Abstract
Afadin, a scaffold protein localized in adherens junctions (AJs), links nectins to the actin cytoskeleton. Nectins are the major cell adhesion molecules of AJs. At the initial stage of cell–cell junction formation, the nectin–afadin interaction plays an indispensable role in AJ biogenesis via recruiting and tethering other components. The afadin PDZ domain (AFPDZ) is responsible for binding the cytoplasmic C-terminus of nectins. AFPDZ is a class II PDZ domain member, which prefers ligands containing a class II PDZ-binding motif, X-Φ-X-Φ (Φ, hydrophobic residues); both nectins and other physiological AFPDZ targets contain this class II motif. Here, we report the first crystal structure of the AFPDZ in complex with the nectin-3 C-terminal peptide containing the class II motif. We engineered the nectin-3 C-terminal peptide and AFPDZ to produce an AFPDZ–nectin-3 fusion protein and succeeded in obtaining crystals of this complex as a dimer. This novel dimer interface was created by forming an antiparallel β sheet between β2 strands. A major structural change compared with the known AFPDZ structures was observed in the α2 helix. We found an approximately 2.5 Å-wider ligand-binding groove, which allows the PDZ to accept bulky class II ligands. Apparently, the last three amino acids of the nectin-3 C-terminus were sufficient to bind AFPDZ, in which the two hydrophobic residues are important.
Keywords: afadin–nectin complex, adherens junction, PDZ domain, sequence specific recognition, crystallography
Introduction
Cell–cell adhesion of epithelia and endothelia serves a barrier function in the body. Cell–cell adhesion machinery, such as adherens junctions (AJs) and tight junctions (TJs), are organized by membrane-incorporated cell adhesion molecules and intracellular components, including scaffold proteins, adaptor proteins, and cytoskeletons. Nectins, afadin, and E-cadherin are the major components of AJs, which play a key role in the initial stage of cell–cell junction formation. Afadin, a major scaffold protein of nectins, directly links nectins to the actin cytoskeleton (F-actin)1,2 and establishes stable AJs via connection to the E-cadherin–catenin complex. In particular, the trans-interaction of nectin cis-dimers is the initial trigger for cell–cell junction formation,3 followed by actin cytoskeleton reorganization. Finally, the E-cadherin–catenin complex is recruited to the nectin–afadin-based cell adhesion site, while afadin organizes their connection by interacting with ponsin, LMO-7, ADIP, or by direct association.4–7 Afadin, bound by activated Rap1, inhibits endocytosis of E-cadherin by interacting with p120ctn, thereby enhancing E-cadherin adhesion activity.8 In epithelial cells, transient interaction between afadin and ZO-1 is required for subsequent TJ formation at the AJ apical site.9 The importance of afadin during development was also reported from studies on afadin deficient mice, in which the organization of cell–cell junctions and cellular polarity were impaired.10,11
The variety of the afadin functions are attributed to its multidomain structure, which comprises several domains: two Ras-association domains, a forkhead-associated domain, a dilute domain, a postsynaptic density-95, disc large, zonula occludens-1 (PDZ) domain, and three proline-rich regions followed by a F-actin binding domain [Fig. 1(A)]. Among them, the PDZ domain binds to nectin C-terminus, whereas other domains interact with the various other proteins during AJ formation.12,13
Figure 1.
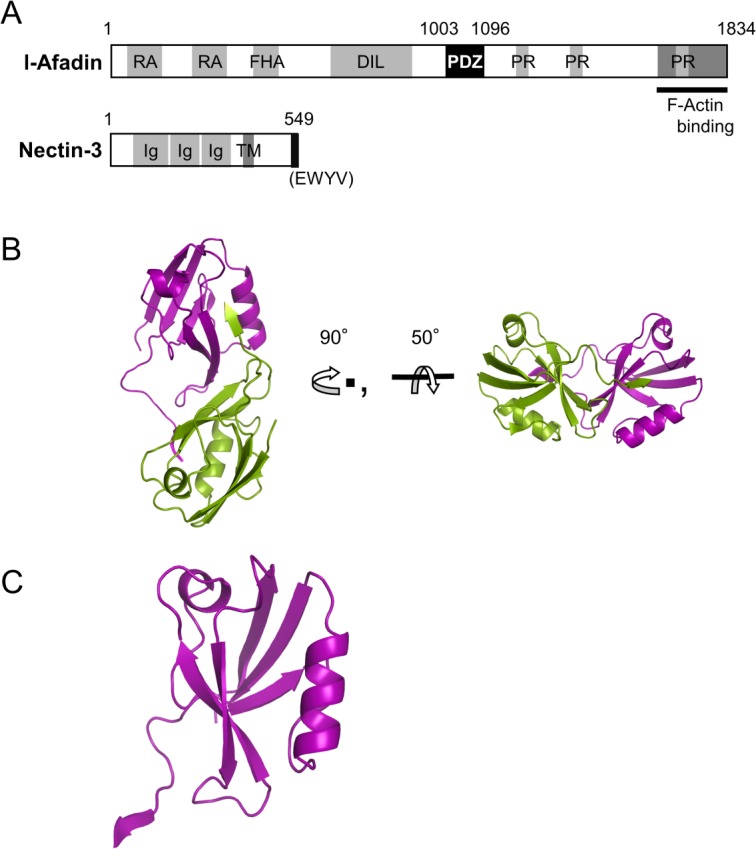
Crystal structure of the AFPDZ–nectin-3 C-terminal peptide complex. (A) Domain architecture of l-afadin and nectin-3. The AFPDZ interacts with the nectin-3 C-terminus. RA, Ras-association; FHA, forkhead-associated; DIL, dilute; PR, proline-rich region; Ig, immunoglobulin-like; TM, transmembrane region. (B) Overall structure of AFPDZ–nectin-3 complex. (C) Close-up view of the monomeric AFPDZ–nectin-3 fusion protein structure.
