Abstract
Chromosome 11q13.5 containing RSF1 (HBXAP), a gene involved in chromatin remodeling, is amplified in several human cancers including ovarian carcinoma. Our previous studies demonstrated requirement of Rsf-1 for cell survival in cancer cells, which contributed to tumor progression; however, its role in tumorigenesis has not yet been elucidated. In this study, we co-immunoprecipitated proteins with Rsf-1 followed by nanoelectrospray mass spectrometry and identified cyclin E1, besides SNF2H, as one of the major Rsf-1 interacting proteins. Like RSF1, CCNE1 is frequently amplified in ovarian cancer, and both Rsf-1 and cyclin E1 were found co-upregulated in ovarian cancer tissues. Ectopic expression of Rsf-1 and cyclin E1 in non-tumorigenic TP53mut RK3E cells led to an increase in cellular proliferation and tumor formation by activating cyclin E1-associated kinase (CDK2). Tumorigenesis was not detected if either cyclin E1 or Rsf-1 was expressed, or they were expressed in a TP53wt background. Domain mapping showed that cyclin E1 interacted with the first 441 amino acids of Rsf-1. Ectopic expression of this truncated domain significantly suppressed G1/S-phase transition, cellular proliferation and tumor formation of RK3E-p53R175H/Rsf-1/cyclin E1 cells. The above findings suggest that Rsf-1 interacts and collaborates with cyclin E1 in neoplastic transformation and TP53 mutations are prerequisite for tumor-promoting functions of RSF/cyclin E1 complex.
Keywords: Rsf-1, HBXAP, cyclin E1, chromatin remodeling, TP53mut, cancer development
Introduction
Chromatin remodeling plays fundamental roles in regulating nucleosome distribution and compactness of chromatin. As such, it participates in diverse genomic events including transcriptional regulation, DNA synthesis, damage repair, methylation and recombination. Cancer cells exhibiting aberration of chromatin remodeling activity due to defects in or aberrant expression of constituent proteins are positively selected during clonal evolution. This is evidenced by frequent somatic mutations in several SWI/SNF chromatin remodeling genes including BRG1 (SMARCA4) in lung cancer [1, 2], PBRM1 (BAF180) in renal cell carcinoma [3], ARID1A (BAF250A) in endometrium-related carcinoma [4–6] and ARID2 in hepatocellular carcinoma [7]. Moreover, analysis of DNA copy number in ovarian high-grade serous carcinoma identified a discrete amplicon at ch11q13.5 [8] and functional screening of genes within this locus demonstrated RSF1 (A.K.A. HBXAP) the most likely “driver” because Rsf-1 knockdown but not other genes within this amplicon was associated with an increased sensitivity of ovarian cancer cells to paclitaxel [9]. RSF1 encodes Rsf-1 that binds to a nucleosome-dependent ATPase, SNF2H, forming the RSF (Remodeling and Spacing Factor) ISWI chromatin remodeling complex [8, 10–12].
Clinical correlation studies found that RSF1 amplification and upregulation are significantly associated with disease aggressiveness and poor clinical outcome in patients with ovarian high-grade serous carcinoma [8, 13]. Similar findings were also reported in patients with ovarian clear cell carcinoma [14], oral squamous carcinoma [15], nasopharyngeal carcinoma [16], gall bladder carcinoma [17], lung cancer [18] and colorectal cancer [19, 20]. Therefore, we have investigated tumor-promoting functions of Rsf-1 and shown its expression essential for cellular survival and proliferation, especially in the presence of cytotoxic chemotherapeutic agents, in ovarian cancer cells with RSF1 amplification/overexpression but not in those without [9, 21]. Ectopic expression of Rsf-1 in SKOV3 ovarian cancer cells, which expressed a low level of endogenous Rsf-1, enhanced tumor growth in tumor xenografts [21]. Furthermore, Rsf-1 overexpression led to DNA damage and was associated with chromosomal instability in vitro and in vivo [22, 23]. These results strongly suggest that RSF1 amplification and its upregulation are critical in the development of ovarian high-grade serous carcinoma.
In order to investigate how Rsf-1 participates in tumor development, we analyzed the proteins that co-immunoprecipitated with Rsf-1 in OVCAR3 ovarian cancer cells. Identification and characterization of the interacting proteins historically provide important clues how proteins of a given pathway form interaction networks that when activated reprogram cellular functions such as differentiation and tumorigenesis. We found cyclin E1 as one of major proteins that complexed with Rsf-1. We demonstrated that Rsf-1 co-upregulated and co-immunoprecipitated with cyclin E1 in ovarian cancer tissues, and determined whether Rsf-1 expression alone or in combination with cyclin E1 promotes tumorigenesis in a mouse xenograft tumor model. Findings from this study provide new insights into molecular collaboration of cancer-associated genes frequently altered in ovarian cancer in promoting tumor development. Our results also suggest molecular cross-talks between molecules involved in chromatin remodeling and cell cycle regulation.
Materials and Methods
Human tumor tissues and immunohistochemistry
A total of 127 high-grade ovarian serous carcinomas were analyzed for expression levels of Rsf-1 and cyclin E1. Tissues were obtained from Department of Pathology at the Johns Hopkins Hospital (Baltimore, MD) under the approval of Johns Hopkins institutional review board. Immunohistochemistry was performed on paraffin sections using following primary antibodies: anti-Rsf-1 (Upstate) [8, 24] and anti-cyclin E1 (Zymed) [25]. Immunoreactivity was developed using an EnVision+System peroxidase kit (DAKO), and the intensity was independently scored by two investigators. For discordant cases, immunointensity was scored by a third investigator and the final intensity score was determined by majority scores.
As Rsf-1 nuclear staining was homogenous, we evaluated immunointensity using a method previously described [8, 24] and scored as negative (0), weakly positive (1+), moderately positive (2+), strongly positive (3+), or intensely positive (4+). Cyclin E1 immunoreactivity was heterogeneous; therefore, we used a combination of the percentage of positively stained cells and the intensity of nuclear staining. The H-score = ∑Pi xi was calculated where i was the intensity of stained tumor cells (0 to 4+), and Pi is the percentage of stained tumor cells for each intensity group (0 to 100%).
