. doi: 10.1038/nchem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, Gonen T, Yeates TO, Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336:1129–1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- Zastrow ML, Peacock AF, Stuckey JA, Pecoraro VL. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PS, Brustad EM, Kannan A, Arnold FH. Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science. 2013;339:307–310. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- Farid TA, Kodali G, Solomon LA, Lichtenstein BR, Sheehan MM, Fry BA, Bialas C, Ennist NM, Siedlecki JA, Zhao Z, Stetz MA, Valentine KG, Anderson JL, Wand AJ, Discher BM, Moser CC, Dutton PL. Elementary tetrahelical protein design for diverse oxidoreductase functions. Nat Chem Biol. 2013;9:826–833. doi: 10.1038/nchembio.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard DJ, Kane KM, Tezcan FA. Re-engineering protein interfaces yields copper-inducible ferritin cage assembly. Nat Chem Biol. 2013;9:169–176. doi: 10.1038/nchembio.1163. [DOI] [PubMed] [Google Scholar]
- Hunter CL, Mauk AG. Engineered metalloregulation of azide binding affinity and reduction potential of horse heart myoglobin. Dalton Trans. 2013;42:3151–3155. doi: 10.1039/c2dt32558f. [DOI] [PubMed] [Google Scholar]
- King NP, Bale JB, Sheffler W, McNamara DE, Gonen S, Gonen T, Yeates TO, Baker D. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oohora K, Hayashi T. Hemoprotein-based supramolecular assembling systems. Curr Opin Chem Biol. 2014;19:154–161. doi: 10.1016/j.cbpa.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Petrik ID, Liu J, Lu Y. Metalloenzyme design and engineering through strategic modifications of native protein scaffolds. Curr Opin Chem Biol. 2014;19:67–75. doi: 10.1016/j.cbpa.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Sono M, Du J, Dawson JH. Evidence of the direct involvement of the substrate TCP radical in functional switching from oxyferrous O2 carrier to ferric peroxidase in the dual-function hemoglobin/dehaloperoxidase from Amphitrite ornata. Biochemistry. 2014;53:4956–4969. doi: 10.1021/bi5002757. [DOI] [PubMed] [Google Scholar]
- Sun S, Sono M, Wang C, Du J, Lebioda L, Dawson JH. Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations. Arch Biochem Biophys. 2014;545:108–115. doi: 10.1016/j.abb.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Goto Y, Hagihara Y, Hamada D, Hoshino M, Nishii I. Acid-induced unfolding and refolding transitions of cytochrome c: a three-state mechanism in H2O and D2O. Biochemistry. 1993;32:11878–11885. doi: 10.1021/bi00095a017. [DOI] [PubMed] [Google Scholar]
- Colón W, Elöve GA, Wakem LP, Sherman F, Roder H. Side chain packing of the N- and C-terminal helices plays a critical role in the kinetics of cytochrome c folding. Biochemistry. 1996;35:5538–5549. doi: 10.1021/bi960052u. [DOI] [PubMed] [Google Scholar]
- Hamada D, Kuroda Y, Kataoka M, Aimoto S, Yoshimura T, Goto Y. Role of heme axial ligands in the conformational stability of the native and molten globule states of horse cytochrome c. J Mol Biol. 1996;256:172–186. doi: 10.1006/jmbi.1996.0075. [DOI] [PubMed] [Google Scholar]
- Marmorino JL, Lehti M, Pielak GJ. Native tertiary structure in an A-state. J Mol Biol. 1998;275:379–388. doi: 10.1006/jmbi.1997.1450. [DOI] [PubMed] [Google Scholar]
- Ptitsyn OB. Protein folding and protein evolution: common folding nucleus in different subfamilies of c-type cytochromes? J Mol Biol. 1998;278:655–666. doi: 10.1006/jmbi.1997.1620. [DOI] [PubMed] [Google Scholar]
- Travaglini-Allocatelli C, Gianni S, Brunori M. A common folding mechanism in the cytochrome c family. Trends Biochem Sci. 2004;29:535–541. doi: 10.1016/j.tibs.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Nakamura S, Sonoyama T, Ohshima A, Kobayashi Y, Takayama SJ, Yamamoto Y, Uchiyama S, Hasegawa J, Sambongi Y. Five amino acid residues responsible for the high stability of Hydrogenobacter thermophilus cytochrome c552: Reciprocal mutation analysis. J Biol Chem. 2005;280:5527–5532. doi: 10.1074/jbc.M412392200. [DOI] [PubMed] [Google Scholar]
