Abstract
The silent mating type information regulation 2 proteins (sirtuins) 1 of class III histone deacetylases (HDACs) have been associated with health span and longevity. SIRT1, the best studied member of the mammalian sirtuins, has a myriad of roles in multiple tissues and organs. However, a significant part of SIRT1’s role that impinges on aging and lifespan may lie in its activities in the central nervous system (CNS) neurons. Systemically, SIRT1 influences energy metabolism and circadian rhythm through its activity in the hypothalamic nuclei. From a cell biological perspective, SIRT1 is a crucial component of multiple interconnected regulatory networks that modulate dendritic and axonal growth, as well as survival against stress. This neuronal cell autonomous activity of SIRT1 is also important for neuronal plasticity, cognitive functions, as well as protection against aging-associated neuronal degeneration and cognitive decline. We discuss recent findings that have shed light on the various activities of SIRT1 in the brain, which collectively impinge on aging-associated disorders and lifespan.
Keywords: aging, cognition, neurodegeneration, SIRT1, metabolism
Introduction
The sirtuin family comprises of class III protein deacetylases, which unlike the class I and II histone deacetylases (HDACs), have an obligatory dependency on nicotinamide adenine dinucleotide (NAD) as a cofactor (Blander and Guarente, 2004). Sirtuins deacetylate a wide range of histone and non-histone targets, and influence multiple aspects of cellular and organismal physiology and pathology (Haigis and Sinclair, 2010; Sebastián et al., 2012; Chang and Guarente, 2014; Herskovits and Guarente, 2014). The most prominent of the latter is health and lifespan, and the notion of sirtuin activities being systemically beneficial to multiple organs, particularly in the later years of life, has attracted tremendous interest in recent years (Tang, 2011). The family’s founding member, Sir2 of S. cerevisiae, mediates transcriptional silencing of the yeast mating loci, telomeres and the ribosomal DNA (Imai et al., 2000). Its suppression of the formation of rDNA circles (Sinclair and Guarente, 1997) could play a role in extending yeast’s replicative, but not chronological lifespan (Kaeberlein et al., 1999; Fabrizio et al., 2005). Importantly, yeast Sir2’s lifespan prolonging activity is also proposed to be biochemically link to, and with its activation mimicking, the effect of caloric restriction/dietary restriction (CR/DR) (Lin et al., 2000; Anderson et al., 2003; Cohen et al., 2004). CR/DR, a regime of moderate reduction of food intake without causing malnutrition, was known to curb metabolic disease as well as extend lifespan in many animal models (Chung et al., 2013). The notion that CR/DR acts through sirtuin has been intensely investigated and debated (Kaeberlein and Powers, 2007; Cantó and Auwerx, 2009a,b; Guarente, 2013; Park et al., 2013). This putative mechanistic link is one reason for the heightened interest in sirtuins as potential drug targets for pharmacological intervention of metabolic disorders and aging (Baur et al., 2012; Chakraborty and Doss, 2013).
Over-expression or activation of Sir2 orthologues was also shown to increase lifespan of invertebrate models such as C. elegans and Drosophila (Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004; Wood et al., 2004). These results have been challenged by more recent works that suggest that the lifespan extension effect was smaller than previously reported (Burnett et al., 2011; Lombard et al., 2011; Viswanathan and Guarente, 2011). The mammalian genome has seven sirtuin paralogues (Michan and Sinclair, 2007) that are differentially localized to the nucleus, cytoplasm and mitochondria. Whether sirtuins are bona fide longevity factors in mammals and humans has been unclear (Naiman and Cohen, 2012; Park et al., 2013). Sirt1, the mammalian paralogue sharing the highest homology to yeast Sir2 and the best studied, could be either pro- or anti-survival (for example, cancer promoting) under different contexts. Unlike the case in yeast, earlier experiments with transgenic over-expression or pharmacological activation of SIRT1 in mice improved metabolic parameters later in life (Baur et al., 2006; Alcendor et al., 2007; Milne et al., 2007; Herranz et al., 2010), but did not significantly increase lifespan in mice (Herranz et al., 2010). However, several recent reports have collectively provided strong evidence that sirtuins could indeed prolong the lifespan of mice. Transgenic over-expression of SIRT6 was shown to increase the lifespan of male (but not female) mice, likely through its influence on Insulin-like growth factor 1 (IGF-1) signaling (Kanfi et al., 2012). Recently, SIRT2 was found to increase lifespan of mice hypomorphic for the mitotic checkpoint kinase BubR1, the level of which declines in aging and aging-associated diseases (North et al., 2014). Importantly, mice with brain-specific transgenic over-expression of SIRT1 (BRASTO) have also recently been shown to have an extended lifespan (Satoh et al., 2013), and the specific SIRT1 activator SRT1720 extends lifespan of mice even when these were fed a standard diet (Mitchell et al., 2014).
The aging-delayed and lifespan extension phenotype of brain-specific Sirt1-overexpressing mice (Satoh et al., 2013) is particularly interesting, and attested to the notion that SIRT1 may effectively influence aging through its activities in the central nervous system (CNS) neurons. SIRT1’s deacetylation of transcription factors such as the forkhead box class O (FoxO) family members (Brunet et al., 2004; Huang and Tindall, 2007), TP53 (Hasegawa and Yoshikawa, 2008) and nuclear factor қB (NF-қB) (Yeung et al., 2004; Chen et al., 2005) modulates key aspects of stress response and survival pathways in postmitotic neurons. In C. elegans, the FoxO orthologue Daf-16 (Ogg et al., 1997) mediates lifespan extension downstream of the IGF-1 receptor/Daf-2 (Kimura et al., 1997) mutant in worms, and restoring Daf-2 expression in worm neurons alone negated the lifespan extension effect of Daf-2 mutants (Wolkow et al., 2000). This suggest that IGF-1 receptor/Daf-2 signaling in neurons could be critical for lifespan and that this could be influenced by sirtuins through FoxOs. As the enzymatic activity of SIRT1 is regulated by NAD+, it is therefore a key nutrient and redox sensor in the energy expensive brain. SIRT1’s role in regulating the activities of key transcription factors (e.g., peroxisome proliferator-activated receptor gamma (PPAR-γ) and its transcriptional co-activator PPARγ coactivator-1α (PGC-1α) and transducer of regulated CREB2 activity 2 (TORC2)) and enzymes (e.g., phosphoglycerate mutase-1) in energy metabolism in peripheral tissues are well known (Cantó and Auwerx, 2009b; Sugden et al., 2010; Li, 2013; Chang and Guarente, 2014). However, metabolic control in mammals is centrally exerted through sensing of energy status in the hypothalamic neurons in the brain, and SIRT1 activities in these neurons influence the development of aging-associated metabolic disorders (Chang and Guarente, 2014; Toorie and Nillni, 2014). SIRT1’s neuroprotective functions are also well known (Tang, 2009; Zhang et al., 2011b). However, by regulating neurite growth and synaptic processes, SIRT1 was recently shown to also play a role in normal cognitive function and synaptic plasticity (Gao et al., 2010; Michán et al., 2010), which is beyond its better known role in countering cognitive decline and neurodegenerative disease in aging.
In the ensuing paragraphs, we highlight recent findings that elucidated the functions of SIRT1 in the brain, and discuss how disruptions of these functions may underlie aging-associated disorders and lifespan.
SIRT1 in CNS neurons—role in neurogenesis, neurite growth neuronal network connections
SIRT1 is ubiquitously expressed, but more targeted investigations have revealed high levels of expression in the developing mouse CNS (Sakamoto et al., 2004; Ogawa et al., 2011), adult mouse and human brain (Ramadori et al., 2008; Zakhary et al., 2010) as well as the porcine brain (Shan et al., 2009). Within the adult brain, SIRT1 expression is found in most brain regions, is prominent in neurons of the hippocampus and hypothalamus (Ramadori et al., 2008; Michán et al., 2010; Zakhary et al., 2010), and is mostly nuclear. Brain SIRT1 levels are decreased in aged neurons (Quintas et al., 2012), and are downregulated by a high fat diet (Wu et al., 2006; Heyward et al., 2012) and various neuropathological conditions (Pallàs et al., 2008; Julien et al., 2009). On the other hand, SIRT1 levels in brain neurons could be increased by CR/DR (Satoh et al., 2010; Quintas et al., 2012) and physical exercise (Falone et al., 2012; Revilla et al., 2014). SIRT1’s high levels in the mammalian CNS and the way these change with physiological and pathological stimuli are indicative of functional importance.
SIRT1 activity is known to affect fate determination of neural progenitor cells (NPCs) during development. SIRT1 activity is dependent on the redox state, and under conditions of oxidative stress, SIRT1 promotes differentiation of NPCs towards the astroglia lineage partly by limiting the levels and activity of the pro-neuronal transcription factor Mash-1 (Prozorovski et al., 2008). Differentiating neural cells from 8-oxoguanine DNA glycosylase knock-out mice spontaneously accumulate mtDNA damage and concomitantly shift their differentiation direction toward an astrocytic lineage, a result of an increased NAD/NADH ratio and SIRT1 activation (Wang et al., 2011). SIRT1 inhibition or silencing was shown by several authors to promote neuronal differentiation (Prozorovski et al., 2008; Zhang et al., 2011a; Liu et al., 2014) and increased neuronal production in both the subventricular zone and the hippocampus (Saharan et al., 2013). Sensory neuron differentiation from progenitors is dependent on SIRT1’s modulation of the acetylation status of Pax3, which acts upstream of Hes1 and Neurog2. SIRT1 silencing increased the level of acetylated Pax3, with consequential decrease in Hes1 (important for stem cell maintenance) and increase in Neurog2 (important for neuronal properties) activities, thus promoting neurogenesis (Ichi et al., 2011). In a different context, however, pro-neurogenic Bcl6 was shown to promote neurogenesis through SIRT1 recruitment, with the silencing of Hes-5 downstream of Notch signaling (Tiberi et al., 2012). Another report has indicated that SIRT1 inactivation in the adult brain resulted in the expansion of oligodendrocyte progenitors that could generate myelinating oligodendrocytes (Rafalski et al., 2013).