Co-immunoprecipitation and nanoelectrospray mass spectrometry
OVCAR3 cells, a line shows an 8-fold increase in DNA copy number at RSF1 locus by quantum dot electrophoretic mobility shift assay [26], were lysed in ice-cold RIPA buffer (Cell Signaling). Soluble lysates were incubated with anti-Rsf-1 antibodies and protein-A gel. After washes, precipitated proteins were separated by SDS-PAGE and visualized by silver staining. Protein bands of interest in gel were excised and subjected to in-gel digestion using a previous method [27]. NanoLC-MS/MS analysis was performed using the Ultimate Capillary LC System (LC Packings) coupled to a QSTARXL quadrupole-time of flight (Q-TOF) mass spectrometer (Applied Biosystems/MDS Sciex). Data acquisition was performed using the Information Dependent Acquisition (Applied Biosystems) software. Ion spectra generated by nanoLC-MS/MS were searched against NCBI databases for exact matches using the ProID (Applied Biosystem/MDS Sciex) and MASCOT programs [28]. A Homo sapiens taxonomy restriction was used, and the mass tolerance of both precursor ions and fragment ions was set to 0.3 Da.
Expression constructs of Rsf-1, cyclin E1 and p53mut, and their tumorigenicity in immunocompromised mice
A Tet-off inducible system was generated in RK3E non-transformed cells by transfecting cells with the tetracycline-controlled transactivator (tTA) vector (Clontech). Full length Rsf-1 (tagged with V5 at C-terminus) and cyclin E1 were cloned into a pBI inducible vector (Clontech), and these expression vectors were subsequently introduced into RK3E-tTA cells. To determine whether TP53mut was required for Rsf-1 to promote tumor development, the above selected clones were further transfected with either a pcDNA4C vector (Invitrogen) or a vector containing a human p53R175H (tagged with Xpress at N-terminus). A total of six transfectants were generated: RK3E-Rsf-1, RK3E-cyclin E1, RK3E-p53R175H/Rsf-1, RK3E-p53R175H/cyclin E1, RK3E-Rsf-1/cyclin E1, and RK3E-p53R175H/Rsf-1/cyclin E1. Cell clones were validated by Western blot analysis and immunostaining. To assess whether Rsf-D1 (1–441 a.a.) disrupts the interaction between Rsf-1 and cyclin E1, the Rsf-D1 segment (tagged with V5 at C-terminus) was cloned into a pcDNA6B/His vector (Invitrogen) and then introduced into RK3E-p53R175H/Rsf-1/cyclin E1 cells. For proliferation assay, cell number was measured at various time points by SYBR green I nucleic acid staining (Molecular Probes).
Equal numbers of cells (5×106 cells per injection) were injected into subcutaneous tissue of immunocompromised (nu/nu) mice. RK3E-p53R175H/Rsf-1/cyclin E1 cells with Rsf-D1 expression or vector control were also injected into mice to evaluate anti-RSF/cyclin E1 complex activity. Six weeks after cell inoculation, mice were sacrificed, and tumors were excised, weighed, and prepared for histological examination and immunohistochemistry.
Kinase activity assay
Protein lysates prepared from cells with Rsf-1/cyclin E1 turned-on or –off were collected with ice-cold RIPA buffer. Kinases in lysates were captured in ELISA plates pre-coated with anti-cyclin E1 (Zymed) or anti-CDK2 (Upsate) antibodies and kinase activity was measured by using an ADP-Glo reagent system (Promega). Six replicates were performed for each treatment and data were presented as mean±S.D.
Results
Rsf-1 interacts with cyclin E1
To identify candidate proteins complexing with Rsf-1, we performed immunoprecipitation in OVCAR3 cells known to amplify and overexpress RSF1 [26]. Silver staining of the immunoprecipitates demonstrated a robust protein with a molecular mass corresponding to SNF2H and several novel bands including one with a molecular mass of approximately 47 kDa (Figure 1A). Mass spectrometry followed by peptide sequencing identified the 47 kDa protein as cyclin E1 (Figure 1B). Reciprocal co-immunoprecipitation indicated the direct interaction between Rsf-1 and cyclin E1 (Figure 1C). Moreover, SNF2H can be also immunoprecipitated with cyclin E1 (Figure 1D), which occurs only when Rsf-1 was turned-on in Rsf-1 inducible SKOV3 cells [9, 21]. These results confirmed the interaction between cyclin E1 and RSF complex via direct binding to Rsf-1, but not SNF2H.
Figure 1.
Cyclin E1 forms a complex with Rsf-1 and SNF2H. (A) Silver stained SDS-PAGE gel shows several novel proteins as well as SNF2H co-immunoprecipitated with Rsf-1. Protein G with an irrelevant antibody was used as the negative control. (B) NanoLC-MS/MS analysis revealed that the cyclin E1 band contained several peptide sequences (bold fonts with underlines) that matched cyclin E1 protein sequence. (C) Reciprocal co-immunoprecipitation demonstrates complex formation between Rsf-1 and cyclin E1. Lane 1: protein G/Ab blank control; lane 2: co-immunoprecipitation; lane 3: cell lysate (5% input). (D) Using Rsf-1 inducible SKOV3 cells, both Rsf-1 and SNF2H co-immunoprecipitated with cyclin E1 when Rsf-1 was turned-on. Lane 1: protein G control; Lane 2: Rsf-1 turned-on; Lane 3: Rsf-1 turned-off. The total input lysates in this experiment have been adjusted so the total amount of SNF2H in each group remains similar.