- Travaglini-Allocatelli C, Gianni S, Dubey VK, Borgia A, Di Matteo A, Bonivento D, Cutruzzola F, Bren KL, Brunori M. An obligatory intermediate in the folding pathway of cytochrome c552 from Hydrogenobacter thermophilus. J Biol Chem. 2005;280:25729–25734. doi: 10.1074/jbc.M502628200. [DOI] [PubMed] [Google Scholar]
- Takahashi YT, Sasaki H, Takayama SJ, Mikami S, Kawano S, Mita H, Sambongi Y, Yamamoto Y. Further enhancement of the thermostability of Hydrogenobacter thermophilus cytochrome c552. Biochemistry. 2006;45:11005–11011. doi: 10.1021/bi061164g. [DOI] [PubMed] [Google Scholar]
- Obuchi M, Kawahara K, Motooka D, Nakamura S, Yamanaka M, Takeda T, Uchiyama S, Kobayashi Y, Ohkubo T, Sambongi Y. Hyperstability and crystal structure of cytochrome c555 from hyperthermophilic Aquifex aeolicus. Acta Crystallogr D Biol Crystallogr. 2009;65:804–813. doi: 10.1107/S0907444909017314. [DOI] [PubMed] [Google Scholar]
- Yamanaka M, Mita H, Yamamoto Y, Sambongi Y. Heme is not required for Aquifex aeolicus cytochrome c555 polypeptide folding. Biosci Biotechnol Biochem. 2009;73:2022–2025. doi: 10.1271/bbb.90220. [DOI] [PubMed] [Google Scholar]
- Oda K, Kodama R, Yoshidome T, Yamanaka M, Sambongi Y, Kinoshita M. Effects of heme on the thermal stability of mesophilic and thermophilic cytochromes c: comparison between experimental and theoretical results. J Chem Phys. 2011;134:025101. doi: 10.1063/1.3519814. [DOI] [PubMed] [Google Scholar]
- Tai H, Irie K, Mikami S, Yamamoto Y. Enhancement of the thermostability of Hydrogenobacter thermophilus cytochrome c552 through introduction of an extra methylene group into its hydrophobic protein interior. Biochemistry. 2011;50:3161–3169. doi: 10.1021/bi200256d. [DOI] [PubMed] [Google Scholar]
- Yamanaka M, Masanari M, Sambongi Y. Conferment of folding ability to a naturally unfolded apocytochrome c through introduction of hydrophobic amino acid residues. Biochemistry. 2011;50:2313–2320. doi: 10.1021/bi101646m. [DOI] [PubMed] [Google Scholar]
- Kamagata K, Kawaguchi T, Iwahashi Y, Baba A, Fujimoto K, Komatsuzaki T, Sambongi Y, Goto Y, Takahashi S. Long-term observation of fluorescence of free single molecules to explore protein-folding energy landscapes. J Am Chem Soc. 2012;134:11525–11532. doi: 10.1021/ja3020555. [DOI] [PubMed] [Google Scholar]
- Masanari M, Wakai S, Ishida M, Kato C, Sambongi Y. Correlation between the optimal growth pressures of four Shewanella species and the stabilities of their cytochromes c5. Extremophiles. 2014;18:617–627. doi: 10.1007/s00792-014-0644-y. [DOI] [PubMed] [Google Scholar]
- Wallace CJ, Clark-Lewis I. Functional role of heme ligation in cytochrome c. Effects of replacement of methionine 80 with natural and non-natural residues by semisynthesis. J Biol Chem. 1992;267:3852–3861. [PubMed] [Google Scholar]
- Bren KL, Gray HB. Structurally engineered cytochromes with novel ligand-binding sites: oxy and carbon monoxy derivatives of semisynthetic horse heart Ala80 cytochrome c. J Am Chem Soc. 1993;115:10382–10383. [Google Scholar]
- Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gotte G, Libonati M, Eisenberg D. Structures of the two 3D domain-swapped RNase A trimers. Protein Sci. 2002;11:371–380. doi: 10.1110/ps.36602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer ME. Protein folding and three-dimensional domain swapping: astrained relationship? Curr Opin Struct Biol. 2002;12:48–53. doi: 10.1016/s0959-440x(02)00288-9. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Schymkowitz JW, Itzhaki LS. The unfolding story of three-dimensional domain swapping. Structure. 2003;11:243–251. doi: 10.1016/s0969-2126(03)00029-7. [DOI] [PubMed] [Google Scholar]
