Abstract
UDP-glucose: anthocyanidin 3-O-glucosyltransferase (UGT78K6) from Clitoria ternatea catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. After the acylation of the 3-O-glucosyl residue, the 3′- and 5′-hydroxyl groups of the product are further glucosylated by a glucosyltransferase in the biosynthesis of ternatins, which are anthocyanin pigments. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85 Å, 2.55 Å, 2.70 Å, and 1.75 Å, respectively. The enzyme recognition of unstable anthocyanidin aglycones was initially observed in this structural determination. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The 3-hydroxyl groups of the acceptor substrates were located at hydrogen-bonding distances to the Nε2 atom of the His17 catalytic residue, supporting a role for glucosyl transfer to the 3-hydroxyl groups of anthocyanidins and flavonols. However, the molecular orientations of these three acceptors are different from those of the known flavonoid glycosyltransferases, VvGT1 and UGT78G1. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180° rotation about the O1–O3 axis of the flavonoid backbones observed in VvGT1 and UGT78G1; consequently, the 5- and 7-hydroxyl groups are protected from glucosylation. These substrate recognition schemes are useful to understand the unique reaction mechanism of UGT78K6 for the ternatin biosynthesis, and suggest the potential for controlled synthesis of natural pigments.
Keywords: glucosylation, glucosyltransferase, anthocyanidin, substrate specificity, crystal structure
Introduction
The world market for dyes and organic pigments is anticipated to grow at 3.6% annually and reach 11 million metric tons by 2018.1 Complex dye and pigment compounds are typically challenging to synthesize chemically, but specifically synthesized as secondary metabolites in plants. Recombinant plant enzymes, including mutant forms based on structural information, can contribute to the development of biosynthesis of novel dyes and organic pigments.
Many small lipophilic compounds in living cells are modified by glycosylation, a process that can regulate the chemical property and bioactivity of those compounds, their intracellular localization, and their metabolism.2 One of the most significant and representative glycosylation reactions in plants is the formation of anthocyanin pigments, a class of flavonoid secondary metabolites. Because of their environmental friendly, nontoxic and biodegradable characteristics, these flavonoids recently have received much attention as high performance pigments, such as dyes or inks, and also as antioxidants for applications in functional foods.3–8 Anthocyanins are water-soluble compounds based on a tricyclic flavonoid core that are known to function as pigments involved in determining the color of flowers, leaves, seeds, and fruits.9,10 The chromophoric aglycones of anthocyanins, anthocyanidins, are red polyhydroxylated flavylium salts. They have limited solubility in water, are rapidly destroyed by alkali, and are very unstable compared with their glycosides (anthocyanins).11 Various anthocyanins modified by species-specific glycosylation and acylation have been identified as flower pigments in a number of plants.12,13 The structural diversity of anthocyanins serves to stabilize the flavonoid aglycones and to produce their characteristic color appearing red, purple, or blue upon pH.14
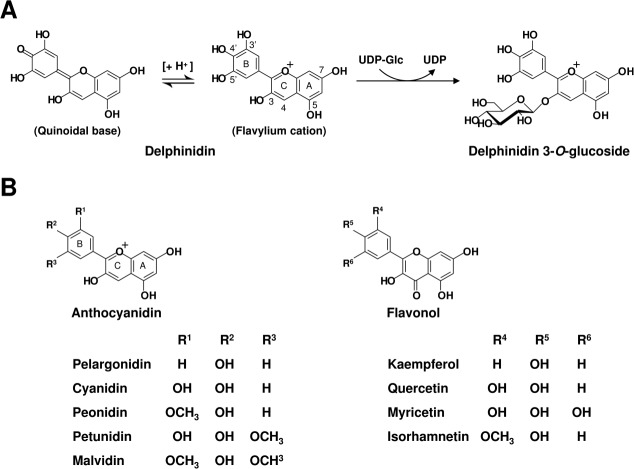
A large group of specific uridine diphosphate glycosyltransferases (UGTs) controls the glycosylation of flavonoids, which is one of the major factors determining the transfer positions of glycosyl groups.15 UGT78K6, previously known as Ct3GT-A, was identified in butterfly pea (Clitoria ternatea) as a UDP-glucose: anthocyanidin 3-O-glucosyltransferase (GenBank accession no. AB185904), which catalyzes glucosyl transfer from UDP-glucose (UDP-Glc) to anthocyanidins such as delphinidin [Fig. 1(A)].16 The glucosylation of delphinidin at the 3-hydroxyl group was proposed as an initial glucosylation step toward the biosynthesis of ternatins, which are blue anthocyanins found in the petals of C. ternatea.17 The putative amino acid sequence of UGT78K6 shows high identity (45%) with that of VvGT1 from a red grape (Vitis vinifera). VvGT1 is a cyanidin 3-O-glycosyltransferase involved in the formation of anthocyanins, with minor activity toward flavonols such as kaempferol.9 The crystal structure of VvGT1, in complex with the acceptor kaempferol and the nontransferable donor UDP-2FGlc, provided the structural basis for understanding its catalytic mechanism and substrate recognition.
Figure 1.
(A) Catalytic reaction by UGT78K6. The glucose moiety is transferred from a UDP-Glc donor to the 3-hydroxyl group of delphinidin. The neutral quinoidal base and the flavylium cation are in equilibrium according to pH. The position numberings, as used in the text, are indicated for the flavylium cation form of delphinidin. (B) Chemical structures of the acceptor substrates examined in this study. Flavonoid backbone structures are composed of two parts: the bicyclic ring system containing the A- and C-rings, and the unicyclic B-ring as indicated for anthocyanidin. The acceptor substrates have a variety of substituents (hydroxyl or methoxyl groups) at the 3′ and 5′ position on the B-ring, in addition to a hydroxyl group at the 4′ position.
We previously determined the three-dimensional structure of recombinant wild-type UGT78K6 and, as expected from its high sequence homology, observed high overall structural similarity with flavonoid UGTs.16 These flavonoid UGTs share the GT-B fold, one of two general folds for the glycosyltransferase superfamily of enzymes, and comprise two N- and C-terminal domains with similar Rossmann-like folds. There is a large cleft between the domains in which the sugar-donor and sugar-acceptor substrates bind.18,19 The His17-Asp114 dyad identified at the catalytic site is widely conserved among the flavonoid UGTs.20 The members also show a common signature motif known as putative secondary plant glycosyltransferase (PSPG) box near the C-terminus that is thought to be involved in binding the UDP moiety of the sugar-donor substrate [Fig. S1, Supporting Information].21
Although the crystal structures of several flavonoid UGTs have been determined,19 the acceptor-substrate complexes are limited to the flavonol-bound forms, VvGT1 in complex with kaempferol or quercetin,9 and UGT78G1 with myricetin,22 presumably because of the instability of anthocyanidins. In solution, anthocyanidins have a positively charged ring system; consequently, deprotonation of the hydroxyl groups is easily facilitated to lead neutral species, quinoidal bases [Fig. 1(A)], which is believed to be of vital importance to flower coloration.23 Structural comparison of UGT78K6 with VvGT1 revealed that several residues employed for binding kaempferol in VvGT1 were not conserved in UGT78K6. The relationship between the primary structures of these enzymes and their acceptor-substrate specificity has not been clarified. Understanding how flavonoid UGTs discriminate between natural plant substrates, including anthocyanidins, flavonols, and isoflavones, will provide key information for flower color modification by genetic engineering and the isolation of novel pigment compounds.