Co-upregulation of Rsf-1 and cyclin E1 in ovarian high-grade serous carcinomas
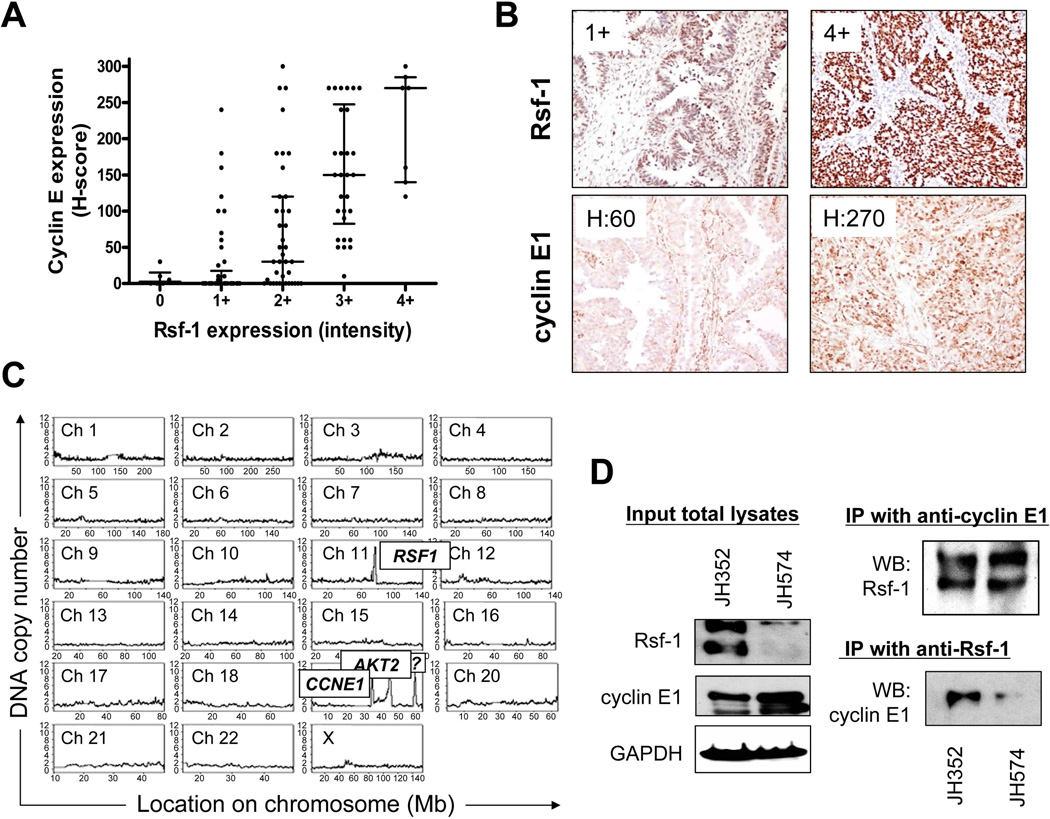
To test if Rsf-1 and cyclin E1 co-upregulated, we correlated expression levels of Rsf-1 and cyclin E1 using immunohistochemistry in 127 ovarian high-grade serous carcinomas. Figures 2A and 2B indicated positive correlation between Rsf-1 and cyclin E1 immunoreactivity with a Spearman’s rank correlation coefficient of 0.661 and P<0.0001. Especially, cancer cells with Rsf-1 immunointensity score ≥3 had higher percentages of cells showing high cyclin E1 immunoreactivity than those with Rsf-1 score ≤2 (Figures 2B). The cutoff of Rsf-1 immunointensity score (≤2 versus ≥3) used in this report was based on our previous study [8] showing that samples with Rsf-1 score ≥3 contained RSF1 amplification in ovarian carcinoma. This cutoff has been independently used in other clinical studies [24, 29]. In fact, genomewide studies by digital karyotyping indicated the existence of co-amplification at both RSF1 and CCNE1 loci in OVCAR3 cells (Figure 2C) [8].
Figure 2.
Expression of Rsf-1 and cyclin E1 in high-grade ovarian serous carcinomas. (A) Scatter plot of Rsf-1 and cyclin E1 immunoreactivity based on immunohistochemistry. The majority of tumors with Rsf-1 overexpression (intensity score ≥ 3) express a high level of cyclin E1. Each symbol represents an individual high-grade serous carcinoma. Error bar: mean ± interquartile range. (B) Rsf-1 and cyclin E1 staining in two representative specimens. One exhibits an intense Rsf-1 staining with an intensity score of 4+ and a high level of H-score for cyclin E1. The other specimen shows weak Rsf-1 immunoreactivity with intensity score of 1+ and demonstrates an H-score of 60 for cyclin E1. (C) Genome-wide study of genetic alterations by digital karyotyping reveals co-amplification of tumor driver genes in OVCAR3 cells, including RSF1 and CCNE1 [26]. (D) Reciprocal co-immunoprecipitations in two ovarian high-grade serous carcinoma specimens, JH352 and JH574.
To further validate the interaction between Rsf-1 and cyclin E1 in ovarian cancer, we performed co-immunoprecipitation in two representative tumor tissues: JH352 with abundant Rsf-1 and cyclin E1, and JH574 with abundant cyclin E1 and modest amount of Rsf-1. In both specimens, Rsf-1 was found co-immunoprecipitated with cyclin E1 (Figure 2D), indicating biological significance of the interaction between Rsf-1 and cyclin E1 in ovarian high-grade serous carcinoma.