PDZ domains are small modular domain of approximately 90 amino acids with a canonical fold of six β-strands (β1–β6) and two α-helices (α1 and α2), in which β2 and α2 form a ligand-binding groove. The ligand interacts with this groove forming a new antiparallel β-sheet against β2. PDZ domains dominantly recognize the C-terminus of target proteins in specific and promiscuous manner and, in some cases, the internal sequence of the target protein. Historically, PDZ domains have been roughly classified into three classes based on their recognition sequences. Class I domains recognize the X-S/T-X-Φ motif, class II domains recognize the X-Φ-X-Φ motif, and class III domains recognize the X-D/E-X-Φ motif, where Φ denotes hydrophobic amino acids. However, some PDZ domains, like the afadin PDZ domain (AFPDZ), belong to multiple classes, and can, therefore, associate with multiple ligands within the different motif classes.
In addition to nectins, AFPDZ interacts with JAM-1, an another AJ resident protein.14 Furthermore, AFPDZ associates with Bcr,15 Jagged-1,16 SPA-1,17 c-Src,18 Eph receptors,19,20 and neurexin.19 There are several previously published NMR-resolved atomic structures of free AFPDZ,21,22 including one AFPDZ–Bcr (class I ligand) complex.23 In addition, an organic compound bound form was solved.22 Nevertheless, the molecular basis for afadin's key physiological function, AFPDZ-binding of class II ligands, still remains unclear. For instance, the molecular modeling study of the AFPDZ–nectin complex based on the existing AFPDZ–Bcr structure failed to explain how AFPDZ recognizes nectins. Specifically, because the third residue from the C-terminus of the nectin-3 was a bulky Trp, a reasonable model could not be built without steric interference.
Because many of the physiological targets of afadin including nectins belong to class II, the ligand recognition mechanism for the class II motif needs to be elucidated. Here, we show the first crystal structure of AFPDZ complexed with the nectin-3 C-terminal peptide as a stabilized dimer using an engineered fusion protein. Our crystal structure reveals the unexpected widening of the AFPDZ ligand-binding groove, which may explain how the bulky Trp residue is recognized.
Results and Discussion
Overall structure of the PDZ–nectin-3 complex
To obtain the PDZ–C-terminal nectin-3 complex crystal, we prepared three PDZ constructs; one consisted of a free PDZ domain, which cocrystallized with nectin-3 peptide, while the other two were PDZ domain–nectin-3 peptide fusion proteins with different linker lengths. Among the latter two, we succeeded in obtaining crystals from the fusion protein with the short linker, named AFPDZ-nec3C. The crystal structure was solved by the molecular replacement method at 2.8 Å resolution in the space group, C2221 (Table I). Two N-terminal residues (Lys1003 and Glu1004) and seven residues derived from the expression vector were disordered in the crystal. The asymmetric unit contained two molecules in the opposite direction. Each molecule formed a symmetric dimer with a crystallographically related molecule [Fig. 1(B)]. This dimer formation was consistent with the physicochemical analysis, in which AFPDZ-nec3C was eluted as a dimer using size-exclusion chromatography (Supporting Information Fig. S1).
Table I.
Data Collection and Refinement Statictics
| Data collection | |
|---|---|
| Space group | C2221 |
| Unit cell parameters (Å) | a = 55.502, b = 90.503, c = 88.765 α = β = γ = 90° |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50–2.8a (2.85–2.80) |
| Total number of unique reflections | 5641 |
| Completeness (%) | 97.4a (94.1) |
| Rmerge (%) | 5.5a (20.2) |
| I/σ | 31.5a (6.2) |
| Redundancy (%) | 5.1a (4.7) |
| Refinement statistics | |
| Resolution range (Å) | 50–2.8a (2.85–2.80) |
| Number of reflections | 5357 |
| Rwork/Rfree | 0.241/0.263 |
| Number of atoms | |
| Protein | 1426 |
| Water | 12 |
| Overall B-factors (Å2) | |
| Protein | 65.73 |
| Water | 69.94 |
| Root mean square deviationsb | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.274 |
| Ramachandran analysis | |
| Favored (%) | 97.89 |
| Allowed (%) | 1.06 |
| Outliers (%) | 1.05 |
Values in parentheses are for the highest resolution shell.
From MolProbity.
Our crystallized AFPDZ structure comprised six β-strands (β1–β6) and two α-helices (α1 and α2), similar to previous reports (PDB code: 1xz9, 1t2m, and 2ain). The nectin-3 peptide binds to the canonical PDZ-peptide binding groove forming a β-strand. Interestingly, only the last three residues at the nectin-3 C-terminus were in contact with the PDZ domain, whereas upstream residues were apart from the PDZ domain surface.