Here we present the crystal structures of UGT78K6 in complex with anthocyanidins, delphinidin, and petunidin, determined to 2.55 and 2.70 Å resolutions, respectively. The flavonol-kaempferol bound form was also determined to 1.85 Å to understand the structural determinants in acceptor-substrate recognition. These structures provide insight into anthocyanidin configurations in enzymes and a different binding scheme for acceptor-substrate recognition as compared with the known flavonoid UGTs.
Results
Glucosylation activity of UGT78K6 against various acceptor substrates
Glucosylation activities were measured using UDP-Glc as a sugar-donor substrate in the presence of various anthocyanidins (cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin) and flavonols (isorhamnetin, kaempferol, myricetin, and quercetin) [Fig. 1(B)]. After normalization of the enzymatic activity of UGT78K6 against delphinidin, the relative activities against other anthocyanidins, malvidin, peonidin, and petunidin, were greater than 70% (Table I). The 3-O-glucosylation of anthocyanidins, particularly delphinidin, was preferably catalyzed by UGT78K6. Significant lower activities (less than 30%) were detected on pelargonidin and cyanidin, suggesting that the different substitution patterns on the B-ring tend to affect activity. Furthermore, UGT78K6 exhibited weak glucosylation activities on a series of flavonols; the relative activities decreased to less than 10% in all cases. These results are consistent with the previous observation that a cyanidin 3-O-glycosyltransferase from V. vinifera showed weak glucosylation activities against flavonols, which have an additional 4-carbonyl group on the flavanol ring system.9
Table I.
Glucosylation Activities for Various Acceptor Substrates
| Specific activity (nkat/mg protein) | Relative activity (%) | |
|---|---|---|
| Anthocyanidin | ||
| Delphinidin | 122.9 | 100 |
| Malvidin | 107.1 | 87 |
| Peonidin | 94.6 | 77 |
| Petunidin | 89.0 | 72 |
| Pelargonidin | 35.7 | 29 |
| Cyanidin | 21.1 | 17 |
| Flavonol | ||
| Isorhamnetin | 10.1 | 8.2 |
| Quercetin | 6.6 | 5.4 |
| Kaempferol | 5.5 | 4.5 |
| Myricetin | 3.0 | 2.4 |
Structure of UGT78K6 with and without the donor substrate
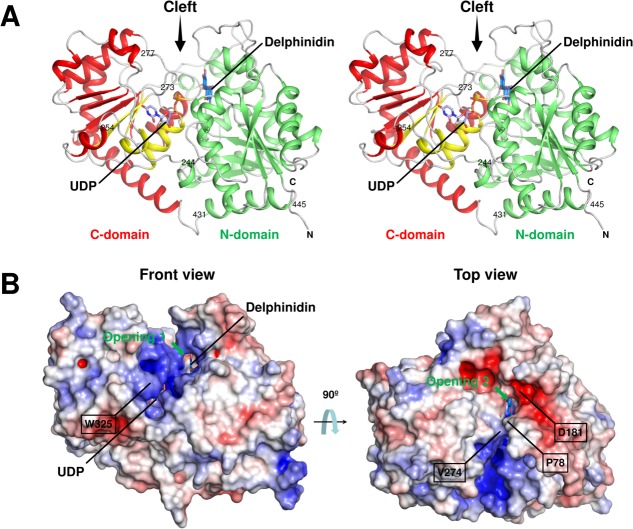
The crystal structures of UGT78K6 in complex with and without the UDP-Glc donor were both determined to a resolution of 1.85 Å. The overall structure shows a typical GT-B fold comprising two Rossmann-like β/α/β domains [Fig. 2(A)], which is conserved in the known flavonoid UGTs.20 The N-terminal β/α/β domain (N-domain) comprising residues 1–244 consists of a central seven-stranded, twisted parallel β-sheet surrounded by eight helices. The C-terminal β/α/β domain (C-domain) is composed of a twisted β-sheet with six strands accompanied by ten helices on its two sides. The N- and C-domains are connected by a flexible loop comprising residues 245–253. There is a cleft between these domains, and its internal space can be largely divided into two cavities that are used as a donor-binding and an acceptor-binding sites [Fig. 2(B)]. The C-terminal helix containing residues 431–445 participates in forming the N-domain after crossing the cleft [Fig. 2(A)].
Figure 2.
(A) Stereo view of the UDP-bound form of UGT78K6. The N-domain (green) and the C-domain (red) are shown with the secondary structures. The PSPG motif comprising residues 325–368 is in yellow. The UDP molecule (light blue carbon) observed in this study is shown as a stick model. The delphinidin molecule was modeled at the acceptor-binding site of the UDP-bound form using the coordinates of the structure in complex with delphinidin (marine blue carbon). Amino acid residues discussed in the text are indicated. (An interactive view is available in the electronic version of the article.) (B) Electrostatic surface potential of UGT78K6. The electrostatic potentials of the protein surface were calculated using the APBS program24 and colored by electrostatic potential isocontours from the potential of +5 kT e−1 (blue) to −5 kT e−1 (red). Front view, same direction as Figure 2(A); top view, rotated ∼90° around the horizontal axis.
The donor-substrate-complex form of UGT78K6 was prepared by soaking the ligand-free crystals in the reservoir solution containing UDP-Glc, as described in Materials and Methods section. The crystal structure was solved to a resolution of 1.85 Å; however, only electron density corresponding to the UDP moiety of UDP-Glc was observed at the donor-binding site [Fig. 3(A)]. Instead of the missing sugar part of UDP-Glc, the residual electron density located adjacent to the UDP moiety was assigned to a glycerol molecule (GOL1002). Similar loss of electron density for the sugar moiety was reported in the UGT71G1 structure in the presence of UDP-galactose and the UGT78G1 structure using UDP-Glc,22,25 which was attribute to enzymatic hydrolysis of the linkage between the β-phosphate and the sugar. The overall structure of the UDP-bound form is nearly identical to that of the unliganded form of UGT78K6, with a root-mean-square deviation (rmsd) of 0.36 Å for all Cα atoms.