Rsf-1 and cyclin E1 promote tumorigenicity in the presence of p53mut
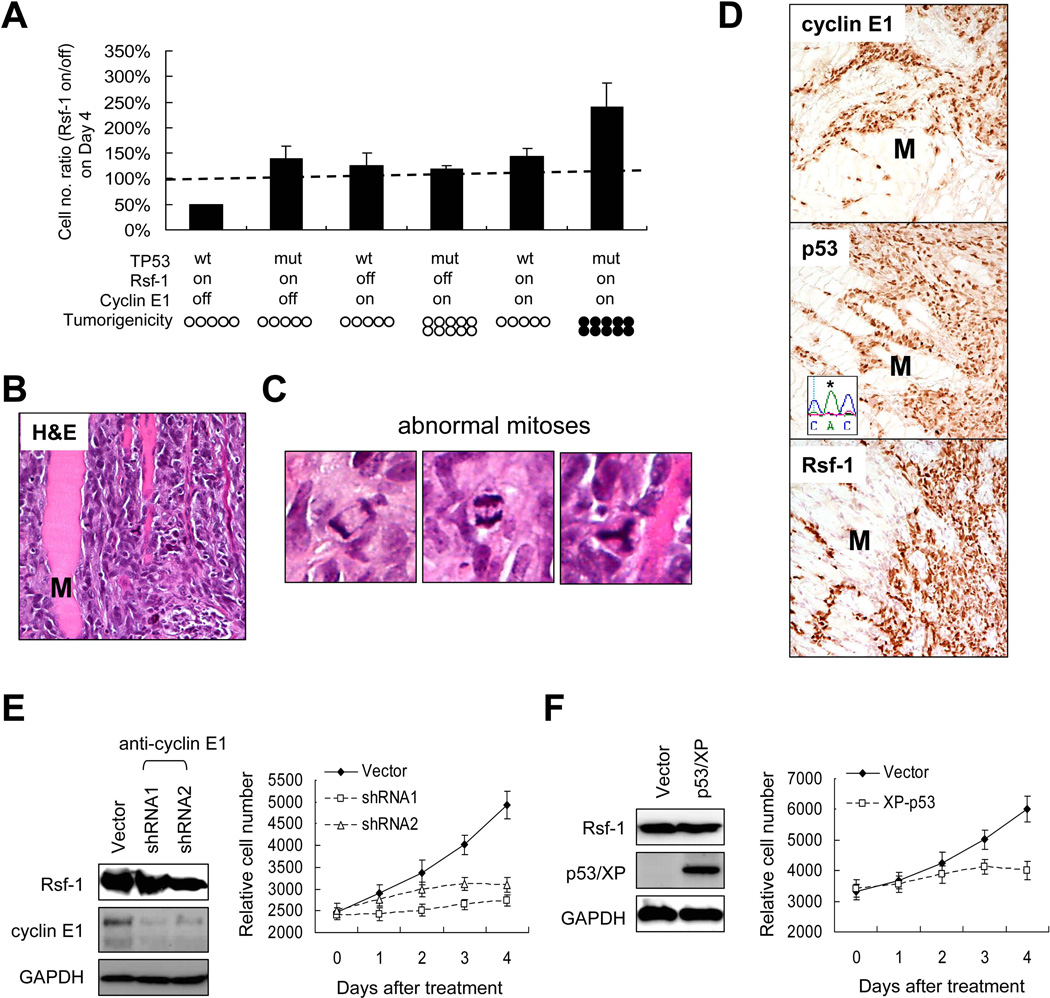
Given the roles of Rsf-1 and cyclin E1 in cancer pathogenesis [25, 30–33] and their upregulation in serous tubal intraepithelial carcinomas, the precursor lesions of ovarian high-grade serous carcinoma [34], we determined if ectopic expression of both proteins could transform non-tumorigenic epithelial cells. Since our recent study demonstrated a prerequisite of p53 functional loss for Rsf-1 to promote tumor progression [22], we reconstituted Rsf-1 expression together with cyclin E1 and human p53R175H in RK3E cells. RK3E cell line, a cellular model widely used for studying transforming ability of potential oncogenes [35–40]. Notably, this cell line was amenable for stable expression after multiple transfections of different constructs as other non-transformed cells and primary cultures were not available for this purpose. Furthermore, our previous study has shown the capability of ectopic Rsf-1 to form RSF chromatin remodeling complex with endogenous rat SNF2H in this cell line [22]. Using Western blot and co-immunoprecipitation analyses, we detected increased levels of Rsf-1 and cyclin E1 in those inducible RK3E clones as early as 6 hours after induction (Supplemental Figure 1A) and ectopic cyclin E1 can interact with RSF complex (Supplemental Figure 1B). Cell growth assay showed that RK3E cells co-expressing Rsf-1, cyclin E1, and p53R175H produced the highest cell numbers in vitro as compared to cells expressing other combinations of those genes (Figure 3A). We then injected individual groups of RK3E cells into subcutaneous tissue of immunocompromised mice and found that cells co-expressing Rsf-1, cyclin E1, and p53R175H produced tumors in all mice tested. By contrast, none of other groups produced grossly detectable tumors (Figure 3A).
Figure 3.
Effects of combined expression of Rsf-1, p53R175H, and cyclin E1 in cellular proliferation and tumorigenicity. (A) A series of stable RK3E cells with different combinations in expressing Rsf-1, p53R175H, and cyclin E1 were generated. In vitro, RK3E cells that expressed Rsf-1, cyclin E1, and p53R175H showed the highest cell number as compared to cells in other groups. The data are expressed as the ratio of cell numbers (gene turned-on/off) at Day 4. Cells co-expressing Rsf-1, p53R175H, and cyclin E1 produced subcutaneous tumors in all 10 mice 4 weeks after injection, whereas none of other groups produced tumors in any mice. Filled circles, mice developing tumors; open circles, mice without tumors. Each circle represents a mouse in the experiment. (B) Microscopically, the RK3E tumors were highly invasive as the tumor cells deeply infiltrated the surrounding skeletal muscle (M) (hematoxylin and eosin stain). (C) Left panel, higher magnification of anaphase bridges. Middle panel, a tri-polar metaphase. Right panel, a tri-polar mitotic figure with anaphase bridges. (D) Immunohistochemistry demonstrated diffuse and intense immunoreactivity of Rsf-1, p53mut, and cyclin E in a representative tumor. Skeletal muscle (M) serves as a negative control. Sequence analysis of genomic DNA purified from the tumor confirms the TP53mut from CGC to CAC (asterisk). Cyclin E1 depletion by specific shRNAs (E) or p53wt restoration (F) caused cell growth suppression in OVCAR3 cells,
Microscopic examination of excised RK3E tumors revealed a highly invasive pattern with tumor cells deeply infiltrating into surrounding normal tissues (Figure 3B). Abnormal mitotic figures including tri-polar or tetra-polar metaphase (8.2% ± 4.8%) and anaphase bridges (2.0% ± 1.8%) were readily observed (Figure 3C). Immunohistochemistry confirmed the expression of Rsf-1, p53R175H, and cyclin E1 in all tumors (Figure 3D). Sequence analysis of genomic DNA purified from mouse tumors showed the introduced human p53R175H (CGC to CAC) coding an amino acid change of R to H at the position 175. To further understand the functional interaction among Rsf-1, cyclin E1 and p53 in cancer, cyclin E1 depletion (Figure 3E) or p53 restoration (Figure 3F) was found sufficient to reduce cell growth in OVCAR3 cells which expressed a high level of Rsf-1, suggesting the essential role of cyclin E1 for Rsf-1 in promoting cell growth in cancer cells with a TP53mut genetic background.