Structural plasticity of ligand-binding groove
We compared the backbone structure of our AFPDZ–nectin-3 to that of the previously published AFPDZ–Bcr complex (Fig. 2). The overall RMSD of the backbone atoms was 2.29 Å and a notable difference was found in the α2 helix, with the exception of the flexible β2–β3 loop. The α2 helix in the AFPDZ–nectin-3 complex was rotated 10° counter-clockwise and finally displaced 2.5 Å away from α2 of the AFPDZ–Bcr complex. As a result, substantial widening of the ligand-binding groove was observed. Finally, a vacancy large enough to accept the bulky Trp side-chain appeared in the middle of the groove (Fig. 3). This portion of the PDZ domain structure appears critical for nectin-3 association, otherwise a part of the Trp indole ring [Trp(−2)] would collide with the PDZ domain in the AFPDZ–Bcr complex.
Figure 2.
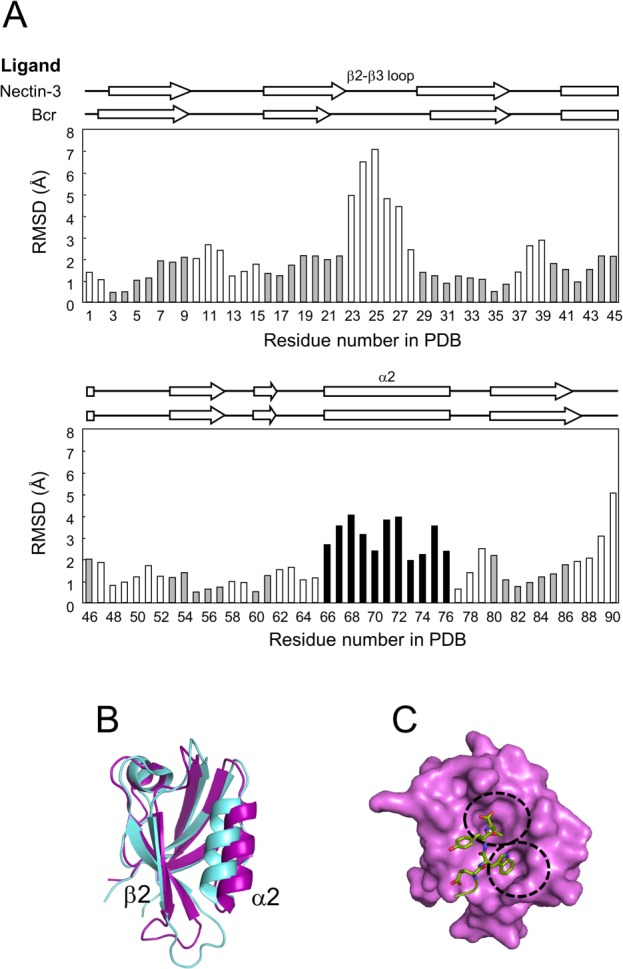
Plasticity of the ligand-binding groove. (A) Backbone structural comparison. The RMSD of α-carbons in the AFPDZ bound by ligand peptides was calculated using MOLMOL.49 Amino acids comprising secondary structures in the AFPDZ–nectin-3 complex are gray in color. The most different element, α2, is highlighted in black. (B) Superposition of the AFPDZ in the AFPDZ–nectin-3 (magenta) and Bcr (light blue) complexes. A ligand-binding pocket is formed between β2 and α2. (C) Surface representation of the AFPDZ–nectin-3 complex. Binding pockets are separately formed for Val(0) and Trp(−2) [dotted circles].
Figure 3.
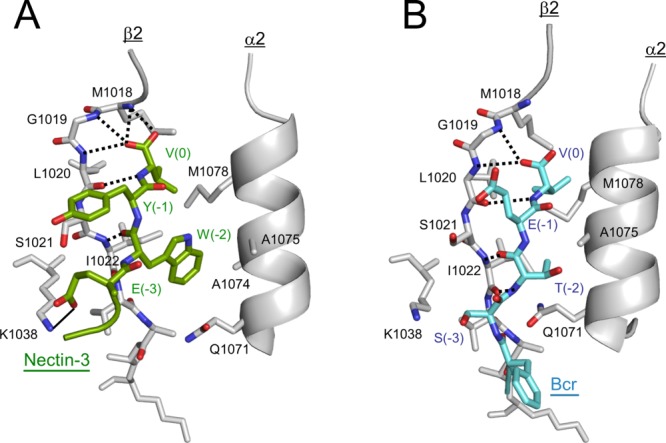
Molecular recognition of ligand peptides by the AFPDZ. Detailed interaction between AFPDZ and nectin-3 (A) or Bcr (B) are shown. Dotted lines indicate hydrogen bonds. The thin line between Lys1038 and Glu(−3) in panel (A) represents a salt bridge. Only β2, α2, and Lys1038 in β3 are shown for AFPDZ for clarity.