- Gotte G, Libonati M. Oligomerization of ribonuclease A: two novel three-dimensional domain-swapped tetramers. J Biol Chem. 2004;279:36670–36679. doi: 10.1074/jbc.M404780200. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–824. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Gronenborn AM. Protein acrobatics in pairs—dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Sendall TJ, Pearce MC, Whisstock JC, Huntington JA. Molecular basis of α1-antitrypsin deficiency revealed by the structure of a domain-swapped trimer. EMBO Rep. 2011;12:1011–1017. doi: 10.1038/embor.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Cao H, Liu Z. Three-dimensional domain swapping in the protein structure space. Proteins. 2012;80:1610–1619. doi: 10.1002/prot.24055. [DOI] [PubMed] [Google Scholar]
- Nagao S, Osuka H, Yamada T, Uni T, Shomura Y, Imai K, Higuchi Y, Hirota S. Structural and oxygen binding properties of dimeric horse myoglobin. Dalton Trans. 2012;41:11378–11385. doi: 10.1039/c2dt30893b. [DOI] [PubMed] [Google Scholar]
- Han H, Kursula P. Periaxin and AHNAK nucleoprotein 2 form intertwined homodimers through domain swapping. J Biol Chem. 2014;289:14121–14131. doi: 10.1074/jbc.M114.554816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gotte G, Libonati M, Eisenberg D. A domain-swapped RNase A dimer with implications for amyloid formation. Nat Struct Biol. 2001;8:211–214. doi: 10.1038/84941. [DOI] [PubMed] [Google Scholar]
- Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- Guo Z, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc Natl Acad Sci USA. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sawaya MR, Eisenberg D. β2-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2011;18:49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, Higuchi Y. Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proc Natl Acad Sci USA. 2010;107:12854–12859. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Matsuo T, Nagao S, Hirota S. Peroxidase activity enhancement of horse cytochrome c by dimerization. Org Biomol Chem. 2011;9:4766–4769. doi: 10.1039/c1ob05552f. [DOI] [PubMed] [Google Scholar]
- Nugraheni AD, Nagao S, Yanagisawa S, Ogura T, Hirota S. Interaction of dimeric horse cytochrome c with cyanide ion. J Biol Inorg Chem. 2013;18:383–390. doi: 10.1007/s00775-013-0982-8. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Nagao S, Osuka H, Komori H, Higuchi Y, Hirota S. Domain swapping of the heme and N-terminal α-helix in Hydrogenobacter thermophilus cytochrome c552 dimer. Biochemistry. 2012;51:8608–8616. doi: 10.1021/bi3011303. [DOI] [PubMed] [Google Scholar]
- Baymann F, Tron P, Schoepp-Cothenet B, Aubert C, Bianco P, Stetter KO, Nitschke W, Schutz M. Cytochromes c555 from the hyperthermophilic bacterium Aquifex aeolicus (VF5). 1. Characterization of two highly homologous, soluble and membranous, cytochromes c555. Biochemistry. 2001;40:13681–13689. doi: 10.1021/bi011201y. [DOI] [PubMed] [Google Scholar]
- Myer YP. Conformation of cytochromes. III. Effect of urea, temperature, extrinsic ligands, and pH variation on the conformation of horse heart ferricytochrome c. Biochemistry. 1968;7:765–776. doi: 10.1021/bi00842a035. [DOI] [PubMed] [Google Scholar]
- Davis LA, Schejter A, Hess GP. Alkaline isomerization of oxidized cytochrome c. Equilibrium and kinetic measurements. J Biol Chem. 1974;249:2624–2632. [PubMed] [Google Scholar]
- Bartsch RG. Cytochromes: bacterial. Methods Enzymol. 1971;23:344–363. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The case for open-source software in drug discovery. Drug Discov Today. 2005;10:213–217. doi: 10.1016/S1359-6446(04)03363-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