Identification of the cyclin E1-binding domain on Rsf-1
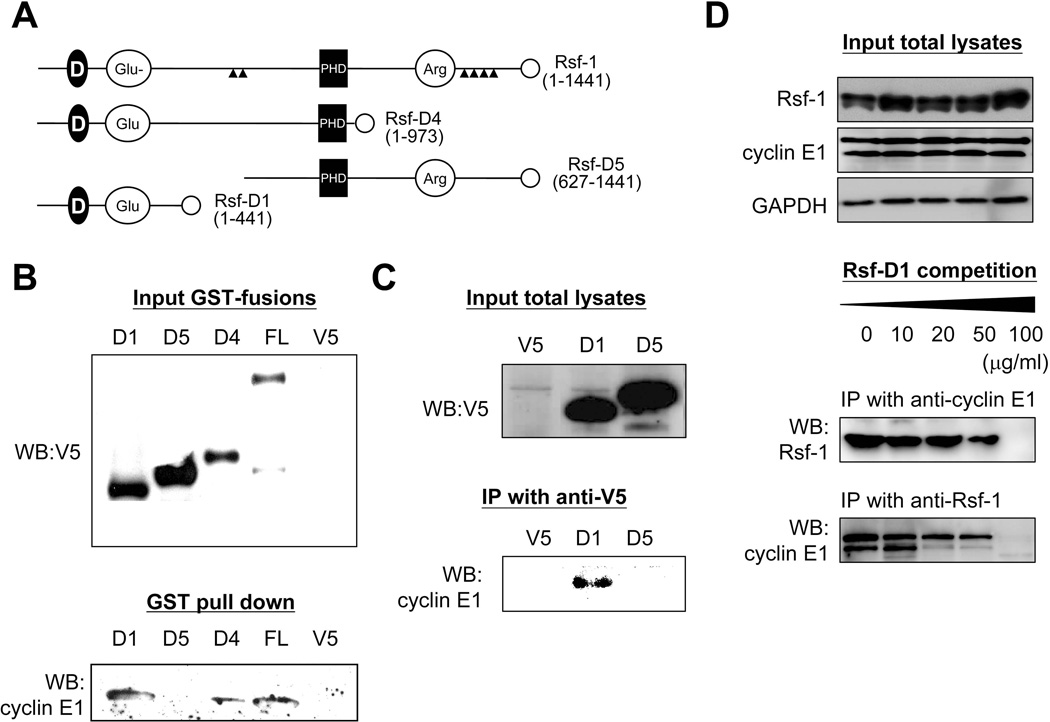
In order to assess biological roles of cyclin E1 in complex formation with Rsf-1, we tried to identify the minimal domain on Rsf-1 binding to cyclin E1. Several Rsf-1 deletion mutants with a V5-tag at their C-termini were fused with a GST-tag and utilized to perform co-immunoprecipitation with cyclin E1 (Figure 4A). GST pull-down assay revealed that cyclin E1 co-immunoprecipitated with Rsf-D1 and Rsf-D4 fragments in addition to the full-length Rsf-1 (Figure 4B). Moreover, using anti-V5 antibodies, we were able to co-immunoprecipitate Rsf-D1/V5 and cyclin E1 in OVCAR3 cells (Figure 4C). More importantly, excessive exogenous Rsf-D1 reduced the interaction between Rsf-1 and cyclin E1 in a dose-dependent manner, indicating a competition between Rsf-D1 and Rsf-1 for cyclin E1 binding (Figure 4D). Thus, we confirmed Rsf-D1 (amino acids 1–441) the cyclin E1-binding domain on Rsf-1. Rsf-D1 contained a DDT domain involved in protein interaction and a Glu-rich transactivating domain for transcription factors, but lacked the PHD and Arg-rich domains (Figure 4A).
Figure 4.
Rsf-D1 (1–441 a.a.) is responsible for interacting with cyclin E1. (A) Schematic presentation of the Rsf-1 deletion mutants tagged with V5 on their C-termini (open circles). D, a conserved DDT domain for DNA-binding homeobox-containing proteins; Glu, glutamine-rich domain; PHD, plant homeodomain; Arg, arginine-rich domain. (B) Expression of various Rsf-1 deletion mutants (top) and GST pull-down of mutant proteins that co-immunoprecipitate with cyclin E1 (bottom). (C) The interaction of Rsf-1 and cyclin E1 was further confirmed by immunoprecipitation of V5 tagged Rsf-1 fragments and Western blot with anti-cyclin E1 antibodies. (D) The interaction between Rsf-1 and cyclin E1 could be blocked by excessive exogenous Rsf-D1.