In the canonical binding mode, amino acid side-chains at positions 0 (the C-terminus) and −2 (the third residue from the C-terminus) direct the ligand to the binding groove. Bulky amino acids often reside at the position −2 in class II ligands, as is the case with the Trp residue of nectin-3. In the AFPDZ–Bcr peptide complex, Thr(-2) is close to forming a van der Waals interaction with Gln1071 in α2 of the AFPDZ,23 whereas Trp(-2) in nectin-3 could not fit into such a narrow ligand-binding groove. Because the width of the free AFPDZ ligand-binding groove more closely resembles that of the AFPDZ–Bcr complex compared with our structure,21,23 the structure of our AFPDZ–nectin-3 complex suggests that AFPDZ can adopt a structure with a wider ligand-binding groove, probably because of its structural plasticity. We also speculate that the α2 helix may be mobile, thus the wide groove may appear by an induced fit.
Such structural plasticity of the AFPDZ ligand-binding groove was observed among the ensemble of 20 AFPDZ–Bcr complex solution structures to some extent (PDB code: 2ain). The distance between Cα-atoms of Ile1022 and Ala1075 varied from 6.8 to 8.1 Å, although they were still narrower than that of the AFPDZ–nectin-3 complex (10.4 Å). Conversely, ligand binding-induced displacement of the α2 helix was reported in other cases of class II PDZ domains (PDB code: 1n7f, 1v1t, and 2ejy).24–26 Therefore, we conclude that the bulky Trp side-chain partially induces widening of the ligand-binding groove.
Recognition of the nectin-3 C-terminus
The nectin-3 peptide forms an antiparallel β-sheet with β2 in a similar manner to the canonical ligand-binding mode, as reported by many PDZ domain structural analyses (Fig. 3). Both carboxylate oxygen atoms of C-terminal Val(0) of nectin-3 were tightly recognized by forming four hydrogen bonds with the backbone amides of Met1018, Gly1019, and Leu1020 of AFPDZ, which belong to the conserved carboxylate-binding loop. Additionally, the amide nitrogen of Val(0) and the carbonyl oxygen of Trp(-2) was involved in hydrogen bonding between the nectin-3 peptide and AFPDZ. However, the main-chain of Glu(-3) and other upstream residues of nectin-3 did not contact the AFPDZ; thus, only a short β sheet was formed. The side-chains of Val(0) and Trp(-2) of nectin-3 were embedded in separate hydrophobic pockets in the AFPDZ ligand-binding groove. Furthermore, the aromatic ring of Tyr(−1) of nectin-3 makes a hydrophobic contact with Ser1021 in β2. Additionally, nectin-3 was further stabilized through a salt bridge formed by Glu(-3) of nectin-3 and Lys1038 in β3 (Supporting Information Fig. S2).
Structural comparison of the ligand recognition mechanism of AFPDZ for nectin-3 and Bcr revealed an increased number of interactions, which encouraged us to speculate that the affinity for nectin-3 is higher than that for Bcr (Fig. 3). Although the β-sheet formed by the Bcr peptide is one residue longer than that of nectin-3, only a single carboxylate oxygen is recognized by two hydrogen bonds. Previous structural and peptide library studies have shown that the C-terminal carboxylate, positions 0 and -2 of the ligand, were crucial for PDZ binding.27–29 Because the C-terminus of both nectin-3 and Bcr contains valine, increased hydrogen bond formation between AFPDZ and nectin-3 should be favored. However, the hydrophobic interaction of Trp(-2) in nectin-3 appeared more important, as it substituted for the van der Waals contact of Thr(-2) with Gln1071 in the AFPDZ–Bcr complex.23 In addition, the salt bridge between the nectin-3 Glu(-3) and Lys1038 is compatible with the hydrogen bond formed between Ser(-3) in Bcr and Lys1038. Thus, we reasoned that nectin-3 may bind to AFPDZ more tightly than Bcr.
We then examined the binding affinity of these peptide ligands using isothermal titration calorimetry (ITC) with the isolated AFPDZ monomer and synthetic 9-mer of the C-terminal peptides (Fig. 4; Table II). The dissociation constants (KD) were 17.8 and 110.9 µM for nectin-3 and Bcr, respectively. The results were consistent with our expectations based on peptide recognition mechanisms; the different affinities may concern a variety of target protein functions. Although the nectin–afadin interaction is responsible for establishing the rigid structure of AJ machinery, Bcr–afadin may be a transient complex for signal transduction.
Figure 4.