Figure 3.
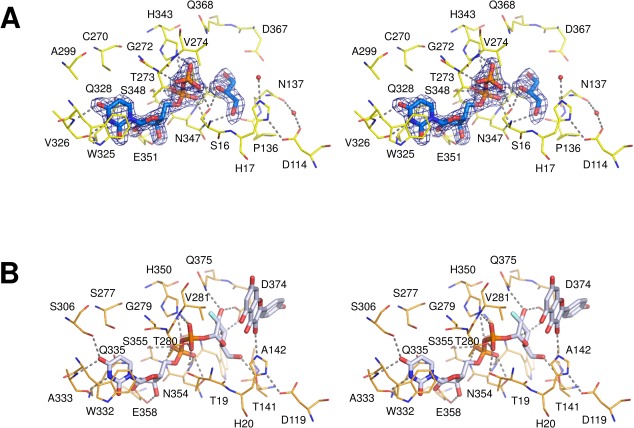
(A) Close-up view of the donor-binding site in UGT78K6. The UDP moiety and a glycerol molecule are shown as stick models. Residues involved in the UDP-binding are shown as yellow lines and labeled. Hydrogen bonds are depicted with dotted lines. The Fo–Fc omit electron density map around the UDP and glycerol molecules are represented as blue meshes, countered at 3.5 σ. (An interactive view is available in the electronic version of the article). (B) Close-up view of the donor-binding site in VvGT1 (PDB ID: 2C1Z). The structure was determined as a complex with the acceptor kaempferol and the nontransferable donor UDP-2FGlc.
The protein surface around the donor-binding site is rich in positive charges [Fig. 2(B)], which would be suitable to attract the anionic diphosphate of UDP-Glc. The donor-binding site is connected to the acceptor-binding site in the protein interior. The acceptor-binding site can be accessed from the solvent through two openings, termed 1 and 2, which are separated by the hydrophobic side-chains of Pro78 from the N-domain and Val274 from the loop region (residues 273–277) in the C-domain. Opening 1, located near the donor-binding site, is surrounded by the relatively positively charged surface. Opening 2 is formed at the negatively charged surface, as shown in Figure 2(B), which may contribute to select the cationic character of the flavylium ring of anthocyanidins. Indeed, an acidic residue, Asp181, located at a polar edge on the lip of opening 2 is involved in the acceptor binding, as described below.
The UDP moiety bound at the donor-binding site shows several specific interactions with the residues in the UGT signature PSPG motif (residues 325–368) of the C-domain [Fig. 3(A)]. The uracil ring of UDP forms a π-stacking interaction with the indole ring of Trp325, which was rotated nearly 180° around the χ2, relative to that observed in the unliganded form of UGT78K6 [Fig. S2, Supporting Information]. The 2′- and 3′-hydroxyl groups of the ribose moiety form hydrogen-bonding interactions with the side-chain carbonyl oxygens (Oε1 and Oε2) of Glu351, which resulted in the rotation of the Asn347 side-chain to form a new hydrogen bond between the Nδ2 atom of Asn347 and the α-phosphate of UDP. Furthermore, the diphosphate part of UDP shows hydrogen bonds with the side-chains of surrounding Ser16, His343, Ser348, and Thr273. This UDP recognition scheme is consistent with previous observations for the flavonoid UGTs complexed with sugar donors.9,22,25 The average temperature factor for the flexible loop (residues from Thr273 to Pro277) was 51.5 Å2 in the unliganded form of UGT78K6; however, it was significantly reduced to 22.7 Å2 after binding of residue Thr273 (the main-chain amide and the side-chain Oγ1 atom) to the β-phosphate group of UDP.
Structures of UGT78K6 in complex with the acceptor substrates
Crystal structures of ligand complexes with different acceptor substrates, delphinidin, petunidin, and kaempferol, were determined to resolutions of 2.55, 2.70, and 1.75 Å, respectively. When the structures were compared with the unliganded form of UGT78K6, the rmsds for all Cα atoms were 0.27, 0.35, and 0.22 Å for the delphinidin, petunidin, and kaempferol complexes, respectively.
The electron density of the delphinidin molecule was clearly visible at the acceptor-binding site [Fig. 4(A)]. The binding orientation was determined based on the electron density of the 4′-hydroxyl oxygen atom on the B-ring. If the acceptor was positioned in an opposite orientation, some uninterpretable electron densities appeared. At the acceptor-binding site, the 4′-hydroxyl group of delphinidin is in hydrogen-bonding distance to the main-chain carbonyl oxygen of Pro78. The 3-, 5-, and 7-hydroxyls of the bicyclic ring form hydrogen bonds with the Nε2 atom of His17, and the side-chain carbonyl oxygens of Asp367 and Asp181, respectively. Some water-mediated hydrogen bonds were also observed at the 5′- and 7-hydroxyls of delphinidin. The rest of the flavylium ring is surrounded by three phenylalanine side-chains from Phe116, Phe192, and Phe365, and hydrophobic side-chains from Pro179 and Leu196 [Fig. S3, Supporting Information]. The B-ring of delphinidin is slightly rotated (9°) from the plane of the bicyclic ring, corresponding to the existence of a flavylium cation form at the acceptor-binding site [Fig. 1(A)].
Figure 4.