Other than neural cell fate, manipulations of SIRT1 levels and activity have also been shown to modulate neuritogenesis—the outgrowth of axons and dendrites. Cytoplasmic SIRT1 enhanced nerve growth factor-induced neuritogenesis in PC12 cells (Sugino et al., 2010). Insulin-induced neurite outgrowth of SH-SY5Y cells is dependent on SIRT1 (Liu et al., 2013b). Primary neurons from transgenic mice overexpressing SIRT1 have enhanced neurite outgrowth and survival apparently via negative regulation of mammalian/mechanistic target of rapamyscin (mTOR) signaling (Guo et al., 2011). SIRT1 has been detected at the axonal growth cone and its activation has a positive influence on both formation and elongation of axons, likely through deacetylation of AKT (Li et al., 2013). Regeneration of peripheral axon after injury is SIRT1-dependent, and SIRT1 targets miR-138, a suppressor of the axonal regeneration process which could in turn reciprocally suppress SIRT1 expression (Liu et al., 2013a). Overexpression of SIRT1 in hippocampal neurons enhanced dendritic arbor complexity, an effect that is mimicked by its activator resveratrol (Codocedo et al., 2012). Brains of Sirt1 knockout mice was also shown to exhibit a decrease in dendritic branching, branch length, and complexity of neuronal dendritic arbors (Michán et al., 2010).
Taken together, these results suggest that SIRT1 could influence neuritogenesis of both axon and dendrite via several targets and mechanisms (Figure 1). This would also mean that SIRT1 is likely to have a role, like those documented for the Class I and II HDACs (Gräff and Tsai, 2013), in the connection and plasticity of the neuronal network.
Figure 1.
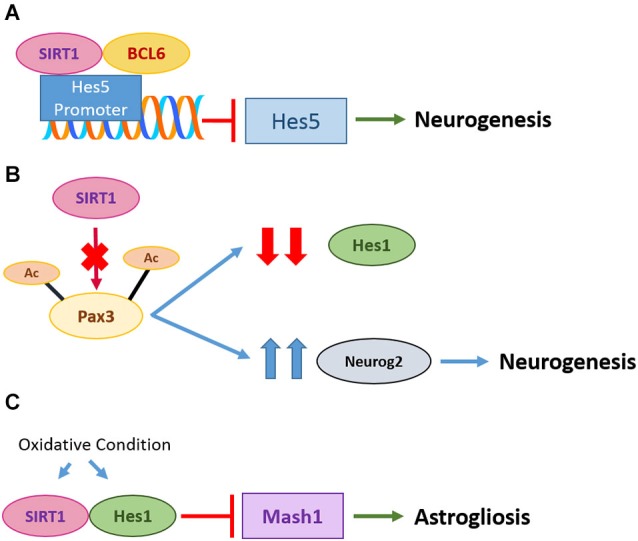
Roles of SIRT1 in neurogenesis and gliogenesis. (A) In neural progenitor cells (NPCs), repression of Notch-dependent Hes5 genes by Bcl6 is important for neurogenesis. Bcl6 triggers exclusion of co-activator Mastermind-like 1 and recruits SIRT1 to inhibit transcription of Hes5, promoting neurogenesis. (B) Pax3 acetylation on C-terminal lysine residues K437 and K475 is critical for regulation of Hes1 and Neurog2. SIRT1 silencing increased acetylation level of Pax3 and subsequently decreased the promoter activity of Hes1 but increased activity of Neurog2, inducing sensory neuron differentiation. (C) Under oxidative condition in NPCs, SIRT1 is upregulated and binds to transcription factor Hes1. This subsequently inhibits pro-neuronal Mash1 and leads to astrogliosis.
SIRT1’s role in synaptic plasticity and cognition
SIRT1 has a widely recognized role in neuroprotection and is antagonistic against progression of neurodegenerative processes (Tang, 2009; Srivastava and Haigis, 2011). Its role in synaptic plasticity and cognitive function was therefore more often associated with a reversal of neuropathological conditions that impairs learning and memory. Whether SIRT1 has a role in the physiologically unperturbed versions of learning and memory was clarified only recently. Sirt1 knockout mice have grossly normal brain anatomy, but exhibited a dendritic development phenotype as noted above (Michán et al., 2010). These mice exhibited defects in synaptic plasticity, although parameters such as basal synaptic transmission are indistinguishable from wild type. Behavioral tests indicated that short-term memory, long term associative memory, as well as spatial learning are all impaired in Sirt1 knock-outs compared to control. The knock-out mice’s hippocampal Schaffer collateral pathway also exhibited a defect in long term potentiation (LTP).
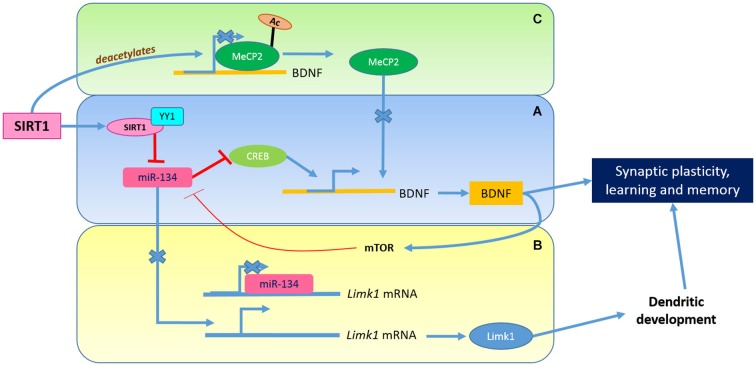
A conditional knockout of SIRT1 in brain (loxP-flanked exon 4 of Sirt1 with Cre recombinase driven by the Nestin promoter) also resulted in a decrease in fear-conditioning and other short-term memory, as well as hippocampal CA1 LTP (Gao et al., 2010). SIRT1-deficient brain had grossly normal anatomy and CA1 neurons had normal basal synaptic transmission. However, the SIRT1-deficient hippocampus had decreased levels of synaptophysin, a marker of the presynaptic termini, and CA1 neurons in these have decreased dendritic spine density. Analysis of known genetic and epigenetic modulators of learning and memory in SIRT1-deficient brain revealed that the levels of a key neurotrophin regulating neural development and synaptic function, the brain-derived neurotrophic factor (BDNF), is reduced. This is explained by a reduction or abolishment of cAMP response element-binding protein (CREB) binding to several BDNF promoters, as the levels of CREB protein (but not mRNA) is downregulated (Figure 2A). The reason why CREB levels are reduced in SIRT1-deficient brain was traced to an upregulation of the levels of miR-134 (which SIRT1 suppresses reciprocally). miR-134 is brain-enriched, and one of its known role is the regulation of dendritic spine size and morphology through modulating the expression of Lim-domain-containing protein kinase 1 (Limk1) (Figure 2B). SIRT1 limits the expression of miR-134 as part of a YYI-containing repressor complex, and promotes BDNF transcription. However, BDNF production may affect mTOR signaling pathway activation, which will also suppress miR-134 activity. The latter also has an effect of relieving miR-134’s repression of Limk1 translation, which promotes dendritic development (Schratt et al., 2006).
Figure 2.
The Role of SIRT1 in synaptic plasticity. SIRT1 modulates synaptic plasticity through the regulation of BDNF. The SIRT1 and YY1 complex limits the expression of miR-134 which affects the CREB-BDNF axis (A) and leading to enhanced Limk1 protein synthesis (B). The release of BDNF activates mTOR signaling pathway which suppress miR-134 activity. MeCP2 deacetylation by SIRT1 could also promote BDNF transcription (C).
SIRT1 could also modulate BDNF expression via the deacetylation of methyl-CpG binding protein 2 (MeCP2; Zocchi and Sassone-Corsi, 2012), a transcription factor mutated in the neurodevelopmental disorder Rett Syndrome (Guy et al., 2011; Liyanage and Rastegar, 2014). BDNF transcription could be elevated by this MeCP2 deacetylation, which promoted its release from the BDNF promoter (Guy et al., 2011; Figure 2C). The presence of high levels of SIRT1, particularly in the hippocampal neurons, may therefore serve a role (amongst others) in synaptic plasticity through the modulation of changes in dendritic spine functions and connections.
From the above discussion, it is easily conceivable that reduction of SIRT1 levels in the aged hippocampus could result in short term memory deficits and cognitive impairment. Aging human adults often exhibit more delocalized brain activity connected to higher order cognitive functions, and recruitment of additional brain areas was proposed as a compensatory mechanism for age-dependent functional decline in the primary areas (Cabeza et al., 2002; Bishop et al., 2010). Aging-associated decline in SIRT1 levels may also impair the plasticity of more distant connections. SIRT1 also has other functions in brain neurons that would contribute to aging and its associated disorders, as further discussed below.
SIRT1 in hypothalamic neurons—control of energy metabolism, circadian rhythm (and lifespan)
The hypothalamus regulates important aspects of metabolism through its neuroendocrine control of the pituitary gland hormones, and regulates body temperature, hunger/satiety and circadian rhythms, amongst others. Major neurons regulating hunger and food intake in the arcuate nucleus of the hypothalamus are the anorexigenic pro-opiomelanocortin (POMC)-expressing neurons and the orexigenic agouti-related peptide (AgRP) expressing neurons (Morton et al., 2006), and recent findings indicate that SIRT1 has major roles in metabolic homeostasis via its activity in these neurons (Table 1). Inhibition or silencing of hypothalamic SIRT1 in rat (by intracerebro-ventricular administration of inhibitor and siRNA) decreased food intake and body wright via a FoxO1-mediated increase in POMC and decrease in AgRP (Cakir et al., 2009). In rats with diet-induce obesity, brain SIRT1 inhibition increased POMC that leads to an increase in α-melanocyte-stimulating hormone (α-MSH), which could in turn increase thyroid releasing hormone and T3 levels (Cyr et al., 2014).
Table 1.