RSF/cyclin E1 complex controls G1/S-phase transition
To study the possible roles of Rsf-1 in cell cycle regulation, we synchronized RK3E-p53R175H/Rsf-1/cyclin E1 cells by the double-thymidine block method. After cells were released from G1/S-phase and entered the S-phase, the expression level of Rsf-1 dramatically reduced and upregulated again when cells were entering G1-phase. The same pattern could also be detected in cyclin E1 (Figure 5A). The above data suggest that Rsf-1 may collaborate with cyclin E1 in regulating G1/S check point. To test this possibility, we starved the cells under Rsf-1/cyclin E1-turned off and -turned on conditions for 24 hrs, and re-stimulated the cells with 10% FBS. Our data demonstrated that co-overexpression of Rsf-1 and cyclin E1 further promoted G1/S-transition after serum stimulation, resulting in higher population of cells at S- and G2/M-phases (Figure 5B). Kinase activity assays also confirmed that the corresponding activity of CDK2 (or cyclin E1-associated kinase complex) could be enhanced by high levels of Rsf-1 and cyclin E1 as compared to the Rsf-1/cyclin E1-turned off control (Figure 5C). As expected, Rsf-D1 expression in Rsf-1/cyclin E1-turned on cells blocked such growth-stimulating signals, resulting in a G1/S-transition rate and CDK2 activity similar to the ones in Rsf-1/cyclin E1-turned off cells.
Figure 5.
Involvement of the RSF/cyclin E1 complex in G1/S-phase transition. (A) Expression levels of Rsf-1 change alone with cell cycle progression with the highest level at G1/S-phase transition and lowest levels at S-phase. Right panel indicates cell population of cell cycle phases at indicated time points after cells were released from double-thymidine block. (B) The G1/S-phase transition rate of treated cells was measured by flow cytometry analyses after 24-hrs serum starvation followed by 10% FBS stimulation. (C) CDK2- or Cyclin E1-associated kinase activity in treated cells was measured by ELISA assay using either anti-CDK2 or anti-cyclin E antibodies to capture kinase complex.
Full-length Rsf-1 is required for tumor-promoting activity of the RSF/cyclin E1 complex
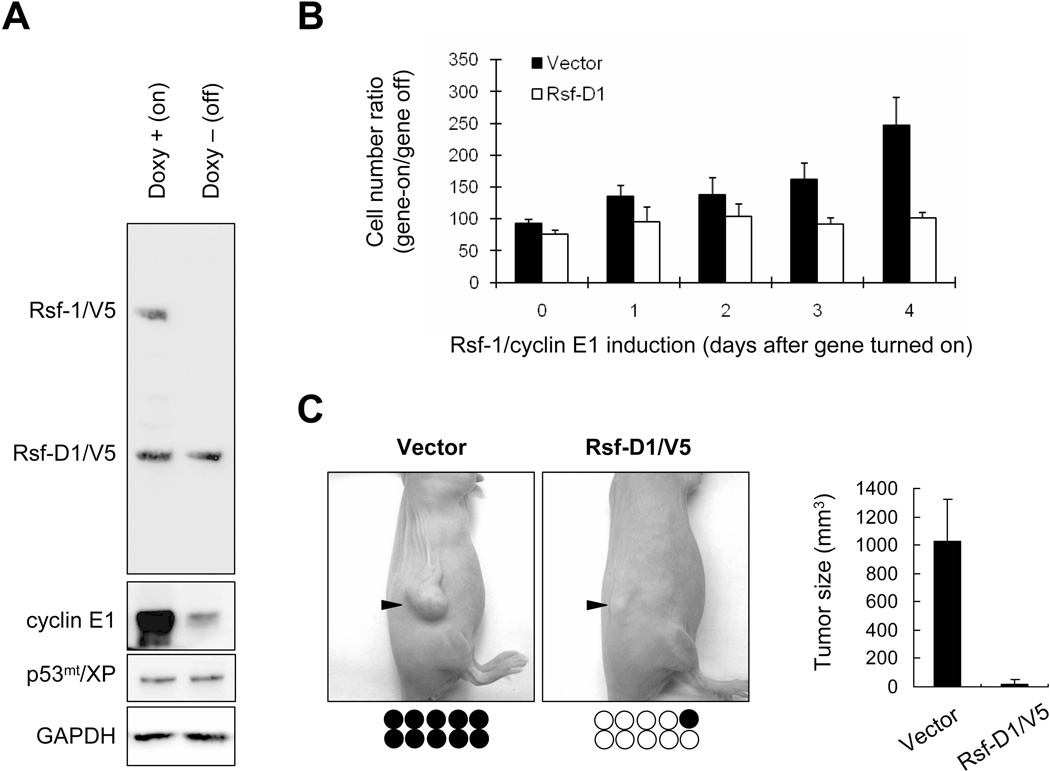
To know if the minimal interacting region determines gross tumor growth, RK3E-p53R175H/Rsf-1/cyclin E1 cells were stably transfected with an Rsf-D1/V5 construct or an empty vector as the negative control. Western blot analysis demonstrated that the selected stable clone constitutively expressed Rsf-D1/V5 and p53R175H/Xpress as detected by anti-V5 and anti-Xpress antibodies, respectively (Figure 6A). Proliferation assays indicated that RK3E-p53R175H/Rsf-1/cyclin E1 cells transfected with the empty vector were highly proliferative when both Rsf-1 and cyclin E1 were turned-on (Figure 6B). By contrast, the growth rate of cells transfected with Rsf-D1 did not significantly change between Rsf-1/cyclin E1 turned-on and turned-off groups. Next, we assessed whether such minimal binding domain affected tumorigenicity of RK3E-p53R175H/Rsf-1/cyclin E1 cells by injecting cells with and without Rsf-D1 expression to subcutaneous tissue of nu/nu mice. We found that the vector control group formed tumors in 10 of 10 (100%) injection sites whereas the Rsf-D1 expressing group formed tumors in only one of 10 (10%) injection sites (Figure 6C). The tumor weight in control group was also significantly higher than that in Rsf-D1 group (p< 0.001).
Figure 6.