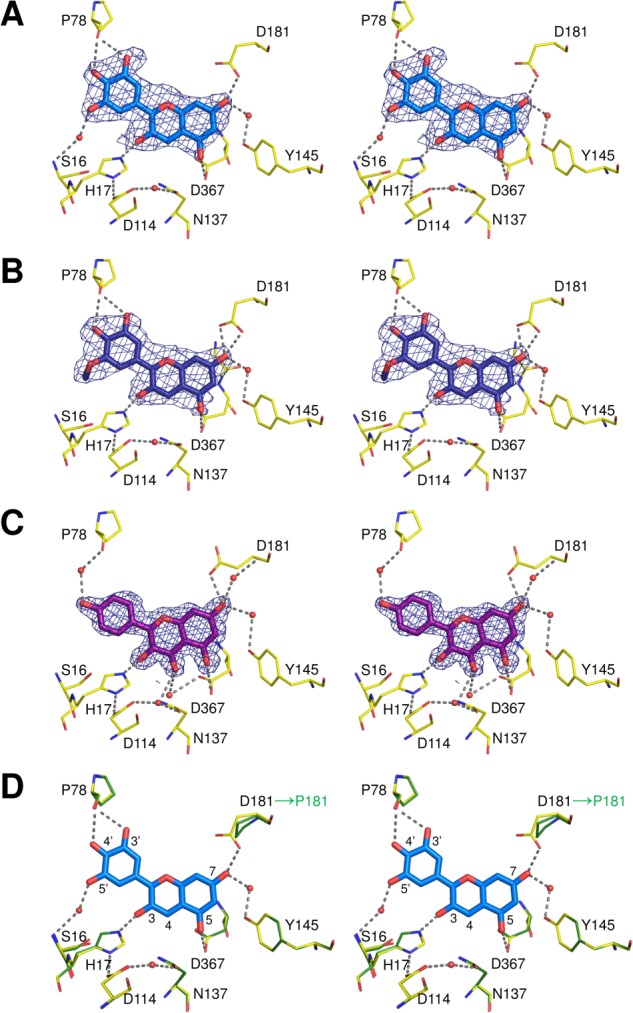
Stereo views of the acceptor-binding sites in the complexed structures with (A) delphinidin, (B) petunidin, and (C) kaempferol. Residues interacting with the acceptor substrates are shown as yellow lines and labeled. Hydrogen bonds are depicted with dotted lines. The Fo–Fc omit electron density maps around the acceptor substrates are represented as blue meshes, countered at 3.0, 2.5, and 3.5 σ, respectively. (An interactive view is available in the electronic version of the article). (D) Superimposition of the delphinidin-bound form of UGT78K6 (yellow lines) and the modeled structure of UGT78K8 (green lines). The UGT78K8 model was generated by the SWISS-MODEL server26 using the delphinidin-bound form of UGT78K6 as a template. The delphinidin molecule is shown as a stick model. Residues involved in the binding of delphinidin are labeled. Hydrogen bonds are depicted with dotted lines. Although the regioselectivities for glucosylation of the acceptors are different, most of the residues involved in acceptor recognition are conserved except for Asp181 of UGT78K6, which is replaced by the nonhydrogen-bonding residue, Pro181, in UGT78K8.
The petunidin-bound form shows that the 5′-methoxy group on the B-ring occupies a position exposed to the bulk solvent, as shown in Figure 4(B). The interaction at the acceptor-binding site is very similar to that of delphinidin. The 3′- and 4′-hydroxyl groups of petunidin are in hydrogen-bonding distance to the main-chain carbonyl oxygen of Pro78. The 3-, 5-, and 7-hydroxyls of the bicyclic ring form hydrogen-bonding interactions with the Nε2 atom of His17, and the side-chain carbonyl oxygens of Asp367 and Asp181, respectively. The B-ring of petunidin is also rotated (12°) from the plane of the bicyclic ring, indicating the existence of a flavylium cation form at the acceptor-binding site [Fig. 1(A)].
The kaempferol molecule is situated in the acceptor-binding site in a manner similar to that seen in the complex structures with delphinidin and petunidin. The 3-hydroxyl group to be glucosylated forms a hydrogen bond to the catalytic residue His17 with a distance of 2.5 Å [Fig. 4(C)]. The 4-carbonyl group on the bicyclic ring makes contact with the Nδ2 atom of Asn137 at a hydrogen-bonding distance of 2.9 Å. Therefore, compared with delphinidin, the flavonol backbone is rotated ∼10° around the C3 carbon atom. Because of the rotation of molecular orientation, the 4′-hydroxyl group on the B-ring no longer interacts with the polypeptide directly. The side-chain carbonyl oxygen atoms of Asp181 and Asp367 are located within hydrogen-bonding distances to the 5- and 7-hydroxyls of kaempferol; however, the side-chain of Asp181 is exposed to a bulk solvent with a different conformation from those in the anthocyanidin complexes. The B- and bicyclic rings of kaempferol are not coplanar (deviating by a 13° angle).
From these observations, it was revealed that UGT78K6 can similarly accommodate the respective acceptor substrates, delphinidin, petunidin, or kaempferol, at its acceptor-binding site. The enzyme primarily interacts with the hydroxyl groups on the bicyclic ring of the acceptors by forming several hydrogen bonds but does not strongly influence on the B-rings, which are located near the opening exposed to the solvent region. Thus, it appears that the different substitution patterns on the B-ring are not recognized positively by this enzyme, although the glucosylation activities against each acceptor were significantly different (Table I).
Discussion
We have identified and characterized three anthocyanidin/anthocyanin UGTs from C. ternatea (UGT78K6, previously called Ct3GT-A; UGT78K7, Ct3GT-B: AB185905 with 92% identity; UGT78K8, Ct3′5′GT: AB115560 with 87% identity) involved in the glucosylation of delphinidin/delphinidin glucoside for the biosynthesis of ternatins [Fig. S1, Supporting Information]. Although the enzymes show high overall sequence identities, UGT78K6 and UGT78K7 catalyze the glucosyl transfer from UDP-Glc to anthocyanidins such as delphinidin (data not shown), and UGT78K8 glucosylates the 3′- and 5′-hydroxyl groups of delphinidin 3-O-(6″-O-malonyl-β-glucoside), which is the later step of the 3-O-glucosylation of delphinidin in the ternatin biosynthesis.27 Of these three enzymes, we succeeded in determining the tertiary structure of UGT78K6 to a resolution of 1.85 Å.16 Enzyme recognition of unstable aglycones, anthocyanidins, in the flavonoid UGTs was initially revealed by this structural analysis of UGT78K6. In describing the structural features of UGT78K6, two important aspects should be noted; one is the structure of the donor-binding site, which is commonly seen in the flavonoid UGTs; the other is the structure of the acceptor-binding site, which recognizes various acceptor substrates to be glycosylated by the UGTs.
Structure of the donor-binding site and the catalytic function of UGT78K6
The structure of the donor-binding site in UGT78K6 was clarified in complex with UDP to a resolution of 1.85 Å resolution, and the structure was compared with that of an enzyme with similar enzymatic function. From the structural comparison obtained using the Dali server,28 we chose the flavonoid UGT VvGT1 from V. vinifera (PDB ID: 2C1Z, rmsd: 1.9 Å for 432 Cα atoms, Dali Z-score: 49.9). When the conserved residues in the PSPG motif of UGT78K6 were overlaid onto those of VvGT1, it was found that the sugar moiety of UDP-2FGlc in VvGT1 can occupy the corresponding position at the donor-binding site of UGT78K6 [Figs. 3(B) and 5(A)]. The Asp374 and Gln375 side-chains interacting with the glucose moiety in VvGT1 are conserved as Asp367 and Gln368 in UGT78K6; however, Thr141 in VvGT1 is replaced by the nonhydrogen bonding residue Pro136. The side-chain of Asn137 in UGT78K6 could participate in the recognition of the glucose moiety instead of Thr141 in VvGT1 because the Asn137 side-chain of UGT78K6 can be located sufficiently close to the 6-hydroxymethyl group of UDP-2FGlc in VvGT1. Therefore, it is considered that the glucose moiety of UDP-Glc would be located at essentially the same position of UDP-2FGlc as that observed for VvGT1.