Summary of SIRT1’s roles in metabolic homeostasis.
| No | Model | Manipulation of SIRT1 | Observation | Reference |
|---|---|---|---|---|
| 1 | POMC expressing neurons | Knockout | Increased α-MSH, increased thyroid hormone and T3 levels, reduced energy expenditure; hence more sensitive to diet-induced obesity | Ramadori et al. (2010, 2011), Cyr et al. (2014) |
| 2 | AgRP expressing neurons | Knockout | Decreased response to hunger-inducing hormone ghrelin, reduced food intake | Dietrich et al. (2010) |
| 3 | Peripheral tissue neurons | Knockout | Increased insulin sensitivity and insulin receptor | Lu et al. (2013) |
| 4 | BSKO mice | Knockout | Defective somatrophic signaling (growth hormone and IGF-1) Defective caloric restriction response | Cohen et al. (2009); Monteserin-Garcia et al. (2013) |
| 5 | AgRP expressing neurons | Over-expression | Reduced food intake | Sasaki et al. (2014) |
| 6 | POMC expressing neurons | Over-expression | Stimulated energy expenditure | Sasaki et al. (2014) |
| 7 | SF-1 expressing neurons | Over-expression | Increased resistance to diet-induced obesity and reduced susceptibility to insulin resistance | Ramadori et al. (2011) |
| 8 | Mediobasal hypothalamus | Activated (by resveratrol) | Improved insulin sensitivity | Knight et al. (2011) |
| 9 | Mouse forebrain | Over-expression | Obesity, impaired glucose tolerance, some defects in motor functions | Wu et al. (2011) |
| 10 | BRASTO mice | Over-expression | Enhanced response to ghrelin hormone | Satoh et al. (2010) |
Hypothalamic SIRT1 protein levels increase on feeding, and SIRT1 suppresses the expression of orexigenic AgRP (Morton et al., 2006; Sasaki et al., 2010). Conditional knock-out of SIRT1 in AgRP neurons (loxP-flanked exon 4 of Sirt1 with Cre recombinase driven by the Agrp promoter) decreased their responses to the hunger inducing hormone ghrelin (Dietrich et al., 2010). Imai and colleagues’ BRASTO mice with transgenic over-expression of SIRT1 in the brain (mouse Sirt1 driven by mouse prion (PrP) promoter) had conversely an enhanced response to ghrelin (Satoh et al., 2010). On the other hand, conditional knockout of SIRT1 in POMC neurons (loxP-flanked exon 4 of Sirt1 with Cre recombinase driven by the Pomc promoter) was shown to reduce energy expenditure and increase susceptibility to diet-induced obesity, as well as signaling processes induced by the satiety hormone leptin (Ramadori et al., 2010). In a recent report in which SIRT1 was conditionally overexpressed in mouse POMC or AgRP neurons (using Rosa26 locus knockin and induction by Pomc- or Agrp-Cre), the authors observed that over-expressed SIRT1 prevented age-associated weight gain in complementary ways (Sasaki et al., 2014). Over-expression of SIRT1 in POMC neurons stimulated energy expenditure, whereas overexpression in AgRP neurons suppressed food intake. Mice with brain-specific SIRT1 knock-out (BSKO: loxP-flanked exon 4 of Sirt1 with Cre recombinase driven by the nestin promoter) have defective somatrophic (growth hormone and IGF-1) signaling (which could be due to Sirt1’s recently documented inhibition of CREB Monteserin-Garcia et al., 2013), as well as CR/DR response (Cohen et al., 2009).
SIRT1’s regulation of energy (glucose and lipid) metabolism in peripheral tissues are well known (Chalkiadaki and Guarente, 2012; Li, 2013), and SIRT1 has been associated with induction of hepatic gluconeogenesis (Rodgers et al., 2005), pancreatic insulin secretion (Bordone et al., 2006), enhanced muscle insulin sensitivity (Schenk et al., 2011) and the “browning” of white adipose tissues (Qiang et al., 2012). The SIRT1 activator resveratrol when acutely administered into the mediobasal hypothalamus (containing the arcuate nucleus) improves insulin sensitivity, and this is negated by SIRT1 inhibition or silencing (Knight et al., 2011). Conditional deletion of SIRT1 in mice in steroidogenic factor 1 (SF1)-expressing neurons (loxP-flanked exon 4 of Sirt1 with Cre recombinase driven by the Sf1 promoter of the ventromedial hypothalamic nucleus (VMH) resulted in heightened susceptibility to diet-induced obesity, and conversely mice overexpressing SIRT1 in these neurons are more resistant to diet-induced obesity and insulin resistance (Ramadori et al., 2011). It appears that SIRT1 activities in the hypothalamus are generally associated with a metabolically favorable phenotype that resembles that observed with CR/DR. On the other hand, it has also been reported that SIRT1 neuron specific knockout mice (with Cre recombinase controlled by of rat synapsin I promoter) had increased insulin sensitivity and increased insulin receptor signaling in peripheral tissues (Lu et al., 2013). Over-expression of SIRT1 in the mouse forebrain was also shown to result in obesity, impaired glucose tolerance and some defects in motor function (Wu et al., 2011). These deviations attest to the underlying complexities of the phenotypes resulting from pharmacological and genetic manipulations, which is dependent on the mice’s genetic background and the exact nature of manipulation.
Another important aspect of SIRT1 activity which is tightly connected to metabolic regulation that has recently come to light is its role in the regulation of circadian rhythm (Maury et al., 2010; Rey and Reddy, 2013). Central control of the mammalian circadian rhythm resides in the hypothalamic suprachiasmatic nucleus (SCN), and is regulated by a feedback loop of transcription factor interactions. Brain and muscle Arnt-like protein 1 (BMAL1) and CLOCK activate the expression of PERIOD 1, 2 and 3 (PER1, 2, 3) and Cryptochrome 1 and 2 (Cry1, 2) genes, with the latter complexing at high levels with the former, repressing their own transcription. The central circadian timer protein CLOCK is a histone acetyltransferase, and SIRT1 deacetylase activity appears to antagonize it in a circadian manner (Nakahata et al., 2009). In peripheral tissues, SIRT1 binds CLOCK-BMAL1 and is thus recruited to the CLOCK-BMAL1 chromatin complex at circadian gene promoters (Nakahata et al., 2008). It could deacetylate BMAL1 to affect its transcriptional activity (Nakahata et al., 2008), and it could also deacetylate and promote the degradation of PER2 (Asher et al., 2008). Clock-Bmal1 regulates the circadian expression of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme in the NAD+ salvage pathway (Nakahata et al., 2009; Ramsey et al., 2009), which is in turn important for SIRT1 activity. While CLOCK promotes NAMPT expression, inhibition of SIRT1, or of NAMPT by FK866, promotes an early onset of circadian peak of circadian genes like Per2 and Dbp (Nakahata et al., 2009). Thus, there appears to be a feedback loop of CLOCK-BMAL1 transcriptional regulation of clock genes involving NAMPT/NAD+ and SIRT1 (Figure 3).
Figure 3.
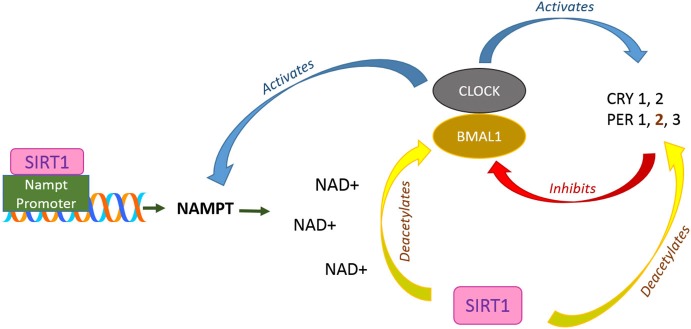
SIRT1 roles in circadian rhythm regulation. In the supraschiasmatic nucleus (SCN) at hypothalamus, CLOCK-BMAL1 activates the transcription of Cryptochrome (CRY) protein 1, 2 and Period (PER) 1, 2, and 3 which in turn, directly repress CLOCK-BMAL1 activity. In addition, CLOCK-BMAL1 also regulates circadian expression of NAMPT. Meanwhile, SIRT1 is recruited to the NAMPT promoter and contributing to the synthesis of its own coenzyme, NAD+, which is important for its deacetylation activity. SIRT1 also directly contributes to circadian rhythms through deacetylation of BMAL1 and PER2.
An important notion that emerges and extended from the above findings is that an essential aspect of SIRT1’s role in aging and aging-associated metabolic diseases lies in its activity at the hypothalamus. As mentioned above, BRASTO mice generated by Imai’s group had metabolic and aging parameters that are superior to age-matched controls, and demonstrated a delayed-aging phenotype and an increased lifespan (Satoh et al., 2013). The phenotype of BRASTO mice was attributed to an enhanced neural activity (as access by c-Fos expression) and the expression of orexin type 2 receptor gene Ox2r at the dorsomedial and lateral hypothalamic nuclei, and not the arcuate and ventromedial hypothalamic nuclei in aged BRASTO mice. Ox2r promoter activity was enhanced by SIRT1 expression through deacetylation of the transcription factor Nk2 homeobox1 (NKX2-1), and Sirt1-NKX2-1-Ox2r was proposed by the authors to form a functional axis responsible for the delayed-aging and increased lifespan phenotype. Interestingly, the phenotypes could not be directly or causally related to SIRT1 expression in the arcuate nuclei, which influences food intake and energy metabolism. Another BRASTO transgenic line with SIRT1 expression that is much higher throughout the brain and homogeneously higher throughout the hypothalamus does not display delayed-aging or increased lifespan (Satoh et al., 2013).
SIRT1’s regulation of the central control of circadian rhythm is affected by aging. At the hypothalamic SCN, neuronal SIRT1 and PGC-1α bind cooperatively and in close proximity at the Bmal1 promoter to regulate Bmal1 expression (Chang and Guarente, 2013). In aged mice, decreased SIRT1 levels in the SCN and reduced Bmal1 and Per2 perturbs the circadian rhythm and impairs the ability to adapt to environmental light/dark changes, a phenotype that is also exhibited by conditional SIRT1 knock-out in young mice. Overexpression of SIRT1 in the brain counters the aging-associated circadian rhythm defects. As impair circadian rhythm contributes critically to metabolic disorders, this result suggests that SIRT1’s activity at the SCN is important for delaying the onset of metabolic dysfunction that result from impaired central control of circadian rhythm.