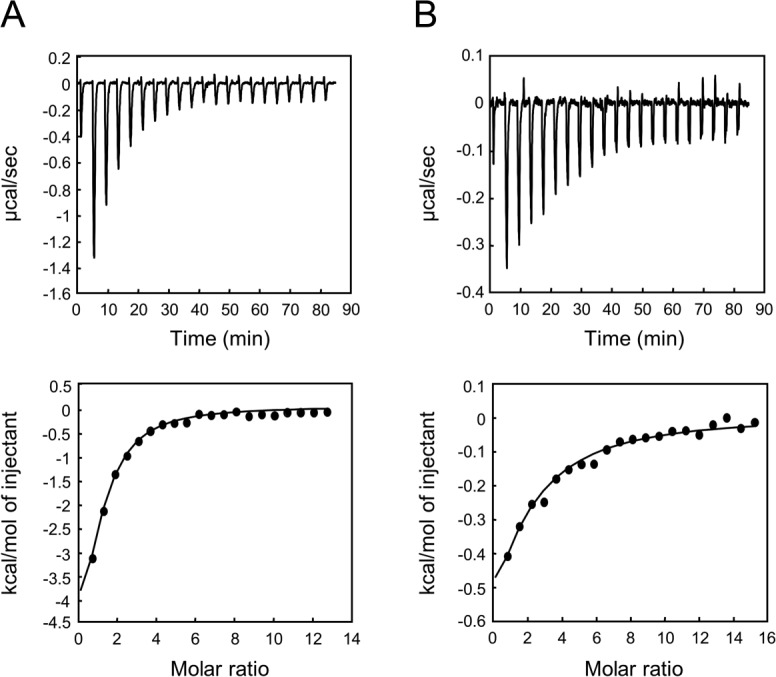
Typical ITC data for AFPDZ ligand binding. The binding constants of ligand peptides to AFPDZ were measured by ITC. (A) Nectin-3 peptide (0.8 mM) titrated into AFPDZ (15 µM). (B) Bcr peptide (2.16 mM) titrated into AFPDZ (31 µM). Raw data (upper panel) and integrated titration curves (lower panel) are shown.
Table II.
Ligand-Binding Affinity of AFPDZ
| Ligand peptide | KD (µM) | Peptide sequences |
|---|---|---|
| −8 −4 0 | ||
| Nectin-3 | 17.80 | VISRREWYV |
| Nectin-3_E3A | 29.37 | VISRRAWYV |
| Nectin-3_R4A | 15.69 | VISRAEWYV |
| Nectin-3_R5A | 13.05 | VISAREWYV |
| Bcr | 110.90 | YSILFSTEV |
| Neurexin | 21.89 | KNKDKEYYV |
In addition to C-terminal four residues of the ligands (positions 0 to -3), which are important for canonical PDZ domain binding, upstream amino acids may also contribute to PDZ recognition.30 We focused on the conserved positively charged residues at the −4 and −5 positions of nectins (Supporting Information Table I). In these positions of nectin-3, we substituted Ala and examined how these substituted peptides bound the AFPDZ. We also substituted Glu(-3) with Ala to verify its involvement in AFPDZ binding and compare the affinity to AFPDZ with those two peptides. The binding affinity was found to be slightly decreased by the Glu(-3) substitution whereas no changes were observed in the substitution of nectin-3 Arg(−4) and Arg(−5), suggesting that these residues are not recognized by AFPDZ. Moreover, we confirmed the importance of the hydrophobic side chain at the -2 position using the neurexin peptide, whose EYYV sequence resembles that of nectin-3 (EWYV). As the KD value of neurexin (21.9 µM) was close to that of nectin-3, this result implies that both the hydrophobic residues at positions -2 and −1 are important in AFPDZ binding. Altogether, both structural and affinity analyses suggest that the interaction between nectin-3 and AFPDZ is primarily mediated by the three C-terminal residues on nectin-3.
Dimer formation with the PDZ domain
We found that AFPDZ-nec3C crystallized into a dimer with the AFPDZ. Two PDZ domains were found to interact at the end of β2 and beginning of the following loop (between β2 and β3), forming an extended β-sheet compared with the ligand-free form [Fig. 5(A)]. Size-exclusion chromatography analyses exhibited that AFPDZ could form dimer by itself (Supporting Information Fig. S1), which indicated the dimerization ability of AFPDZ. However, we cannot exclude the possibility that the fused nectin-3 peptide caused and stabilized the dimer structure of AFPDZ-nec3C. Recently, it has been reported that some PDZ domains are responsible for the functional dimerization of the proteins containing them.24,31–35 It has been hypothesized that at AJ, the extracellular region of nectins forms a cis-homodimer and then clusters laterally to form a cell–cell junction.36,37 The physiologically active form of afadin remains elusive; however, at AJ, each afadin molecules come closer to one another at intracellular sites. Thus, we speculated that afadin can dimerize through the PDZ domain, thereby stabilizing the nectin-3 cis-homodimer in a manner similar to the crystal structure.
Figure 5.
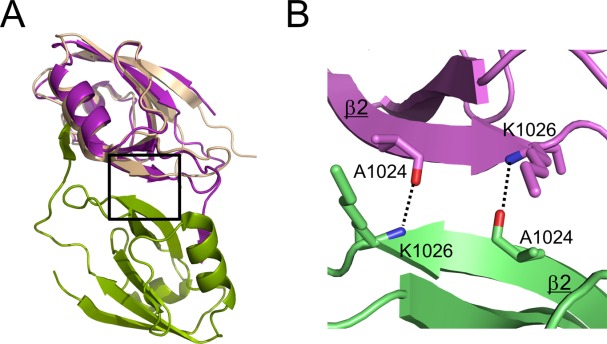
AFPDZ domain dimer formation. Ala1024 and Lys1026 both formed hydrogen bonds with their associated partners in the other monomer. These interactions serve the dimer interface at the end of extended β2. (A) Superposition of the ligand-free form (1t2m) [wheat] on the AFPDZ-nec3C. The box indicates the area shown in (B). (B) Close-up view of the dimer interface.