Rsf-D1 (1–441 a.a.) decreases tumor-promoting activity of RSF/cyclin E1 complex. Rsf-D1/V5 gene was introduced into the tumorigenic RK3E cells co-expressing p53R175H, Rsf-1 and cyclin E1. (A) Western blot analysis demonstrated the selected stable clone constitutively expressing Rsf-D1 (anti-V5) and p53R175H (anti-Xpress). GAPDH served as the loading control. (B) Rsf1-D1 overexpression attenuates RSF/cyclin E1 proliferation-enhancing activity. RK3E-p53R175H/Rsf-1/cyclin E1 cells transfected with the empty vector showed high proliferative activity when both Rsf-1 and cyclin E1 were turned-on. By contrast, growth rate of cells transfected with Rsf-D1 vector was not significantly changed when Rsf-1 and cyclin E1 were turned-off. The data are expressed as the ratio of cell numbers (gene turned-on/off). (C) Both vector and Rsf-D1 transfected cells were injected into immunocompromised (nu/nu) mice, with two injection sites per mouse (five mice per group). Vector treated cells produced tumors in 100% (10/10) of sites with larger tumor size, as compared to Rsf-D1 expressing cells which produced tumors in 10% (1/10) of sites. Filled circles, injection sites developing tumors; open circles, injection sites without tumors. Each circle represents an individual injection site.
Discussion
Our study provides clear evidence that cyclin E1 directly interacts with RSF ISWI complex through binding to Rsf-1, a finding suggestive of a molecular cross-talk between chromatin remodeling and cell cycle regulation. This finding is interesting because only SNF2H has been reproducibly reported so far as the binding partner of Rsf-1. Since RSF chromatin remodeling participates in a variety of nuclear activity including regulation of gene expression [41], it is likely that RSF complex interacts with other nuclear proteins to exert its context-dependent chromatin remodeling functions. With the recent finding of cyclin D1 in assembling/stabilizing RAD51 DNA repair complex [42], our study supports a non-catalytic function for cyclin E1 besides a cell cycle regulator. It is plausible that excessive Rsf-1 recruits and directs cyclin E1 to specific subchromosomal regions involving active chromatin remodeling where DNA synthesis and repair take place, although the detailed mechanisms await further investigation. The molecular collaboration between Rsf-1 and CCNE1, two frequently amplified oncogenes in ovarian cancer, provides a new opportunity in understanding the role of ISWI chromatin remodeling in cancer.
Complex formation between Rsf-1 and cyclin E1 suggests that they are co-upregulated in cancer. In fact, Rsf-1 induction is associated with increased cyclin E1 in Rsf-1-nducible SKOV3 cells, probably due to complex stabilization (Supplementary Figure 2). Furthermore, a significant positive correlation between Rsf-1 and cyclin E1 expression was found in ovarian high-grade serous carcinomas. Excessive Rsf-1 and cyclin E1 proteins may collaborate to promote oncogenesis by increasing cellular proliferation that overrides negative regulatory mechanisms operating in non-transformed cells. To test this hypothesis, we engineered RK3E cells to express different combinations of Rsf-1, cyclin E1, and p53mut. We constitutively express cyclin E1 because upregulation of cyclin E1 by Rsf-1 may not achieve the level as high as that in in ovarian cancer cells. Mutant p53 was introduced because our recent study showed TP53mut essential for non-transformed cells to survive following ectopic expression of Rsf-1, which otherwise would trigger DNA strand breaks followed by DNA damage response leading to cell cycle arrest and apoptosis [22]. Cell cycle analysis revealed that the interaction between Rsf-1 and cyclin E1 enhances the activity of CDK2/cyclin E complex and promotes G1/S transition rate in cells with p53mut. Indeed, Rsf-1 could increase the efficiency of tumor formation in mice only if the cells had also been engineered to express cyclin E1 and p53mut, supporting molecular collaboration of Rsf-1, cyclin E1 and TP53mut in tumor initiation. Expression of Rsf-1 deletion mutants (Rsf-D1 and Rsf-D4) that contain the binding domain to cyclin E1 abolishes growth stimulating effects and tumorigenicity in mice. Although Rsf-D1 and Rsf-D4 may have effects on cancer cells other than binding to cyclin E1, our finding suggests Rsf-1-cyclin E1 interaction a prerequisite for their oncogenic collaboration.
Several molecular models have been developed to simulate tumorigenesis of ovarian high-grade serous carcinoma, but their biological significance remains elusive, mainly because genes like SV40 T antigen and H-RAS [43, 44] introduced into those cell models appear biologically irrelevant. By contrast, our approach might provide a more real scenario of how cancer develops in human ovary, especially for ovarian high-grade serous carcinoma. First, we overexpressed both cyclin E1 and Rsf-1 because genomic loci harboring CCNE1 and RSF1 are among the most frequently amplified chromosomal regions in this tumor type. Furthermore, both genes are among the top 60 “driver” genes that show the most significant correlation between DNA copy number and RNA expression level by analyzing whole ovarian cancer genome from the Tumor Genome Atlas (TCGA) data set [45]. Clinically, upregulation of both genes is associated with a worse clinical outcome in patients [8, 25, 29, 31, 46, 47], and biologically, expression of both RSF1 and CCNE1 are able to stimulate cellular growth in vitro and propel tumor progression in vivo [18, 25, 46].
Similarly, TP53mut was introduced in this model because of its high prevalence (>85%) in high-grade ovarian serous carcinomas [48]. More importantly, somatic TP53 mutations and upregulation of Rsf-1 and cyclin E1 occur very early in tumor progression as observed in non-invasive (in situ) precursor lesions, serous tubal intraepithelial carcinoma, but not in the adjacent normal tubal epithelium [34, 49]. Thus, we proposed the defined molecular alterations sufficient for tumorigenicity based on co-expression of three early and prevalent molecular changes. Future engineered mouse models that express Rsf-1, cyclin E1, and p53mut in ovary and fallopian tube are required to determine if this is the case in developing ovarian cancer.
In summary, our study demonstrates a role for interaction between cyclin E1 and RSF chromatin remodeling complex which collaborates with p53mut in promoting cell growth and tumorigenicity of non-transformed cells. During cancer development, the interaction between full-length Rsf-1 and cyclin E1 is required for tumor growth, or otherwise the tumorigenicity can be reduced. The biological relevance of those molecular alterations comes from a high correlation among Rsf-1 expression, cyclin E1 expression and TP53 mutations in clinical tumor specimens. The findings reported here could have significant implications for future studies aimed at elucidating the pathogenesis of ovarian high-grade serous carcinoma.