SIRT1’s role in CNS neuroprotection and neurodegeneration
SIRT1 has been associated with a neuroprotective function in a myriad of neuronal injury and neurodegeneration paradigms (Gan and Mucke, 2008; Pallàs et al., 2008; Tang, 2009; Donmez and Outeiro, 2013; Herskovits and Guarente, 2014). A large number of studies using genetic and pharmacological manipulation of SIRT1 activity in invertebrates and mice models of neuronal injuries and disorders have been reported. Interestingly, both activation and inhibition of SIRT1 have been shown to be beneficial. It should also be noted that SIRT1 activities are not always pro-survival (Li et al., 2008; Ng and Tang, 2013; Sansone et al., 2013). SIRT1 activation by resveratrol (Raval et al., 2006) or transgenic overexpression of the protein (Hernández-Jiménez et al., 2013) was shown to protect against brain ischemic damages, as well as neuronal death in traumatic brain injury (Zhao et al., 2012). On the other hand, the SIRT1 inhibitor nicotinamide could also protect neurons against excitotoxicity and cerebral ischemia (Liu et al., 2009). An early study showed that resveratrol protected both C. elegans and mouse neurons against the cytotoxicity of mutant huntingtin (Parker et al., 2005). Later genetic manipulations in mice appeared to confirm SIRT1’s beneficial effect in Huntington’s disease (HD; Jeong et al., 2011; Jiang et al., 2011), but the SIRT1 inhibitor selisistat was recently shown to alleviate HD pathology in Drosophila and mouse models (Smith et al., 2014). The beneficial effect of SIRT1 elevation or activation for models of amyotrophic lateral sclerosis is more consistent (Kim et al., 2007; Song et al., 2014). SIRT1 activity is also generally shown to protect against toxic α-synuclein aggregates in worm and mice models (van Ham et al., 2008; Donmez et al., 2012), and protected dopaminergic neurons in the MPTP mouse model of Parkinson’s disease (Mudò et al., 2012).
As far as aging and aging-associated disorder is concerned, however, the main neurodegenerative disease of interest would be Alzheimer’s disease (AD; Ballard et al., 2011; Bonda et al., 2011) and related dementias, which are discussed below.
Multiple evidences indicate that SIRT1 elevation or activation is beneficial in AD and dementia (Braidy et al., 2012; Lalla and Donmez, 2013). SIRT1’s attenuation of NF-қB signaling in microglia was shown to be protective against neuronal death induced by Aβ peptides (Chen et al., 2005). This role of SIRT1 in AD is apparently connected to CR/DR, which reduces Aβ generation and amyloid plaque deposition in the brain of Tg2576 transgenic AD mice (Wang et al., 2005) and primate (Qin et al., 2006a). SIRT1 activation by resveratrol was shown to protect against toxicity induced by the cyclin-dependent kinase 5 (Cdk5) activator p25, and mutant Cu/Zn superoxide dismutase 1 (SOD1) in culture neurons and transgenic mice (Kim et al., 2007). Furthermore, direct injection of SIRT1-expressing lentivirus into the hippocampal CA1 region of p25 transgenic mice protected against aging associated neurodegeneration (Kim et al., 2007). In another AD mice model (bearing App(swe) and Psen1 dE9 transgenes), transgenic over-expression of SIRT1 in the brain markedly reduced amyloid plaque formation, gliosis and decline in learning and memory capabilities, while brain SIRT1 specific knock-out had greatly attenuated lifespan likely due to juvenile onset of an AD-like disease (Donmez et al., 2010). However, contrasting the above, the SIRT1 inhibitor nicotinamide was also shown to attenuate/delay cognitive effects of 3xTg-AD mice (triple transgenic for mutant Psen1, App(swe) and Tau) via SIRT1 inhibition and reduction of Tau phosphorylation (Green et al., 2008).
The mechanisms underlying the protective effect of SIRT1 and its activity in AD models are complex and multifaceted. SIRT1’s reduction of Aβ generation appears unique to AD pathology, and has thus been the focused of several studies that yielded interesting and useful insights. Aβ production requires the sequential action of β-secretase BACE1 and the γ-secretase complex on the amyloid precursor protein (APP). If APP is first cleaved by an α-secretase, a member of the A Disintegrin and metalloproteinase (ADAM) family, its BACE1 recognition site is loss. Aβ production is effectively prevented (Bandyopadhyay et al., 2007), and the product of α-secretase cleavage, the soluble N-terminal fragment of APP (sAPPα), is neuroprotective (Corrigan et al., 2012). SIRT1 over-expression in primary neuron cultures from Tg2576 mouse reduced Aβ secretion resulted in elevated levels of sAPPα and decreased Rho-activated kinase 1 (ROCK1) expression. This mimicked the phenotype elicited by caloric restriction (Qin et al., 2006b). Incidentally, blood cholesterol-lowering inhibitors of 3-hydroxy-3-methyl-glutaryl (HMG)-CoA reductase (statins) have also been shown to improve cognitive function and amyloid phenotype of AD models in multiple studies (Tong et al., 2009; Kurata et al., 2011). Statins also increased sAPPα production (Pedrini et al., 2005), which likely occurs via inhibition of ROCK signaling, as statins also inhibit isoprenoid biosynthesis that would affect the activity of Rho family GTPases upstream of ROCK (Ma et al., 2009). A constitutively-active ROCK1, which inhibited statin-stimulated sAPPα shedding, could in fact attenuate the SIRT1-mediated response (Qin et al., 2006b). Recent studies have indicated that the influence of Rho/ROCK signaling on amyloidogenesis is complex, involving the phosphorylation of APP and modulation of BACE1 activity (Herskovits and Guarente, 2014).
A more recent report has shown that SIRT1 could directly activate the transcription of the α-secretase ADAM10, likely via deacetylation of retinoic acid receptor β (RARβ), which is known to activate ADAM10 transcription (Donmez et al., 2010; Figure 4). SIRT1 over-expression in mouse neural N2a cells expressing mutant APPswe have significantly elevated ADAM10 and sAPPα levels, and cilostazol (an inhibitor of type III phosphodiesterase) appears to suppress Aβ production in a SIRT1-RAR-ADAM10-dependent manner (Lee et al., 2014). SIRT1’s protective effect of AD-susceptible neurons may be preceded by enhanced α-secretase-mediated non-amyloidogenic APP processing, and a decline in SIRT1 levels in the aged brain would therefore predispose its neurons to amyloidogenic APP processing and AD. Not much yet is known about how cholesterol and isoprenoid metabolism affects development and progression of AD (Sun et al., 2014; Wood et al., 2014). However, other connections between SIRT1, cholesterol metabolism and AD exist. The apolipoprotein E ε4 allele (ApoE4), a major risk factor for AD (Hauser and Ryan, 2013), binds to APP and significantly reduces sAPPα secretion, sAPPα/Aβ, and sAPPα/sAPPβ ratios. In cell culture and AD postmortem tissue, ApoE4 expression is shown to be associated with a decreased level of SIRT1 and increased level of SIRT2 (which is associated with neurodegeneration Liu et al., 2012; Outeiro et al., 2007), resulting in a decrease in SIRT1/SIRT2 ratio (Theendakara et al., 2013). The decreased SIRT1/SIRT2 ratio is perhaps indicative of a neuroprotective state that has been reversed by ApoE4 expression.
Figure 4.
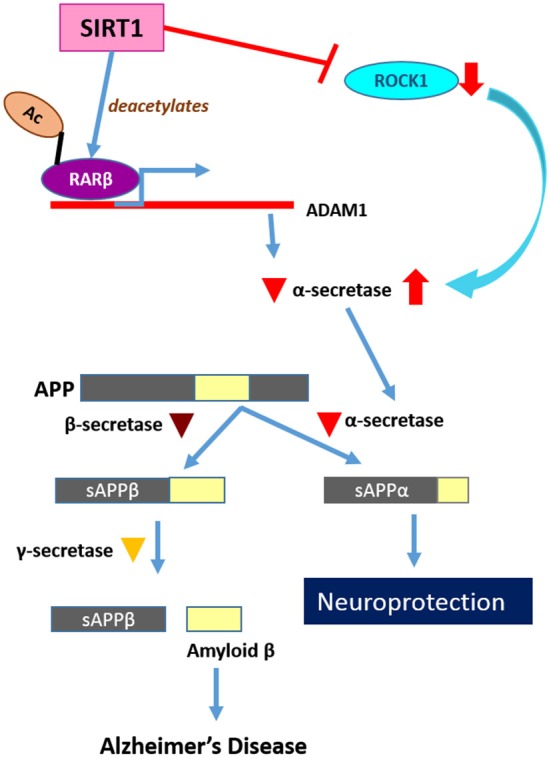
SIRT1’s roles in neuroprotection and neurodegeneration. SIRT1 has a neuroprotective function through suppression of amyloid β production. SIRT1 activates RARβ as a transcription factor of ADAM10, which encodes α-secretase. The APP cleaved by α-secretase is no longer able to generate Amyloid β peptides and this non-amyloidogenic cleavage is therefore neuroprotective.
Epilogue
In the discussions above, we highlighted known and recently revealed roles and activities of SIRT1 in the brain. Many previous reports have attested to an improved metabolic status and health at late stages of life with SIRT1 elevation and activation (reviewed in Cantó and Auwerx, 2009a; Chaudhary and Pfluger, 2009; Chang and Guarente, 2014), but evidence for actual lifespan extension by SIRT1 is only recently reported (Satoh et al., 2013; Mercken et al., 2014; Mitchell et al., 2014). We expounded on the notion that SIRT1’s role at the late stages of life and lifespan itself are linked with, and causally related to, its activities in the brain. In particular, two key aspects of late stages of life that are dependent on SIRT1 are normally loss, namely cognitive function and metabolic regulation. The three strongest lines of evidence in this regard are (1) SIRT1 is important for certain aspects of neural connectivity and synaptic plasticity, (2) Loss or decline in CNS SIRT1 activity occurs during aging, and this deficit is directly associated with defined neurophysiological and neuropathological mechanisms of cognitive decline and metabolic dysfunction; and (3) SIRT1 overexpression in certain neurons in the brain increased lifespan.