Figure 5.
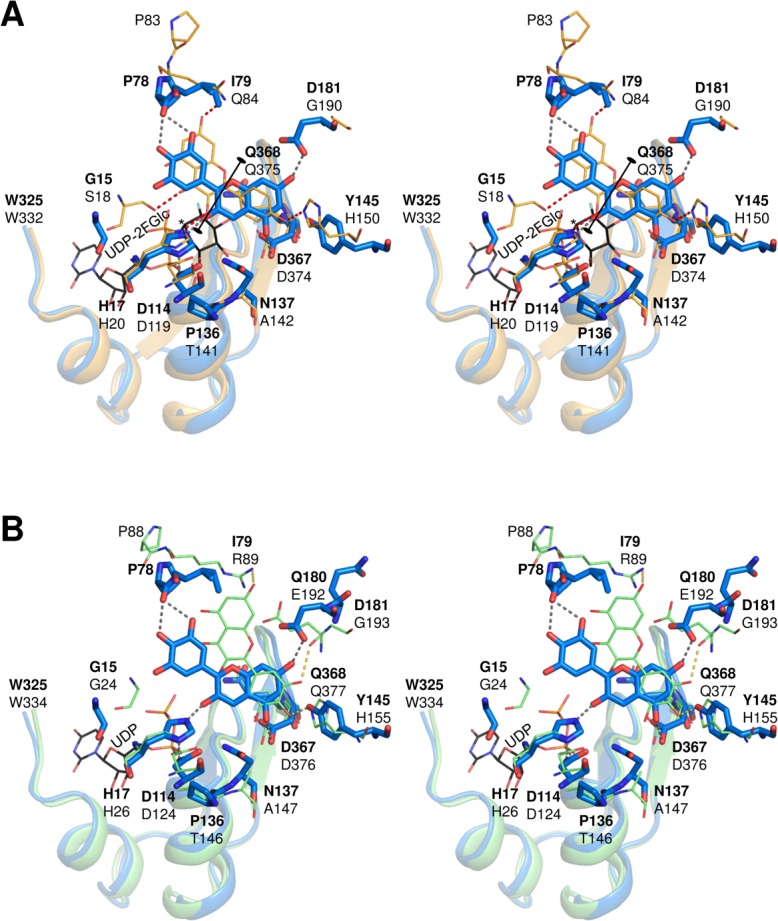
Comparison of the acceptor-binding schemes. (A) The UGT78K6 structure in complex with delphinidin (blue sticks labeled with bold letters) was superimposed with the VvGT1 structure bound with kaempferol (orange lines labeled with black letters) and UDP-2FGlc (black lines labeled with black letter) based on the conserved UDP-binding motifs (residues 325–367 in UGT78K6 and residues 332–374 in VvGT1). Hydrogen bonds are depicted with dotted lines. The anomeric C1 carbon to be attacked by a nucleophile is indicated by an asterisk. (B) Superimposition of the UGT78K6 structure in complex with delphinidin (blue sticks labeled with bold letters) and UGT78G1 bound with myricetin (green lines labeled with black letters) and UDP (black lines labeled with black letter) based on the conserved UDP-binding motifs (residues 325–367 in UGT78K6 and residues 334–376 in UGT78G1).
Furthermore, it was found that the location of the catalytic residues (His17 and Asp114) were essentially the same in each structure. Since the hydrogen bond between His17 and Asp114 is indispensable for effective deprotonation of His17 through the charge relay system with the negatively charged Asp114,20 the glucosylation by UGT78K6 is likely to follow a typical inverting mechanism shared among flavonoid UGTs [Fig. 6]. The distance between the Nε2 atom of His17 and the anomeric C1 carbon of glucose is 5.2 Å in UGT78K6, which is comparable to the distance 5.1 Å in the VvGT1 complex structure [Fig. 5(A)]. His17 would serve as a general base that deprotonates the 3-hydroxyl group of acceptor substrates, and then the anomeric C1 carbon on the glucose is attacked by the nucleophilic oxyanion.21 In the transition state, the negative charge on the leaving phosphate may be stabilized by the positively charged residue, His343 (hydrogen bonded to the β-phosphate group of UDP with a distance of 2.9 Å) in UGT78K6, followed by displacement of the UDP moiety.
Figure 6.
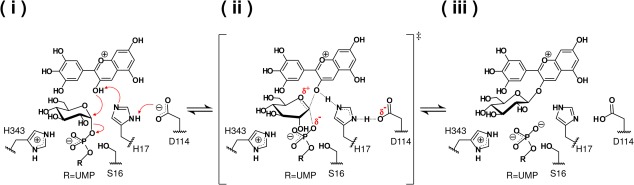
Proposed catalytic mechanism for the conversion of delphinidin to delphinidin 3-O-glucoside. (i) The His17-Asp114 catalytic dyad serves as a general base that deprotonates the nucleophile hydroxyl group of the acceptor substrate. (ii) The generated negative charge on the departing phosphate can be stabilized by the positive amino acid, His343, or the hydroxyl group of Ser16 which is adjacent to both the β-phosphate of UDP and the O5 oxygen of glucose. (iii) The resulting glucoside product and UDP are released from the active site.
Acceptor-binding site in complex with various acceptor substrates
In the structure of UGT78K6 in complex with delphinidin, the 3′-, 4′-, 5-, and 7-hydroxyl groups of delphinidin are recognized by the proton acceptors, the main-chain carbonyl of Pro78, and the aspartate side-chains of Asp181 and Asp367, respectively. Because the crystals were prepared under mild acidic conditions (pH 5.6), the hydrogen bonds with the aspartate residues at the 5- and 7-hydroxyls imply that the delphinidin molecule has a flavylium cation form, which is in equilibrium with neutral quinoidal base forms in solution [Fig. 1(A)].23,29 The quinoidal base forms usually exist as a mixture, since the pKa values of the 4′-, 5-, and 7-hydroxyl group of anthocyanidins are very similar.30 The resolution of the structure determination may not be sufficient to discuss the keto-enol tautomers of anthocyanidins in detail; however, such transformations at the hydroxyl groups of the acceptor substrates appear to affect the glucosylation activity of UGT78K6. The enzyme showed significantly lower glucosylation activities against pelargonidin and cyanidin compared with delphinidin, as shown in Table I. These two acceptor substrates have a higher acidity at the 5- or 7-hydroxyl group than the 4′-hydroxyl group; consequently, they tend to adopt the neutral 5- or 7-quinoidal base form under the experimental conditions,31,32 which would be unfavorable for interaction with the deprotonated aspartate residues of Asp181 and Asp367 at the acceptor-binding site of UGT78K6. Thus, the enzyme can potentially discriminate between the flavylium cation and the neutral quinoidal forms of anthocyanidins in solution.