Recently, the crystal structure of the extracellular region of nectin-1 was revealed by our group and others.38,39 In our crystal, nectin-1 formed a V-shaped cis-dimer via interactions between the first immunoglobulin (Ig)-like domains; thus, the third Ig-like domains were 170 Å apart. At the onset of AJ formation, clustering of nectin–afadin complexes at cell–cell contact sites were observed.37 In this case, not only the extracellular region but also the intracellular region of nectins must be tethered to each other. On dimerization at the end of β2, two AFPDZ assemble obliquely. The ligand-binding grooves located at the middle of the lateral faces of the dimer while the inlets of the peptide binding grooves are aranged at the same upper side of the dimer, like a two-hole electric power socket (Fig. 1B and Supporting Information Fig. S3). Such coordination might improve the catching efficiency of nectin C-terminus and promote afadin–nectin interactions near the cytoplasmic tails of membrane-incorporated nectins. Moreover, dimerization with AFPDZ would increase its affinity for the nectin homodimer, which synergistically enhances afadin–nectin complex formation through a tethering effect. Because the KD between the nectin-3 C-terminal peptide and the AFPDZ monomer was 17.8 µM, and appears too weak to maintain the rigid structure of AJ, the dimer–dimer interaction is probable.
Thus, our finding provides molecular evidence that intracellular regions derived from the neighboring nectin cis-dimers may be closely situated on nectin–PDZ dimerization. In this case, the dimerized PDZ domain is able to interact with multiple nectin cis-dimers (i.e., by binding one nectin molecule from a nectin cis-dimer and one nectin molecule from another neighboring nectin cis-dimer); then, when afadin and nectin dimerize inside and outside of the plasma membrane, a belt-like structure of nectin clusters is created, adding to the strength and rigidity of AJ. In agreement with the proposed dimer–dimer interaction, both the afadin–nectin and nectin–nectin interaction, together with Necl-5 at the cell–cell contact site, are co-operatively enhanced.40 These hypotheses must be experimentally confirmed in future.
Materials and Methods
Protein expression and purification
The DNA fragment encoding the mouse AFPDZ (1003–1095) fused with the nectin-3-derived C-terminal hexapeptide (RRWEYV) was amplified by PCR and cloned into the pGEX-6P3-PRESAT vector.41 The protein was expressed in Escherichia coli BL21(DE3) and purified using DEAE Sepharose (GE Healthcare) and GST-accept (Nakarai tesque). After extensively washing the glutathione-S-transferase (GST) column, the GST-tag was cleaved using PreScission protease (GE Healthcare) at 4°C. The PDZ–nectin-3 fusion protein was eluted and concentrated, then the protein solution was loaded on a Superdex 75 HR 26/60 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5 at 4°C) and 150 mM NaCl. The purified protein was concentrated at 3.5 mg/mL for the stock and further concentrated before crystallization.
Crystallization and structure determination
The fusion protein crystal was grown using the hanging drop vapor diffusion method at 20°C. One microliter of 15–22 mg/mL protein solution was mixed with 1 µL of mother liquor containing 100 mM Hepes-NaOH (pH 7.5) and 2M sodium formate. Crystals were transferred to a cryoprotectant containing 100 mM Hepes-NaOH (pH 7.5), 20 mM Tris-HCl (pH 7.5 at 4°C), 2.2M sodium formate, 150 mM NaCl, and 16% ethylene glycol and flash frozen in liquid nitrogen. Diffraction data were collected at the Photon Factory, BL-17A (Japan). Images were processed using the HKL2000 package.42 The structure was solved by molecular replacement using the Balbes program43 and the second PDZ domain of human Dlg3 (PDB code: 2FE5) as a search model. Density modification and initial model building were performed with Parrot,44 Buccaneer,45 and Coot,46 and structural refinement was performed using Refmac 547 and Coot.46 Refinement cycles included rigid body refinement and restrained refinement with translation/libration/screw (TLS) algorithm. Stereochemical properties were evaluated by MolProbity.48 Structure figures were prepared using PyMOL (Schrödinger).
Isothermal titration calorimetry
ITC experiments were performed at 22°C in 25 mM Hepes-NaOH (pH 7.5) and 150 mM NaCl using MCS-ITC (Malvern Instruments, UK). Peptides (0.8–2.16 mM) were titrated into AFPDZ (15–31 µM) in the sample cell using a 250-µL syringe. Each experiment comprised a preliminary 3-µL injection followed by 20 subsequent 13-µL injections with 4-min intervals. Data were analyzed using Origin software (Malvern Instruments) using fixed stoichiometry, n = 1.
Structure coordinate
The coordinate has been deposited to the Protein Data Bank with accession code: 3AXA.
Acknowledgments
We thank Drs. Takuji Oyama and Daizo Hamada for helpful advice on structure determination.