A more comprehensive understanding of SIRT1’s role in CNS neurons, particularly those of the hypothalamus and hippocampus, would hopefully aid future therapeutic interventions. However, much remains to be learned. Mouse experiments already calls for caution in analysis and interpretation. A constitutive and global elevation of brain SIRT1 (by whatever means) is unlikely to be helpful in an all-encompassing manner in reversing cognitive and metabolic dysfunctions. Notably, overexpression of brain SIRT1 in mice has been reported to result in metabolic (Wu et al., 2011) and cognitive (Kakefuda et al., 2009) defects. Our understanding of SIRT1’s regulation of circadian rhythm is at an early stage, and the roles of other nuclear sirtuins in this regard, such as SIRT6, are being revealed (Masri and Sassone-Corsi, 2014). SIRT6 also interacts with CLOCK-BMAL1, but probably not in the same complex as SIRT1. Unlike SIRT1, it operates directly at the transcription level by recruiting the clock machinery to chromatin. An important role for SIRT6 is circadian chromatin recruitment of SREBP-1, thus impinging on the circadian regulation of a host of genes in fatty acid and cholesterol metabolism (Masri et al., 2014). A thorough understanding of how SIRT1 and SIRT6 differentially or synergistically regulate the circadian mechanism would be of fundamental importance (Masri and Sassone-Corsi, 2014).
An other important aspect of note is that beyond its role in regulating gene expression via deacetylation of transcription factors, Sirt1 is also a regulator of epigenetic changes. Sirt1 is a histone deacetylase (Hayakawa et al., 2015), and could alter DNA methylation through its activity on factors such as MeCP2 (Zocchi and Sassone-Corsi, 2012). A recent report in mice showed that microglial SIRT1 deficiency influences both aging-associated and tau-mediated memory deficits via the pro-inflammatory cytokine IL-1β (Cho et al., 2015). Activation of IL-1β transcription by SIRT1 deficiency appears to result from IL-1β proximal promoter hypomethylation, a state that is strongly associated with elevated IL-1β expression and aging. A more defined understanding of the epigenetic impact of SIRT1, either direct or indirect, on organismal aging via the neurons would be highly desirable. On the other hand, molecules linking SIRT1 to learning and memory are not yet fully explored. The histone H2A variant, H2A.Z, a known mediator of thermosensory response, was recently shown to play a role in memory consolidation (Zovkic et al., 2014). Interestingly, at least in some tissues, SIRT1 level and activity negatively affect H2A.Z expression (Baptista et al., 2013). It is therefore plausible that SIRT1’s role in learning and memory is connected to its effect on H2A.Z expression, a notion that requires further confirmation.
The mitochondrial SIRT3, SIRT4 and SIRT5 could also be key regulators of metabolism (Hirschey et al., 2010; Laurent et al., 2013a,b; Rardin et al., 2013; Weir et al., 2013), and the interplay or crosstalk between nuclear SIRT1 and mitochondrial SIRT3 in metabolic regulation and disorder is not yet understood in detail. Sirtuin-mediated lifespan extension has now been observed for SIRT1, SIRT6 (Kanfi et al., 2012), and under specific circumstances, for SIRT2 (North et al., 2014). An understanding of how mammalian sirtuins interact in complex traits like lifespan would require much more work.
SIRT1’s neuroprotective function has also been extended by recent findings, and it appears to play a role in the maintenance of genomic stability in postmitotic neurons (Dobbin et al., 2013). SIRT1 deacetylates and activates HDAC1 in the repair of double stranded DNA breaks through the nonhomologous end-joining pathway. In as far as learning and memory is concern, SIRT1 and HDACs may have, albeit indirect, opposite functions. While SIRT1 activation preserves memory and cognition (Gao et al., 2010; Michán et al., 2010), HDAC inhibition enhances cognition (Gräff and Tsai, 2013). HDAC inhibitors are known to have varying effects on the levels of sirtuins, and trichostatin A (TSA) and butyrate downregulate Sirt1 (Kyrylenko et al., 2003). How sirtuins interact functionally with HDACs in their myriad neuronal functions would also be an interesting and important pursuit in the near future.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Acknowledgments
BLT was funded by a bench-to-bedside grant (NUHSRO/2011/009/STB/B2B-05) from the National University Health System (NUHS), and supported by NUS Graduate School for Integrative Sciences and Engineering. The authors declare no conflict of interest. We thank the reviewers for their constructive criticisms and suggestions, which improved the manuscript.
References
- Alcendor R. R., Gao S., Zhai P., Zablocki D., Holle E., Yu X., et al. (2007). Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 100, 1512–1521. 10.1161/01.res.0000267723.65696.4a [DOI] [PubMed] [Google Scholar]
- Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003). Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423, 181–185. 10.1038/nature01578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., et al. (2008). SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328. 10.1016/j.cell.2008.06.050 [DOI] [PubMed] [Google Scholar]
- Ballard C., Gauthier S., Corbett A., Brayne C., Aarsland D., Jones E. (2011). Alzheimer’s disease. Lancet 377, 1019–1031. 10.1016/S0140-6736(10)61349-9 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Goldstein L. E., Lahiri D. K., Rogers J. T. (2007). Role of the APP non-amyloidogenic signaling pathway and targeting alpha-secretase as an alternative drug target for treatment of Alzheimer’s disease. Curr. Med. Chem. 14, 2848–2864. 10.2174/092986707782360060 [DOI] [PubMed] [Google Scholar]
- Baptista T., Graça I., Sousa E. J., Oliveira A. I., Costa N. R., Costa-Pinheiro P., et al. (2013). Regulation of histone H2A.Z expression is mediated by sirtuin 1 in prostate cancer. Oncotarget 4, 1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., et al. (2006). Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342. 10.1038/nature05354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur J. A., Ungvari Z., Minor R. K., Le Couteur D. G., de Cabo R. (2012). Are sirtuins viable targets for improving healthspan and lifespan? Nat. Rev. Drug Discov. 11, 443–461. 10.1038/nrd3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N. A., Lu T., Yankner B. A. (2010). Neural mechanisms of ageing and cognitive decline. Nature 464, 529–535. 10.1038/nature08983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G., Guarente L. (2004). The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. 10.1146/annurev.biochem.73.011303.073651 [DOI] [PubMed] [Google Scholar]
- Bonda D. J., Lee H. G., Camins A., Pallàs M., Casadesus G., Smith M. A., et al. (2011). The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. Lancet Neurol. 10, 275–279. 10.1016/s1474-4422(11)70013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., et al. (2006). Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 4:e31. 10.1371/journal.pbio.0040031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidy N., Jayasena T., Poljak A., Sachdev P. S. (2012). Sirtuins in cognitive ageing and Alzheimer’s disease. Curr. Opin. Psychiatry 25, 226–230. 10.1097/YCO.0b013e32835112c1 [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015. 10.1126/science.1094637 [DOI] [PubMed] [Google Scholar]
- Burnett C., Valentini S., Cabreiro F., Goss M., Somogyvári M., Piper M. D., et al. (2011). Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485. 10.1038/nature10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeza R., Anderson N. D., Locantore J. K., McIntosh A. R. (2002). Aging gracefully: compensatory brain activity in high-performing older adults. Neuroimage 17, 1394–1402. 10.1006/nimg.2002.1280 [DOI] [PubMed] [Google Scholar]
- Cakir I., Perello M., Lansari O., Messier N. J., Vaslet C. A., Nillni E. A. (2009). Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS One 4:e8322. 10.1371/journal.pone.0008322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Auwerx J. (2009a). Caloric restriction, SIRT1 and longevity. Trends Endocrinol. Metab. 20, 325–331. 10.1016/j.tem.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Auwerx J. (2009b). PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 20, 98–105. 10.1097/MOL.0b013e328328d0a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Doss C. G. P. (2013). Sirtuins family–recent development as a drug target for aging, metabolism and age related diseases. Curr. Drug Targets 14, 666–675. 10.2174/1389450111314060008 [DOI] [PubMed] [Google Scholar]
- Chalkiadaki A., Guarente L. (2012). Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296. 10.1038/nrendo.2011.225 [DOI] [PubMed] [Google Scholar]
- Chang H. C., Guarente L. (2013). SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153, 1448–1460. 10.1016/j.cell.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Guarente L. (2014). SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 25, 138–145. 10.1016/j.tem.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N., Pfluger P. T. (2009). Metabolic benefits from Sirt1 and Sirt1 activators. Curr. Opin. Clin. Nutr. Metab. Care 12, 431–437. 10.1097/MCO.0b013e32832cdaae [DOI] [PubMed] [Google Scholar]
- Chen J., Zhou Y., Mueller-Steiner S., Chen L. F., Kwon H., Yi S., et al. (2005). SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-κB signaling. J. Biol. Chem. 280, 40364–40374. 10.1074/jbc.m509329200 [DOI] [PubMed] [Google Scholar]
- Cho S. H., Chen J. A., Sayed F., Ward M. E., Gao F., Nguyen T. A., et al. (2015). SIRT1 deficiency in microglia contributes to cognitive decline in aging and neurodegeneration via epigenetic regulation of IL-1β. J. Neurosci. 35, 807–818. 10.1523/JNEUROSCI.2939-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. W., Kim D. H., Park M. H., Choi Y. J., Kim N. D., Lee J., et al. (2013). Recent advances in calorie restriction research on aging. Exp. Gerontol. 48, 1049–1053. 10.1016/j.exger.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Codocedo J. F., Allard C., Godoy J. A., Varela-Nallar L., Inestrosa N. C. (2012). SIRT1 regulates dendritic development in hippocampal neurons. PLoS One 7:e47073. 10.1371/journal.pone.0047073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., et al. (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392. 10.1126/science.1099196 [DOI] [PubMed] [Google Scholar]