In terms of biosynthesis, the anthocyanidins are divided into only three types; delphinidin, pelargonidin, and cyanidin.33 Delphinidin and its derivatives, petunidin and malvidin, generally tend to show blue/violet color under physiological condition.34 If the cyanidin derivative, peonidin, become expressed in the petal of C. ternatea by genetic engineering, the red/purple color pigment could be accumulated due to the activity of UGT78K6. To synthesize new pigments based on pelargonidin, which is a source of orange/red color, the mutations at Asp181 and Asp367 of UGT78K6 would be required for the effective production.
Comparison of the acceptor recognition with the known flavonoid UGTs
The UGT78K6 structures showed that both anthocyanidin and flavonol substrates bind to the acceptor-binding site in the same manner, although the glucosylation activities against flavonols were significantly low (Table I), supporting that the activities depend on the deprotonation efficiency of the hydroxyl group to be glucosylated.9 On the other hand, the superposition of the UGT78K6 structure on the VvGT1 (with the closest sequence similarity to UGT78K6) shows that the acceptor-substrate recognition scheme of UGT78K6 is quite different from that of VvGT1.9 In the scheme of VvGT1, the flavonol kaempferol lies at the acceptor-binding site in a reverse direction from that seen in UGT78K6, by rotating 180° about the O1-O3 axis of the acceptor substrate [Fig. 5(A)]. At the binding site, the 4′-hydroxyl group of the B-ring and the 7-hydroxyl group of the flavonol backbone form hydrogen bonds with the imidazole ring of His150 and the side-chain amide of Gln84, respectively. The 3-hydroxyl group to be glycosylated is located near the catalytic His20. This binding orientation was also observed in the UGT78G1 structure in complex with myricetin (PDB ID: 3HBF),22 in which two basic side-chains of Arg89 and His155, and the main-chain carbonyl of Glu192 form hydrogen bonds with the hydroxyls of the acceptor [Fig. 5(B)]. In both VvGT1 and UGT78G1, the three important residues, Gln84/Arg89, His150/His155, and Gly190/Gly193, are substituted to Ile79, Tyr145, and Asp181 in UGT78K6, respectively. Thus, the rotated binding mode for the acceptors observed in UGT78K6 is presumably caused by the absence of direct hydrogen-bonding interaction with the substrate because of substitution of Gln84 to Ile79, by the positional shift in the acceptor due to the His145 to Tyr145 substitution, and by the formation of a strong hydrogen-bonding interaction between the acceptor and UGT78K6 by the substitution of Gly190 to Asp181.
Acceptor-binding sites of the enzymes involved in ternatin biosynthesis
It is known that the ternatin biosynthesis in C. ternatea requires at least three flavonoid UGTs, UGT78K6, UGT78K7, and UGT78K8. Comparing the residues involved in acceptor recognition in UGT78K6, UGT78K7, and UGT78K8 based on structural modeling, the active site structure of UGT78K7 was essentially the same as that of UGT78K6 [Fig. S1, Supporting Information]. Within the same plant, UGT78K7 may be a homolog of UGT78K6. In UGT78K8, most of the residues forming the acceptor-binding site were conserved; however, Asp181 of UGT78K6 was replaced by Pro181 [Fig. 4(D)]. This substitution suggests that the residue Asp181 is a key candidate for determining the binding orientation at the acceptor-binding site, and plays an important role in the differentiation of the regioselective glucosylations by UGT78K6 and UGT78K8. It is further supported by the report that the site-directed mutagenesis at the acceptor-binding site close to Asp181 (Phe148 to Val or Tyr 202 to Ala in UGT71G1) resulted in a drastic change of regioselectivity for quercetin glycosylation.35
Various anthocyanins modified by species-specific glycosylation and acylation have been found as flower pigments in a number of plants.12 The structural diversity of anthocyanins serves not only to stabilize the flavonoid aglycones but also to produce their characteristic coloring, which depends on the copigmentation phenomenon, the complexation with metals, and the pH values under physiological conditions.36 Ternatins isolated from C. ternatea are anthocyanins, all of which are delphinidin derivatives that are glucosylated regioselectively at the 3-, 3′-, and 5′-hydroxyls of the delphinidin aglycone.27,37,38 This study indicates that the 5- or 7-hydroxyl group of delphinidin cannot approach to the catalytic His17 at the acceptor-binding site and that delphinidin 5-O-glucoside or 7-O-glucoside would not be accommodated as an acceptor substrate because of the large substituent. Consequently, the enzyme may contribute to the determination of the positions to which glucosyl residues are transferred, and the unique glycosides are predominantly produced through the ternatin biosynthetic pathway of C. ternatea.
Materials and Methods
Protein expression and purification
The gene encoding UGT78K6 (GenBank accession no. AB185904) was PCR-amplified using the sense primer 5′-GACGACGACAAGATGAAAAACAAGCAG CATGTTGC-3′ and the antisense primer 5′-GAGGAGAAGCCCGGTTTAGCTAGAGGAAATCACT TC-3′, and the obtained product was ligated into a pET-30 Ek/LIC vector (Novagen).16 From the resultant plasmid, the cDNA fragment with an enterokinase cleavage site was isolated by digestion with BglII and XhoI and subcloned into the BamHI/SalI digested pQE31 vector (Qiagen). The recombinant protein was over-expressed in Escherichia coli XL1-Blue cells (Stratagene) by inducing with 1 mM isopropyl-β-D-galactoside for 20 h at 298 K. The cells were harvested by centrifugation and resuspended in a buffer containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 20 mM imidazole, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride. After disrupting the cells by sonication, the cell debris was removed by centrifugation, and the supernatant was applied to a Ni-Sepharose column (GE Healthcare). The eluted fraction containing UGT78K6 was dialyzed against 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, and 2 mM CaCl2. The N-terminal His-tag was removed by digestion using recombinant enterokinase (Novagen). Then, cation exchange chromatography was performed on an SP-5PW column (Tohso, Japan) to purify the enzyme to homogeneity.
Glucosylation activity
To examine the acceptor-substrate specificity of UGT78K6, six types of anthocyanidins (cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin chloride) were purchased from Funakoshi (Tokyo, Japan). Four types of flavonols (isorhamnetin, kaempferol, myricetin, and quercetin) and UDP-Glc were purchased from Wako Pure Chemicals (Kyoto, Japan).