Glossary
Abbreviations:
- AFPDZ
afadin PDZ domain
- Bcr
breakpoint cluster region
- disc large
zonula occludens-1
- PDZ
postsynaptic density-95
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- Mandai K, Nakanishi H, Satoh A, Obaishi H, Wada M, Nishioka H, Itoh M, Mizoguchi A, Aoki T, Fujimoto T, Matsuda Y, Tsukita S, Takai Y. Afadin: a novel actin filament–binding protein with one PDZ domain localized at Cadherin-based cell-to-cell adherens junction. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Shimizu K, Kawakatsu T, Yasumi M, Shingai T, Fukuhara A, Ozaki-Kuroda K, Irie K, Nakanishi H, Takai Y. Antagonistic and agonistic effects of an extracellular fragment of nectin on formation of E-cadherin-based cell-cell adhesion. Genes Cells. 2003;8:51–63. doi: 10.1046/j.1365-2443.2003.00616.x. [DOI] [PubMed] [Google Scholar]
- Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, Mizoguchi A, Takai Y. Ponsin/SH3P12: an l-afadin—and vinculin-binding protein localized at cell–cell and cell–matrix adherens junctions. J Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooshio T, Irie K, Morimoto K, Fukuhara A, Imai T, Takai Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J Biol Chem. 2004;279:31365–31373. doi: 10.1074/jbc.M401957200. [DOI] [PubMed] [Google Scholar]
- Asada M, Irie K, Morimoto K, Yamada A, Ikeda W, Takeuchi M, Takai Y. ADIP, a novel Afadin- and alpha-actinin-binding protein localized at cell-cell adherens junctions. J Biol Chem. 2003;278:4103–4111. doi: 10.1074/jbc.M209832200. [DOI] [PubMed] [Google Scholar]
- Takai Y, Ikeda W, Ogita H, Rikitake Y. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu Rev Cell Dev Biol. 2008;24:309–342. doi: 10.1146/annurev.cellbio.24.110707.175339. [DOI] [PubMed] [Google Scholar]
- Hoshino T, Sakisaka T, Baba T, Yamada T, Kimura T, Takai Y. Regulation of E-cadherin endocytosis by nectin through afadin, Rap1, and p120ctn. J Biol Chem. 2005;280:24095–24103. doi: 10.1074/jbc.M414447200. [DOI] [PubMed] [Google Scholar]
- Ooshio T, Kobayashi R, Ikeda W, Miyata M, Fukumoto Y, Matsuzawa N, Ogita H, Takai Y. Involvement of the interaction of afadin with ZO-1 in the formation of tight junctions in Madin-Darby canine kidney cells. J Biol Chem. 2010;285:5003–5012. doi: 10.1074/jbc.M109.043760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda W, Nakanishi H, Miyoshi J, Mandai K, Ishizaki H, Tanaka M, Togawa A, Takahashi K, Nishioka H, Yoshida H, Mizoguchi A, Nishikawa S, Takai Y. Afadin: a key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J Cell Biol. 1999;146:1117–1131. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhadanov AB, Provance DW, Speer CA, Coffin JD, Goss D, Blixt JA, Reichert CM, Mercer JA. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr Biol. 1999;9:880–888. doi: 10.1016/s0960-9822(99)80392-3. [DOI] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Ogita H, Rikitake Y, Miyoshi J, Takai Y. Cell adhesion molecules nectins and associating proteins: implications for physiology and pathology. Proc Jpn Acad Ser B. 2010;86:621–629. doi: 10.2183/pjab.86.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K, Schulz CU, Meyer Zu Brickwedde MK, Pendl GG, Vestweber D. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J Biol Chem. 2000;275:27979–27988. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- Radziwill G, Erdmann RA, Margelisch U, Moelling K. The Bcr kinase downregulates Ras signaling by phosphorylating AF-6 and binding to its PDZ domain. Mol Cell Biol. 2003;23:4663–4672. doi: 10.1128/MCB.23.13.4663-4672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano JM, Beverly LJ, Capobianco AJ. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J Biol Chem. 2003;278:8771–8779. doi: 10.1074/jbc.M211427200. [DOI] [PubMed] [Google Scholar]
- Su L, Hattori M, Moriyama M, Murata N, Harazaki M, Kaibuchi K, Minato N. AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem. 2003;278:15232–15238. doi: 10.1074/jbc.M211888200. [DOI] [PubMed] [Google Scholar]
- Radziwill G, Weiss A, Heinrich J, Baumgartner M, Boisguerin P, Owada K, Moelling K. Regulation of c-Src by binding to the PDZ domain of AF-6. EMBO J. 2007;26:2633–2644. doi: 10.1038/sj.emboj.7601706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock B, Böhme B, Karn T, Yamamoto T, Kaibuchi K, Holtrich U, Holland S, Pawson T, Rübsamen-Waigmann H, Strebhardt K. PDZ-domain-mediated interaction of the Eph-related receptor tyrosine kinase EphB3 and the ras-binding protein AF6 depends on the kinase activity of the receptor. Proc Natl Acad Sci USA. 1998;95:9779–9784. doi: 10.1073/pnas.95.17.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert M, Schneider S, Meskenaite V, Adams MT, Canaani E, Baechi T, Moelling K, Hovens CM. The junction-associated protein AF-6 interacts and clusters with specific Eph receptor tyrosine kinases at specialized sites of cell-cell contact in the brain. J Cell Biol. 1999;144:361–371. doi: 10.1083/jcb.144.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu Y, Yang Y, Huang A, Wu J, Shi Y. Solution structure of AF-6 PDZ domain and its interaction with the C-terminal peptides from Neurexin and Bcr. J Biol Chem. 2005;280:13841–13847. doi: 10.1074/jbc.M411065200. [DOI] [PubMed] [Google Scholar]