- Cohen D. E., Supinski A. M., Bonkowski M. S., Donmez G., Guarente L. P. (2009). Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 23, 2812–2817. 10.1101/gad.1839209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan F., Vink R., Blumbergs P. C., Masters C. L., Cappai R., van den Heuvel C. (2012). sAPPα rescues deficits in amyloid precursor protein knockout mice following focal traumatic brain injury. J. Neurochem. 122, 208–220. 10.1111/j.1471-4159.2012.07761.x [DOI] [PubMed] [Google Scholar]
- Cyr N. E., Steger J. S., Toorie A. M., Yang J. Z., Stuart R., Nillni E. A. (2014). Central Sirt1 regulates body weight and energy expenditure along with the POMC-derived peptide α-MSH and the processing enzyme CPE production in diet-induced obese male rats. Endocrinology 155, 2423–2435. 10.1210/en.2013-1998 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dietrich M. O., Antunes C., Geliang G., Liu Z. W., Borok E., Nie Y., et al. (2010). Agrp neurons mediate Sirt1’s action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J. Neurosci. 30, 11815–11825. 10.1523/JNEUROSCI.2234-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin M. M., Madabhushi R., Pan L., Chen Y., Kim D., Gao J., et al. (2013). SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 16, 1008–1015. 10.1038/nn.3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G., Arun A., Chung C. Y., McLean P. J., Lindquist S., Guarente L. (2012). SIRT1 protects against α-synuclein aggregation by activating molecular chaperones. J. Neurosci. 32, 124–132. 10.1523/JNEUROSCI.3442-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Donmez G., Outeiro T. F. (2013). SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 5, 344–352. 10.1002/emmm.201302451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G., Wang D., Cohen D. E., Guarente L. (2010). SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142, 320–332. 10.1016/j.cell.2010.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fabrizio P., Gattazzo C., Battistella L., Wei M., Cheng C., McGrew K., et al. (2005). Sir2 blocks extreme life-span extension. Cell 123, 655–667. 10.1016/j.cell.2005.08.042 [DOI] [PubMed] [Google Scholar]
- Falone S., D’Alessandro A., Mirabilio A., Cacchio M., Di Ilio C., Di Loreto S., et al. (2012). Late-onset running biphasically improves redox balance, energy- and methylglyoxal-related status, as well as SIRT1 expression in mouse hippocampus. PLoS One 7:e48334. 10.1371/journal.pone.0048334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L., Mucke L. (2008). Paths of convergence: sirtuins in aging and neurodegeneration. Neuron 58, 10–14. 10.1016/j.neuron.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang W. Y., Mao Y. W., Gräff J., Guan J. S., Pan L., et al. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109. 10.1038/nature09271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J., Tsai L. H. (2013). The potential of HDAC inhibitors as cognitive enhancers. Annu. Rev. Pharmacol. Toxicol. 53, 311–330. 10.1146/annurev-pharmtox-011112-140216 [DOI] [PubMed] [Google Scholar]
- Green K. N., Steffan J. S., Martinez-Coria H., Sun X., Schreiber S. S., Thompson L. M., et al. (2008). Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J. Neurosci. 28, 11500–11510. 10.1523/JNEUROSCI.3203-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2013). Calorie restriction and sirtuins revisited. Genes Dev. 27, 2072–2085. 10.1101/gad.227439.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Qian L., Zhang J., Zhang W., Morrison A., Hayes P., et al. (2011). Sirt1 overexpression in neurons promotes neurite outgrowth and cell survival through inhibition of the mTOR signaling. J. Neurosci. Res. 89, 1723–1736. 10.1002/jnr.22725 [DOI] [PubMed] [Google Scholar]
- Guy J., Cheval H., Selfridge J., Bird A. (2011). The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 27, 631–652. 10.1146/annurev-cellbio-092910-154121 [DOI] [PubMed] [Google Scholar]
- Haigis M. C., Sinclair D. A. (2010). Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295. 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Yoshikawa K. (2008). Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J. Neurosci. 28, 8772–8784. 10.1523/JNEUROSCI.3052-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser P. S., Ryan R. O. (2013). Impact of apolipoprotein E on Alzheimer’s disease. Curr. Alzheimer Res. 10, 809–817. 10.2174/15672050113109990156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T., Iwai M., Aoki S., Takimoto K., Maruyama M., Maruyama W., et al. (2015). SIRT1 suppresses the senescence-associated secretory phenotype through epigenetic gene regulation. PLoS One 10:e0116480. 10.1371/journal.pone.0116480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Jiménez M., Hurtado O., Cuartero M. I., Ballesteros I., Moraga A., Pradillo J. M., et al. (2013). Silent information regulator 1 protects the brain against cerebral ischemic damage. Stroke 44, 2333–2337. 10.1161/STROKEAHA.113.001715 [DOI] [PubMed] [Google Scholar]
- Herranz D., Muñoz-Martin M., Cañamero M., Mulero F., Martinez-Pastor B., Fernandez-Capetillo O., et al. (2010). Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat. Commun. 1:3. 10.1038/ncomms1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits A. Z., Guarente L. (2014). SIRT1 in neurodevelopment and brain senescence. Neuron 81, 471–483. 10.1016/j.neuron.2014.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyward F. D., Walton R. G., Carle M. S., Coleman M. A., Garvey W. T., Sweatt J. D. (2012). Adult mice maintained on a high-fat diet exhibit object location memory deficits and reduced hippocampal SIRT1 gene expression. Neurobiol. Learn. Mem. 98, 25–32. 10.1016/j.nlm.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., et al. (2010). SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125. 10.1038/nature08778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tindall D. J. (2007). Dynamic FoxO transcription factors. J. Cell Sci. 120, 2479–2487. 10.1242/jcs.001222 [DOI] [PubMed] [Google Scholar]
- Ichi S., Boshnjaku V., Shen Y. W., Mania-Farnell B., Ahlgren S., Sapru S., et al. (2011). Role of Pax3 acetylation in the regulation of Hes1 and Neurog2. Mol. Biol. Cell 22, 503–512. 10.1091/mbc.e10-06-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800. 10.1038/35001622 [DOI] [PubMed] [Google Scholar]
- Jeong H., Cohen D. E., Cui L., Supinski A., Savas J. N., Mazzulli J. R., et al. (2011). Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat. Med. 18, 159–165. 10.1038/nm.2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Wang J., Fu J., Du L., Jeong H., West T., et al. (2011). Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat. Med. 18, 153–158. 10.1038/nm.2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien C., Tremblay C., Emond V., Lebbadi M., Salem N., Bennett D. A., et al. (2009). Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J. Neuropathol. Exp. Neurol. 68, 48–58. 10.1097/NEN.0b013e3181922348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580. 10.1101/gad.13.19.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R. W. (2007). Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res. Rev. 6, 128–140. 10.1016/j.arr.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Kakefuda K., Fujita Y., Oyagi A., Hyakkoku K., Kojima T., Umemura K., et al. (2009). Sirtuin 1 overexpression mice show a reference memory deficit, but not neuroprotection. Biochem. Biophys. Res. Commun. 387, 784–788. 10.1016/j.bbrc.2009.07.119 [DOI] [PubMed] [Google Scholar]
- Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. 10.1038/nature10815 [DOI] [PubMed] [Google Scholar]
- Kim D., Nguyen M. D., Dobbin M. M., Fischer A., Sananbenesi F., Rodgers J. T., et al. (2007). SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 26, 3169–3179. 10.1038/sj.emboj.7601758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. (1997). daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946. 10.1126/science.277.5328.942 [DOI] [PubMed] [Google Scholar]
- Knight C. M., Gutierrez-Juarez R., Lam T. K. T., Arrieta-Cruz I., Huang L., Schwartz G., et al. (2011). Mediobasal hypothalamic SIRT1 is essential for resveratrol’s effects on insulin action in rats. Diabetes 60, 2691–2700. 10.2337/db10-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T., Miyazaki K., Kozuki M., Panin V. L., Morimoto N., Ohta Y., et al. (2011). Atorvastatin and pitavastatin improve cognitive function and reduce senile plaque and phosphorylated tau in aged APP mice. Brain Res. 1371, 161–170. 10.1016/j.brainres.2010.11.067 [DOI] [PubMed] [Google Scholar]
- Kyrylenko S., Kyrylenko O., Suuronen T., Salminen A. (2003). Differential regulation of the Sir2 histone deacetylase gene family by inhibitors of class I and II histone deacetylases. Cell. Mol. Life Sci. 60, 1990–1997. 10.1007/s00018-003-3090-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalla R., Donmez G. (2013). The role of sirtuins in Alzheimer’s disease. Front. Aging Neurosci. 5:16. 10.3389/fnagi.2013.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G., de Boer V. C. J., Finley L. W. S., Sweeney M., Lu H., Schug T. T., et al. (2013a). SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation. Mol. Cell. Biol. 33, 4552–4561. 10.1128/MCB.00087-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G., German N. J., Saha A. K., de Boer V. C. J., Davies M., Koves T. R., et al. (2013b). SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 50, 686–698. 10.1016/j.molcel.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. R., Shin H. K., Park S. Y., Kim H. Y., Lee W. S., Rhim B. Y., et al. (2014). Cilostazol suppresses β-amyloid production by activating a disintegrin and metalloproteinase 10 via the upregulation of SIRT1-coupled retinoic acid receptor-β. J. Neurosci. Res. 92, 1581–1590. 10.1002/jnr.23421 [DOI] [PubMed] [Google Scholar]
- Li X. (2013). SIRT1 and energy metabolism. Acta Biochim. Biophys. Sin. (Shanghai) 45, 51–60. 10.1093/abbs/gms108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. H., Chen C., Tu Y., Sun H. T., Zhao M. L., Cheng S. X., et al. (2013). Sirt1 promotes axonogenesis by deacetylation of Akt and inactivation of GSK3. Mol. Neurobiol. 48, 490–499. 10.1007/s12035-013-8437-3 [DOI] [PubMed] [Google Scholar]