The enzymatic reactions were carried out by adding 2 µg of the purified protein to a 20 µL reaction mixture containing 0.1M potassium phosphate (pH 7.4), 0.4 mM of each acceptor substrate and 1 mM UDP-Glc as a sugar-donor substrate. To determine the specific activity expressed in nkat (the amount of enzyme catalyzing the glucosylation of 1 nmol of substrate per second), the reaction mixtures were incubated for 30, 60, and 90 s at 303 K. The reactions were then terminated by adding 4 µL of 1M HCl solution for anthocyanidins or 1M HCl in 50% methanol for flavonols. After addition of an equal volume (24 μL) of 0.05M trifluoroacetic acid in 5% acetonitrile for anthocyanidins or 0.05M trifluoroacetic acid in 80% acetonitrile for flavonols, 5 μL aliquots of each reaction mixture were analyzed by reverse-phase HPLC equipped with a Develosil C30-UG5 column (Nomura Chemical, Japan).
Crystallization and structure determination
The recombinant wild-type UGT78K6 was crystallized by the hanging-drop vapor-diffusion method as reported previously.16 After mixing equal volumes of the protein solution (20 mg mL−1) and the reservoir solution containing 0.1M sodium citrate tribasic dihydrate (pH 5.6), 0.2M ammonium acetate, and 26% (w/v) polyethylene glycol 4,000, the mixture was equilibrated against the reservoir solution at 293 K. After growing the ligand-free crystals up to 0.05 × 0.05 × 0.5 mm in size, the acceptor substrates (delphinidin, petunidin, and kaempferol) in dimethyl sulfoxide or UDP-Glc in the reservoir solution were added to each drop up to concentrations of 0.5 or 20 mM. The crystals were then soaked in a cryoprotectant solution containing 25% (v/v) glycerol in addition to the reservoir. X-ray diffraction data were collected under a stream of gaseous nitrogen at 100 K at beamline BL6A at the Photon Factory (Tsukuba, Japan) or at beamline BL38B1 at the SPring-8 facility (Harima, Japan). All data sets were processed and scaled using the HKL2000 software package.39 The crystal of the unliganded form of UGT78K6 belongs to the space group P21 with cell dimensions of a = 50.2 Å, b = 55.2 Å, c = 86.2 Å, and β = 105.1°. The substrate-bound crystals were sufficiently isomorphous with the unliganded form; therefore, its coordinates (PDB ID: 3WC4) were used as a starting model for each refinement with the PHENIX software.40 After several rounds of iterative manual rebuilding of the protein coordinates, each ligand was constructed into 2Fo–Fc and Fo–Fc electron density maps. Model building and water picking were conducted using Coot41 and verified manually. PROCHECK42 was employed to evaluate the final molecular models. The statistics for data collection and structure refinement are summarized in Table II. All graphic images of molecular structure were generated using the PyMOL molecular visualization system.43
Table II.
Data Collection and Refinement Statistics of the UGT78K6 Structures in Complex with UDP, Delphinidin, Petunidin, and Kaempferol
| Unliganded | UDP | Delphinidin | Petunidin | Kaempferol | |
|---|---|---|---|---|---|
| Data collectiona | |||||
| Space group | P21 | ||||
| Cell dimensions | |||||
| a, b, c (Å) | 50.2, 55.2, 86.2 | 50.1, 55.3, 85.5 | 50.0, 55.1, 86.0 | 49.9, 55.3, 85.8 | 50.0, 55.1, 85.9 |
| β (°) | 105.1 | 105.0 | 104.6 | 104.6 | 104.92 |
| Wavelength (Å) | 0.978 | 1.000 | 0.978 | 0.978 | 1.000 |
| Resolution (Å) | 1.85 (1.92–1.85) | 1.85 (1.92–1.85) | 2.55 (2.64–2.55) | 2.70 (2.8–2.7) | 1.75 (1.81–1.75) |
| Observed reflections | 139,758 | 142,408 | 53,965 | 46,602 | 165,969 |
| Rmerge | 9.9 (42.2) | 8.3 (37.4) | 9.9 (32.3) | 9.9 (40.2) | 6.0 (36.7) |
| I/σ(I) | 24.7 (4.6) | 15.2 (2.1) | 20.9 (6.3) | 16.2 (4.0) | 21.5 (2.0) |
| Completeness (%) | 99.1 (99.2) | 99.6 (97.5) | 99.5 (99.9) | 98.9 (98.4) | 99.7 (97.8) |
| Redundancy | 3.6 | 3.7 | 3.6 | 3.7 | 3.7 |
| Refinement | |||||
| Unique reflections | 39,179 | 38,545 | 14,854 | 12,437 | 45,271 |
| Rwork/Rfree | 0.170/0.211 | 0.151/0.187 | 0.181/0.234 | 0.182/0.241 | 0.159/0.195 |
| Number of atoms | |||||
| Protein | 3,436 | 3,437 | 3,437 | 3,437 | 3,437 |
| Water/Others | 436/16 | 486/41 | 127/40 | 75/47 | 408/47 |
| B-factors | |||||
| Protein | 18.9 | 15.1 | 32.4 | 32.6 | 18.3 |
| Water/Others | 27.1/42.7 | 25.7/22.5 | 30.2/41.7 | 26.2/34.3 | 27.9/34.0 |
| Rmsds | |||||
| Bond lengths (Å) | 0.013 | 0.006 | 0.003 | 0.003 | 0.006 |
| Bond angles (º) | 1.5 | 1.1 | 0.7 | 0.7 | 1.1 |
| Ramachandran plot (%) | |||||
| Favored/Allowed | 98.4/1.6 | 97.1/2.9 | 96.4/3.4 | 96.0/3.8 | 96.9/2.9 |
| Outliers | 0.0 | 0.0 | 0.2 | 0.2 | 0.2 |
| PDB ID | 3WC4 | 4WHM | 4REM | 4REN | 4REL |
Numbers in parentheses represent statistics in highest resolution shell.
Accession numbers
The atomic coordinates of each substrate-bound forms of UGT78K6 have been deposited in the RCSB Protein Data Bank (PDB), codes 4WHM (UDP bound), 4REM (delphinidin bound), 4REN (petunidin bound), and 4REL (kaempferol bound).
Acknowledgments
The authors thank the beam-line staff at the Photon Factory (proposal no. 2009G033) and the SPring-8 (proposal no. 2009A1557) synchrotron facility for their support on data collection. They are also gratefully acknowledge Professor P. I. Mackenzie (Flinders University, Australia) for the nomenclature of the UGT numbers described in the text. None of the authors have any conflict of interest to declare.