- Joshi M, Vargas C, Boisguerin P, Diehl A, Krause G, Schmieder P, Moelling K, Hagen V, Schade M, Oschkinat H. Discovery of low-molecular-weight ligands for the AF6 PDZ domain. Angew Chem Int Ed. 2006;45:3790–3795. doi: 10.1002/anie.200503965. [DOI] [PubMed] [Google Scholar]
- Chen Q, Niu X, Xu Y, Wu J, Shi Y. Solution structure and backbone dynamics of the AF-6 PDZ domain/Bcr peptide complex. Protein Sci. 2007;16:1053–1062. doi: 10.1110/ps.062440607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Park SH, Rho S-H, Lee JH, Kang GB, Sheng M, Kim E, Eom SH. Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization. J Biol Chem. 2003;278:8501–8507. doi: 10.1074/jbc.M212263200. [DOI] [PubMed] [Google Scholar]
- Grembecka J, Cierpicki T, Devedjiev Y, Derewenda U, Kang BS, Bushweller JH, Derewenda ZS. The binding of the PDZ tandem of syntenin to target proteins. Biochemistry. 2006;45:3674–3683. doi: 10.1021/bi052225y. [DOI] [PubMed] [Google Scholar]
- Kusunoki H, Kohno T. Structural insight into the interaction between the p55 PDZ domain and glycophorin C. Biochem Biophys Res Commun. 2007;359:972–978. doi: 10.1016/j.bbrc.2007.05.215. [DOI] [PubMed] [Google Scholar]
- Lee H-J, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Wiedemann U, Boisguerin P, Leben R, Leitner D, Krause G, Moelling K, Volkmer-Engert R, Oschkinat H. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J Mol Biol. 2004;343:703–718. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- Luck K, Charbonnier S, Travé G. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 2012;586:2648–2661. doi: 10.1016/j.febslet.2012.03.056. [DOI] [PubMed] [Google Scholar]
- Im YJ, Lee JH, Park SH, Park SJ, Rho S-H, Kang GB, Kim E, Eom SH. Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization. J Biol Chem. 2003;278:48099–48104. doi: 10.1074/jbc.M306919200. [DOI] [PubMed] [Google Scholar]
- Wu J, Yang Y, Zhang J, Ji P, Du W, Jiang P, Xie D, Huang H, Wu M, Zhang G, Wu J, Shi Y. Domain-swapped dimerization of the second PDZ domain of ZO2 may provide a structural basis for the polymerization of claudins. J Biol Chem. 2007;282:35988–35999. doi: 10.1074/jbc.M703826200. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Lye MF, Anderson JM, Lavie A. Domain swapping within PDZ2 is responsible for dimerization of ZO proteins. J Biol Chem. 2007;282:37710–37716. doi: 10.1074/jbc.M707255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Pan L, Wei Z, Zhao Y, Zhang M. Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites. EMBO J. 2008;27:2113–2123. doi: 10.1038/emboj.2008.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yu J, Jia Y, Pan L, Shen C, Xia J, Zhang M. Redox-regulated lipid membrane binding of the PICK1 PDZ domain. Biochemistry. 2010;49:4432–4439. doi: 10.1021/bi100269t. [DOI] [PubMed] [Google Scholar]
- Momose Y, Honda T, Inagaki M, Shimizu K, Irie K, Nakanishi H, Takai Y. Role of the second immunoglobulin-like loop of nectin in cell-cell adhesion. Biochem Biophys Res Commun. 2002;293:45–49. doi: 10.1016/S0006-291X(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Miyahara M, Nakanishi H, Takahashi K, Satoh-horikawa K, Tachibana K, Takai Y. Interaction of nectin with afadin is necessary for its clustering at cell-cell contact sites but not for its cis dimerization or trans interaction. J Biol Chem. 2000;275:613–618. doi: 10.1074/jbc.275.1.613. [DOI] [PubMed] [Google Scholar]
- Narita H, Yamamoto Y, Suzuki M, Miyazaki N, Yoshida A, Kawai K, Iwasaki K, Nakagawa A, Takai Y, Sakisaka T. Crystal structure of the cis-dimer of nectin-1: implications for the architecture of cell-cell junctions. J Biol Chem. 2011;286:12659–12669. doi: 10.1074/jbc.M110.197368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Vendome J, Brasch J, Jin X, Hong S, Katsamba PS, Ahlsen G, Troyanovsky RB, Troyanovsky SM, Honig B, Shapiro L. Nectin ectodomain structures reveal a canonical adhesive interface. Nat Struct Mol Biol. 2012;19:906–915. doi: 10.1038/nsmb.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita S, Ogita H, Takai Y. Cooperative role of the nectin-nectin and nectin-afadin interactions in formation of nectin-based cell-cell adhesion. J Biol Chem. 2011;286:36297–36303. doi: 10.1074/jbc.M111.261768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda N, Tenno T, Takasu H, Hiroaki H, Shirakawa M. The PRESAT-vector: asymmetric T-vector for high-throughput screening of soluble protein domains for structural proteomics. Protein Sci. 2004;13:652–658. doi: 10.1110/ps.03439004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr Sect D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. Recent developments in classical density modification. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. The buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