- Li Y., Xu W., McBurney M. W., Longo V. D. (2008). SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 8, 38–48. 10.1016/j.cmet.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. J., Defossez P. A., Guarente L. (2000). Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128. 10.1126/science.289.5487.2126 [DOI] [PubMed] [Google Scholar]
- Liu L., Arun A., Ellis L., Peritore C., Donmez G. (2012). Sirtuin 2 (SIRT2) enhances 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigrostriatal damage via deacetylating forkhead box O3a (Foxo3a) and activating Bim protein. J. Biol. Chem. 287, 32307–32311. 10.1074/jbc.c112.403048 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu D., Gharavi R., Pitta M., Gleichmann M., Mattson M. P. (2009). Nicotinamide prevents NAD+ depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD+ consumption by SIRT1 may endanger energetically compromised neurons. Neuromolecular Med. 11, 28–42. 10.1007/s12017-009-8058-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. J., Hammer D., Komlos D., Chen K. Y., Firestein B. L., Liu A. Y. C. (2014). SIRT1 knockdown promotes neural differentiation and attenuates the heat shock response. J. Cell. Physiol. 229, 1224–1235. 10.1002/jcp.24556 [DOI] [PubMed] [Google Scholar]
- Liu C. M., Wang R. Y., Saijilafu , Jiao Z. X., Zhang B. Y., Zhou F. Q. (2013a). MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 27, 1473–1483. 10.1101/gad.209619.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yao Z., Zhang L., Zhu H., Deng W., Qin C. (2013b). Insulin induces neurite outgrowth via SIRT1 in SH-SY5Y cells. Neuroscience 238, 371–380. 10.1016/j.neuroscience.2013.01.034 [DOI] [PubMed] [Google Scholar]
- Liyanage V. R. B., Rastegar M. (2014). Rett syndrome and MeCP2. Neuromolecular Med. 16, 231–264. 10.1007/s12017-014-8295-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D. B., Pletcher S. D., Cantó C., Auwerx J. (2011). Ageing: longevity hits a roadblock. Nature 477, 410–411. 10.1038/477410a [DOI] [PubMed] [Google Scholar]
- Lu M., Sarruf D. A., Li P., Osborn O., Sanchez-Alavez M., Talukdar S., et al. (2013). Neuronal Sirt1 deficiency increases insulin sensitivity in both brain and peripheral tissues. J. Biol. Chem. 288, 10722–10735. 10.1074/jbc.M112.443606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Zhao Y., Kwak Y. D., Yang Z., Thompson R., Luo Z., et al. (2009). Statin’s excitoprotection is mediated by sAPP and the subsequent attenuation of calpain-induced truncation events, likely via rho-ROCK signaling. J. Neurosci. 29, 11226–11236. 10.1523/JNEUROSCI.6150-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S., Rigor P., Cervantes M., Ceglia N., Sebastian C., Xiao C., et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672. 10.1016/j.cell.2014.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S., Sassone-Corsi P. (2014). Sirtuins and the circadian clock: bridging chromatin and metabolism. Sci. Signal. 7:re6. 10.1126/scisignal.2005685 [DOI] [PubMed] [Google Scholar]
- Maury E., Ramsey K. M., Bass J. (2010). Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ. Res. 106, 447–462. 10.1161/CIRCRESAHA.109.208355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercken E. M., Hu J., Krzysik-Walker S., Wei M., Li Y., McBurney M. W., et al. (2014). SIRT1 but not its increased expression is essential for lifespan extension in caloric-restricted mice. Aging Cell 13, 193–196. 10.1111/acel.12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michán S., Li Y., Chou M. M. H., Parrella E., Ge H., Long J. M., et al. (2010). SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 30, 9695–9707. 10.1523/JNEUROSCI.0027-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S., Sinclair D. (2007). Sirtuins in mammals: insights into their biological function. Biochem. J. 404, 1–13. 10.1042/bj20070140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., et al. (2007). Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716. 10.1038/nature06261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. J., Martin-Montalvo A., Mercken E. M., Palacios H. H., Ward T. M., Abulwerdi G., et al. (2014). The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. 6, 836–843. 10.1016/j.celrep.2014.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteserin-Garcia J., Al-Massadi O., Seoane L. M., Alvarez C. V., Shan B., Stalla J., et al. (2013). Sirt1 inhibits the transcription factor CREB to regulate pituitary growth hormone synthesis. FASEB J. 27, 1561–1571. 10.1096/fj.12-220129 [DOI] [PubMed] [Google Scholar]
- Morton G. J., Cummings D. E., Baskin D. G., Barsh G. S., Schwartz M. W. (2006). Central nervous system control of food intake and body weight. Nature 443, 289–295. 10.1038/nature05026 [DOI] [PubMed] [Google Scholar]
- Mudò G., Mäkelä J., Di Liberto V., Tselykh T. V., Olivieri M., Piepponen P., et al. (2012). Transgenic expression and activation of PGC-1α protect dopaminergic neurons in the MPTP mouse model of Parkinson’s disease. Cell. Mol. Life Sci. 69, 1153–1165. 10.1007/s00018-011-0850-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiman S., Cohen H. Y. (2012). The contentious history of sirtuin debates. Rambam Maimonides Med. J. 3:e0022. 10.5041/RMMJ.10093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., et al. (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. 10.1016/j.cell.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657. 10.1126/science.1170803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng F., Tang B. L. (2013). When is Sirt1 activity bad for dying neurons? Front. Cell. Neurosci. 7:186. 10.3389/fncel.2013.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B. J., Rosenberg M. A., Jeganathan K. B., Hafner A. V., Michan S., Dai J., et al. (2014). SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 33, 1438–1453. 10.15252/embj.201386907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Wakai C., Saito T., Murayama A., Mimura Y., Youfu S., et al. (2011). Distribution of the longevity gene product, SIRT1, in developing mouse organs. Congenit. Anom. (Kyoto) 51, 70–79. 10.1111/j.1741-4520.2010.00304.x [DOI] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., et al. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C elegans. Nature 389, 994–999. 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- Outeiro T. F., Kontopoulos E., Altmann S. M., Kufareva I., Strathearn K. E., Amore A. M., et al. (2007). Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 317, 516–519. 10.1126/science.1143780 [DOI] [PubMed] [Google Scholar]
- Pallàs M., Pizarro J. G., Gutierrez-Cuesta J., Crespo-Biel N., Alvira D., Tajes M., et al. (2008). Modulation of SIRT1 expression in different neurodegenerative models and human pathologies. Neuroscience 154, 1388–1397. 10.1016/j.neuroscience.2008.04.065 [DOI] [PubMed] [Google Scholar]
- Park S., Mori R., Shimokawa I. (2013). Do sirtuins promote mammalian longevity? A critical review on its relevance to the longevity effect induced by calorie restriction. Mol. Cells 35, 474–480. 10.1007/s10059-013-0130-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. A., Arango M., Abderrahmane S., Lambert E., Tourette C., Catoire H., et al. (2005). Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet. 37, 349–350. 10.1038/ng1534 [DOI] [PubMed] [Google Scholar]
- Pedrini S., Carter T. L., Prendergast G., Petanceska S., Ehrlich M. E., Gandy S. (2005). Modulation of statin-activated shedding of Alzheimer APP ectodomain by ROCK. PLoS Med. 2:e18. 10.1371/journal.pmed.0020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., et al. (2008). Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 10, 385–394. 10.1038/ncb1700 [DOI] [PubMed] [Google Scholar]
- Qiang L., Wang L., Kon N., Zhao W., Lee S., Zhang Y., et al. (2012). Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 150, 620–632. 10.1016/j.cell.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Chachich M., Lane M., Roth G., Bryant M., de Cabo R., et al. (2006a). Calorie restriction attenuates Alzheimer’s disease type brain amyloidosis in Squirrel monkeys (Saimiri sciureus). J. Alzheimers Dis. 10, 417–422. [DOI] [PubMed] [Google Scholar]
- Qin W., Yang T., Ho L., Zhao Z., Wang J., Chen L., et al. (2006b). Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J. Biol. Chem. 281, 21745–21754. 10.1074/jbc.m602909200 [DOI] [PubMed] [Google Scholar]
- Quintas A., de Solís A. J., Díez-Guerra F. J., Carrascosa J. M., Bogónez E. (2012). Age-associated decrease of SIRT1 expression in rat hippocampus: prevention by late onset caloric restriction. Exp. Gerontol. 47, 198–201. 10.1016/j.exger.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Rafalski V. A., Ho P. P., Brett J. O., Ucar D., Dugas J. C., Pollina E. A., et al. (2013). Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat. Cell Biol. 15, 614–624. 10.1038/ncb2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G., Fujikawa T., Anderson J., Berglund E. D., Frazao R., Michán S., et al. (2011). SIRT1 deacetylase in SF1 neurons protects against metabolic imbalance. Cell Metab. 14, 301–312. 10.1016/j.cmet.2011.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G., Fujikawa T., Fukuda M., Anderson J., Morgan D. A., Mostoslavsky R., et al. (2010). SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 12, 78–87. 10.1016/j.cmet.2010.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G., Lee C. E., Bookout A. L., Lee S., Williams K. W., Anderson J., et al. (2008). Brain SIRT1: anatomical distribution and regulation by energy availability. J. Neurosci. 28, 9989–9996. 10.1523/jneurosci.3257-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., et al. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654. 10.1126/science.1171641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin M. J., He W., Nishida Y., Newman J. C., Carrico C., Danielson S. R., et al. (2013). SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933. 10.1016/j.cmet.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval A. P., Dave K. R., Pérez-Pinzón M. A. (2006). Resveratrol mimics ischemic preconditioning in the brain. J. Cereb. Blood Flow Metab. 26, 1141–1147. 10.1038/sj.jcbfm.9600262 [DOI] [PubMed] [Google Scholar]