Glossary
Abbreviations:
- PSPG
putative secondary plant glycosyltransferase
- rmsd
root-mean-square deviation
- UDP-Glc
uridine diphosphate glucose
- UGT
UDP glycosyltransferase.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- Study #2508. World dyes & organic pigments: industry study with forecasts for 2013 & 2018. Cleveland, OH: Freedonia Group; 2009. [Google Scholar]
- Lim EK, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004;23:2915–2922. doi: 10.1038/sj.emboj.7600295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti MM, Wrolstad RE. Acylated anthocyanins from edible sources and their applications in food systems. Biochem Eng J. 2003;14:217–225. [Google Scholar]
- Del Pozo-Insfran D, Brenes CH, Talcott ST. Phytochemical composition and pigment stability of Açai (Euterpe oleracea Mart.) J Agric Food Chem. 2004;52:1539–1545. doi: 10.1021/jf035189n. [DOI] [PubMed] [Google Scholar]
- Garzón GA, Wrolstad RE. Major anthocyanins and antioxidant activity of Nasturtium flowers (Tropaeolum majus. Food Chem. 2009;114:44–49. [Google Scholar]
- Zyoud A, Zaatar N, Saadeddin I, Helal MH, Campet G, Hakim M, Park DH, Hilal HS. Alternative natural dyes in water purification: anthocyanin as TiO2-sensitizer in methyl orange photo-degradation. Solid State Sci. 2011;13:1268–1275. [Google Scholar]
- Buraidah MH, Teo LP, Yusuf SNF, Noor MM, Kufian MZ, Careem MA, Majid SR, Taha RM, Arof AK. TiO2/chitosan-NH4I(+I2)-BMII-based dye-sensitized solar cells with anthocyanin dyes extracted from black rice and red cabbage. 2011. pp. 1–11. Int J Photoenergy 2011 Article ID 273683:
- Ozuomba JO, Okoli LU, Ekpunobi AJ. The performance and stability of anthocyanin local dye as a photosensitizer for DSSCs. Adv Appl Sci Res. 2013;4:60–69. [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Dao LT, Takeoka GR, Edwards RH, Berrios JDJ. Improved method for the stabilization of anthocyanidins. J Agric Food Chem. 1998;46:3564–3569. [Google Scholar]
- Yoshida K, Mori M, Kondo T. Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat Prod Rep. 2009;26:884–915. doi: 10.1039/b800165k. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F, Kalc G, Senior M, Dyson B, Nakamura N, Katsumoto Y, Chandler S. Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives. Biosci Biotechnol Biochem. 2010;74:1760–1769. doi: 10.1271/bbb.100358. [DOI] [PubMed] [Google Scholar]
- He F, Liang NN, Mu L, Pan QH, Wang J, Reeves MJ, Duan CQ. Anthocyanins and their variation in red wines I. Monomeric anthocyanins and their color expression. Molecules. 2012;17:1571–1601. doi: 10.3390/molecules17021571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X. Crystal structure of Medicago truncatula UGT85H2–insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase. J Mol Biol. 2007;370:951–963. doi: 10.1016/j.jmb.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Hiromoto T, Honjo E, Tamada T, Noda N, Kazuma K, Suzuki M, Kuroki R. Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. J Synchrotron Radiat. 2013;20:894–898. doi: 10.1107/S0909049513020712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuma K, Kogawa K, Noda N, Kato N, Suzuki M. Identification of delphinidin 3-O-(6″-O-malonyl)-β-glucoside-3′-O-β-glucoside, a postulated intermediate in the biosynthesis of ternatin C5 in the blue petals of Clitoria ternatea (butterfly pea) Chem Biodiv. 2004;1:1762–1770. doi: 10.1002/cbdv.200490132. [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Wang X. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 2009;583:3303–3309. doi: 10.1016/j.febslet.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Breton C, Fournel-Gigleux S, Palcic MM. Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol. 2012;22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Modolo LV, Li L, Pan H, Blount JW, Dixon RA, Wang X. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J Mol Biol. 2009;392:1292–1302. doi: 10.1016/j.jmb.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Havsteen BH. The biochemistry and medical significance of the flavonoids. Pharmacol Ther. 2002;96:67–202. doi: 10.1016/s0163-7258(02)00298-x. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2008;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Kogawa K, Kato N, Kazuma K, Noda N, Suzuki M. Purification and characterization of UDP-glucose: anthocyanin 3′,5′-O-glucosyltransferase from Clitoria ternatea. Planta. 2007;226:1501–1509. doi: 10.1007/s00425-007-0584-1. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez L, Mosquera RA. Molecular structure and antioxidant properties of delphinidin. J Phys Chem A. 2008;112:10614–10623. doi: 10.1021/jp8043237. [DOI] [PubMed] [Google Scholar]
- Mazza G, Brouillard R. Recent developments in the stabilization of anthocyanins in food products. Food Chem. 1987;25:207–225. [Google Scholar]
- Estévez L, Mosquera RA. A density functional theory study on pelargonidin. J Phys Chem. 2007;A111:11100–11109. doi: 10.1021/jp074941a. [DOI] [PubMed] [Google Scholar]
- Leopoldini M, Russo N, Toscano M. The molecular basis of working mechanism of natural polyphenolic antioxidants. Food Chem. 2011;125:288–306. [Google Scholar]
- Tanaka Y, Brugliera F, Chandler S. Recent progress of flower colour modification by biotechnology. Int J Mol Sci. 2009;10:5350–5369. doi: 10.3390/ijms10125350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananga A, Georgiev V, Ochieng J, Phills B, Tsolova V. Production of anthocyanins in grape cell cultures: a potential source of raw material for pharmaceutical, food, and cosmetic industries. In: Poljuha D, Sladonja B, editors. The Mediterranean Genetic code—grapevine and olive. Croatia: InTech; 2013. pp. 247–−287. . In:, Eds. ( [Google Scholar]
- He XZ, Wang X, Dixon RA. Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation. J Biol Chem. 2006;281:34441–34447. doi: 10.1074/jbc.M605767200. [DOI] [PubMed] [Google Scholar]
- Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A, Tao GQ, Nehra NS, Lu CY, Dyson BK, Tsuda S, Ashikari T, Kusumi T, Mason JG, Tanaka Y. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007;48:1589–1600. doi: 10.1093/pcp/pcm131. [DOI] [PubMed] [Google Scholar]
- Saito N, Abe K, Honda T, Timberlake CF, Bridle P. Acylated delphinidin glucosides and flavonols from Clitoria ternatea. Phytochemistry. 1985;24:1583–1586. [Google Scholar]
- Terahara N, Toki K, Saito N, Honda T, Toki K, Osajima Y. Acylated anthocyanins of Clitoria ternatea flowersd and their acyl moieties. Phytochemistry. 1990;29:949–953. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information