- Revilla S., Suñol C., García-Mesa Y., Giménez-Llort L., Sanfeliu C., Cristòfol R. (2014). Physical exercise improves synaptic dysfunction and recovers the loss of survival factors in 3xTg-AD mouse brain. Neuropharmacology 81, 55–63. 10.1016/j.neuropharm.2014.01.037 [DOI] [PubMed] [Google Scholar]
- Rey G., Reddy A. B. (2013). Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 23, 234–241. 10.1016/j.tcb.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118. 10.1038/nature03354 [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S. L. (2004). Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U S A 101, 15998–16003. 10.1073/pnas.0404184101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharan S., Jhaveri D. J., Bartlett P. F. (2013). SIRT1 regulates the neurogenic potential of neural precursors in the adult subventricular zone and hippocampus. J. Neurosci. Res. 91, 642–659. 10.1002/jnr.23199 [DOI] [PubMed] [Google Scholar]
- Sakamoto J., Miura T., Shimamoto K., Horio Y. (2004). Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 556, 281–286. 10.1016/s0014-5793(03)01444-3 [DOI] [PubMed] [Google Scholar]
- Sansone L., Reali V., Pellegrini L., Villanova L., Aventaggiato M., Marfe G., et al. (2013). SIRT1 silencing confers neuroprotection through IGF-1 pathway activation. J. Cell. Physiol. 228, 1754–1761. 10.1002/jcp.24334 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Kikuchi O., Shimpuku M., Susanti V. Y., Yokota-Hashimoto H., Taguchi R., et al. (2014). Hypothalamic SIRT1 prevents age-associated weight gain by improving leptin sensitivity in mice. Diabetologia 57, 819–831. 10.1007/s00125-013-3140-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Kim H. J., Kobayashi M., Kitamura Y. I., Yokota-Hashimoto H., Shiuchi T., et al. (2010). Induction of hypothalamic Sirt1 leads to cessation of feeding via agouti-related peptide. Endocrinology 151, 2556–2566. 10.1210/en.2009-1319 [DOI] [PubMed] [Google Scholar]
- Satoh A., Brace C. S., Ben-Josef G., West T., Wozniak D. F., Holtzman D. M., et al. (2010). SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 30, 10220–10232. 10.1523/jneurosci.1385-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Brace C. S., Rensing N., Cliften P., Wozniak D. F., Herzog E. D., et al. (2013). Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 18, 416–430. 10.1016/j.cmet.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S., McCurdy C. E., Philp A., Chen M. Z., Holliday M. J., Bandyopadhyay G. K., et al. (2011). Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J. Clin. Invest. 121, 4281–4288. 10.1172/JCI58554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. M., Tuebing F., Nigh E. A., Kane C. G., Sabatini M. E., Kiebler M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Sebastián C., Satterstrom F. K., Haigis M. C., Mostoslavsky R. (2012). From sirtuin biology to human diseases: an update. J. Biol. Chem. 287, 42444–42452. 10.1074/jbc.r112.402768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Wang Y., Wu T., Liu C., Guo J., Zhang Y., et al. (2009). Porcine sirtuin 1 gene clone, expression pattern and regulation by resveratrol. J. Anim. Sci. 87, 895–904. 10.2527/jas.2008-1344 [DOI] [PubMed] [Google Scholar]
- Sinclair D. A., Guarente L. (1997). Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91, 1033–1042. 10.1016/s0092-8674(00)80493-6 [DOI] [PubMed] [Google Scholar]
- Smith M. R., Syed A., Lukacsovich T., Purcell J., Barbaro B. A., Worthge S. A., et al. (2014). A potent and selective Sirtuin 1 inhibitor alleviates pathology in multiple animal and cell models of Huntington’s disease. Hum. Mol. Genet. 23, 2995–3007. 10.1093/hmg/ddu010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Chen L., Zhang X., Li J., Le W. (2014). Resveratrol ameliorates motor neuron degeneration and improves survival in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Biomed. Res. Int. 2014:483501. 10.1155/2014/483501 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Srivastava S., Haigis M. C. (2011). Role of sirtuins and calorie restriction in neuroprotection: implications in Alzheimer’s and Parkinson’s diseases. Curr. Pharm. Des. 17, 3418–3433. 10.2174/138161211798072526 [DOI] [PubMed] [Google Scholar]
- Sugden M. C., Caton P. W., Holness M. J. (2010). PPAR control: it’s SIRTainly as easy as PGC. J. Endocrinol. 204, 93–104. 10.1677/joe-09-0359 [DOI] [PubMed] [Google Scholar]
- Sugino T., Maruyama M., Tanno M., Kuno A., Houkin K., Horio Y. (2010). Protein deacetylase SIRT1 in the cytoplasm promotes nerve growth factor-induced neurite outgrowth in PC12 cells. FEBS Lett. 584, 2821–2826. 10.1016/j.neures.2010.07.1100 [DOI] [PubMed] [Google Scholar]
- Sun J. H., Yu J. T., Tan L. (2014). The role of cholesterol metabolism in Alzheimer’s disease. Mol. Neurobiol. [Epub ahead of print]. 10.1007/s12035-014-8749-y [DOI] [PubMed] [Google Scholar]
- Tang B. L. (2009). Sirt1’s complex roles in neuroprotection. Cell. Mol. Neurobiol. 29, 1093–1103. 10.1007/s10571-009-9414-2 [DOI] [PubMed] [Google Scholar]
- Tang B. L. (2011). Sirt1’s systemic protective roles and its promise as a target in antiaging medicine. Transl. Res. 157, 276–284. 10.1016/j.trsl.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Theendakara V., Patent A., Peters Libeu C. A., Philpot B., Flores S., Descamps O., et al. (2013). Neuroprotective Sirtuin ratio reversed by ApoE4. Proc. Natl. Acad. Sci. U S A 110, 18303–18308. 10.1073/pnas.1314145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberi L., van den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., et al. (2012). BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat. Neurosci. 15, 1627–1635. 10.1038/nn.3264 [DOI] [PubMed] [Google Scholar]
- Tissenbaum H. A., Guarente L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230. 10.1038/35065638 [DOI] [PubMed] [Google Scholar]
- Tong X. K., Nicolakakis N., Fernandes P., Ongali B., Brouillette J., Quirion R., et al. (2009). Simvastatin improves cerebrovascular function and counters soluble amyloid-beta, inflammation and oxidative stress in aged APP mice. Neurobiol. Dis. 35, 406–414. 10.1016/j.nbd.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Toorie A. M., Nillni E. A. (2014). Minireview: central Sirt1 regulates energy balance via the melanocortin system and alternate pathways. Mol. Endocrinol. 28, 1423–1434. 10.1210/me.2014-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham T. J., Thijssen K. L., Breitling R., Hofstra R. M. W., Plasterk R. H. A., Nollen E. A. A. (2008). C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 4:e1000027. 10.1371/journal.pgen.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M., Guarente L. (2011). Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature 477, E1–E2. 10.1038/nature10440 [DOI] [PubMed] [Google Scholar]
- Wang W., Esbensen Y., Kunke D., Suganthan R., Rachek L., Bjørås M., et al. (2011). Mitochondrial DNA damage level determines neural stem cell differentiation fate. J. Neurosci. 31, 9746–9751. 10.1523/jneurosci.0852-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ho L., Qin W., Rocher A. B., Seror I., Humala N., et al. (2005). Caloric restriction attenuates beta-amyloid neuropathology in a mouse model of Alzheimer’s disease. FASEB J. 19, 659–661. 10.1096/fj.04-3182fje [DOI] [PubMed] [Google Scholar]
- Weir H. J. M., Lane J. D., Balthasar N. (2013). SIRT3: a central regulator of mitochondrial adaptation in health and disease. Genes Cancer 4, 118–124. 10.1177/1947601913476949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow C. A., Kimura K. D., Lee M. S., Ruvkun G. (2000). Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147–150. 10.1126/science.290.5489.147 [DOI] [PubMed] [Google Scholar]
- Wood W. G., Li L., Müller W. E., Eckert G. P. (2014). Cholesterol as a causative factor in Alzheimer’s disease: a debatable hypothesis. J. Neurochem. 129, 559–572. 10.1111/jnc.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. G., Rogina B., Lavu S., Howitz K., Helfand S. L., Tatar M., et al. (2004). Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689. 10.1038/nature02789 [DOI] [PubMed] [Google Scholar]
- Wu D., Qiu Y., Gao X., Yuan X. B., Zhai Q. (2011). Overexpression of SIRT1 in mouse forebrain impairs lipid/glucose metabolism and motor function. PLoS One 6:e21759. 10.1371/journal.pone.0021759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Ying Z., Gomez-Pinilla F. (2006). Oxidative stress modulates Sir2alpha in rat hippocampus and cerebral cortex. Eur. J. Neurosci. 23, 2573–2580. 10.1111/j.1460-9568.2006.04807.x [DOI] [PubMed] [Google Scholar]
- Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., et al. (2004). Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380. 10.1038/sj.emboj.7600244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhary S. M., Ayubcha D., Dileo J. N., Jose R., Leheste J. R., Horowitz J. M., et al. (2010). Distribution analysis of deacetylase SIRT1 in rodent and human nervous systems. Anat. Rec. (Hoboken) 293, 1024–1032. 10.1002/ar.21116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Chen G., Fan D., Deng M. (2011a). Inhibition of Sirt1 promotes neural progenitors toward motoneuron differentiation from human embryonic stem cells. Biochem. Biophys. Res. Commun. 404, 610–614. 10.1016/j.bbrc.2010.12.014 [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang S., Gan L., Vosler P. S., Gao Y., Zigmond M. J., et al. (2011b). Protective effects and mechanisms of sirtuins in the nervous system. Prog. Neurobiol. 95, 373–395. 10.1016/j.pneurobio.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Luo P., Guo Q., Li S., Zhang L., Zhao M., et al. (2012). Interactions between SIRT1 and MAPK/ERK regulate neuronal apoptosis induced by traumatic brain injury in vitro and in vivo. Exp. Neurol. 237, 489–498. 10.1016/j.expneurol.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Zocchi L., Sassone-Corsi P. (2012). SIRT1-mediated deacetylation of MeCP2 contributes to BDNF expression. Epigenetics 7, 695–700. 10.4161/epi.20733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic I. B., Paulukaitis B. S., Day J. J., Etikala D. M., Sweatt J. D. (2014). Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515, 582–586. 10.1038/nature13707 [DOI] [PMC free article] [PubMed] [Google Scholar]