Supplementary Material
Acknowledgements
This study was supported by grants from NIH/NCI (R01CA129080 and RO1CA103937), National Science Council, Taiwan (NSC98-2320-B-039-033-MY3), Department of Health, Taiwan (DOH101-TD-C-111-005) and China Medical University Hospital, Taiwan (DMR98-031). The authors thank Dr. D.-S. Chen at China Medical University, Taiwan for technical assistance.
Footnotes
Disclosure of potential conflicts of interest:
The authors have no potential conflicts of interest to disclose.
Author contribution statement
JS, FT and CH collected the clinical samples and patient information. JS, JC, BG and ML carried out the experiments. JS, TW and IS designed the experiments and analyzed the data. JS and IS organized the figures/tables and wrote the paper. All authors had final approval of the submitted manuscript.
List of online Supporting information
Figure S1. Induction of Rsf-1 and cyclin E1 expression in RK3E cells.
Figure S2. Co-upregulation of Rsf-1, SNF2H and cyclin E expression in ovarian carcinoma cells.
References
- 1.Medina PP, Romero OA, Kohno T, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Nieto S, Canada A, Pros E, et al. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Hum Mutat. 2011;32:E1999–E2017. doi: 10.1002/humu.21415. [DOI] [PubMed] [Google Scholar]
- 3.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, Wang TL, Shih Ie M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan B, Mao TL, Panuganti PK, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Zhao H, Zhang X, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nature Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih Ie M, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JH, Sheu JJ, Guan B, et al. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69:1407–1415. doi: 10.1158/0008-5472.CAN-08-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeRoy G, Loyola A, Lane WS, et al. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 11.Loyola A, Huang J-Y, LeRoy G, et al. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23:6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loyola A, LeRoy G, Wang YH, et al. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 2001;15:2837–2851. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih Ie M, Davidson B. Pathogenesis of ovarian cancer: clues from selected overexpressed genes. Future Oncol. 2009;5:1641–1657. doi: 10.2217/fon.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda D, Chen X, Guan B, et al. Rsf-1 (HBXAP) expression is associated with advanced stage and lymph node metastasis in ovarian clear cell carcinoma. Int J Gynecol Pathol. 2011;30:30–35. doi: 10.1097/PGP.0b013e3181e9a319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang FM, Li CF, Huang HY, et al. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. Am J Pathol. 2011;178:2407–2415. doi: 10.1016/j.ajpath.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai HC, Huang HY, Lee SW, et al. Associations of Rsf-1 overexpression with poor therapeutic response and worse survival in patients with nasopharyngeal carcinoma. J Clin Pathol. 2012;65:248–253. doi: 10.1136/jclinpath-2011-200413. [DOI] [PubMed] [Google Scholar]
- 17.Chen TJ, Huang SC, Huang HY, et al. Rsf-1/HBXAP overexpression is associated with disease-specific survival of patients with gallbladder carcinoma. APMIS. 2011;119:808–814. doi: 10.1111/j.1600-0463.2011.02808.x. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Dong Q, Wang E. Rsf-1 is overexpressed in non-small cell lung cancers and regulates cyclinD1 expression and ERK activity. Biochem Biophys Res Commun. 2012;420:6–10. doi: 10.1016/j.bbrc.2012.02.095. [DOI] [PubMed] [Google Scholar]
- 19.Lin CY, Tian YF, Wu LC, et al. Rsf-1 expression in rectal cancer: with special emphasis on the independent prognostic value after neoadjuvant chemoradiation. J Clin Pathol. 2012 doi: 10.1136/jclinpath-2012-200786. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Dong Q, Wang E. Rsf-1 overexpression correlates with poor prognosis and cell proliferation in colon cancer. Tumour Biol. 2012 doi: 10.1007/s13277-012-0399-y. [DOI] [PubMed] [Google Scholar]
- 21.Sheu JJ, Choi JH, Yildiz I, et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68:4050–4057. doi: 10.1158/0008-5472.CAN-07-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheu JJ, Guan B, Choi JH, et al. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285:38260–38269. doi: 10.1074/jbc.M110.138735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kshirsagar M, Jiang W, Shih Ie M. DNA damage response is prominent in ovarian high-grade serous carcinomas, especially those with Rsf-1 (HBXAP) overexpression. J Oncol. 2012;2012:621685. doi: 10.1155/2012/621685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao TL, Hsu CY, Yen MJ, et al. Expression of Rsf-1, a chromatin-remodeling gene, in ovarian and breast carcinoma. Hum Pathol. 2006;37:1169–1175. doi: 10.1016/j.humpath.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Davidson B, Skrede M, Silins I, et al. Low-molecular weight forms of cyclin E differentiate ovarian carcinoma from cells of mesothelial origin and are associated with poor survival in ovarian carcinoma. Cancer. 2007;110:1264–1271. doi: 10.1002/cncr.22918. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Liu KJ, Wang TL, et al. Mapping DNA quantity into electrophoretic mobility through quantum dot nanotethers for high-resolution genetic and epigenetic analysis. ACS Nano. 2012;6:858–864. doi: 10.1021/nn204377k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terry DE, Umstot E, Desiderio DM. Optimized sample-processing time and peptide recovery for the mass spectrometric analysis of protein digests. J Am Soc Mass Spectrom. 2004;15:784–794. doi: 10.1016/j.jasms.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Hirosawa M, Hoshida M, Ishikawa M, et al. MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming. Comput Appl Biosci. 1993;9:161–167. doi: 10.1093/bioinformatics/9.2.161. [DOI] [PubMed] [Google Scholar]
- 29.Davidson B, Trope CG, Wang TL, et al. Expression of the chromatin remodeling factor Rsf-1 is upregulated in ovarian carcinoma effusions and predicts poor survival. Gynecol Oncol. 2006;103:814–819. doi: 10.1016/j.ygyno.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Etemadmoghadam D, deFazio A, Beroukhim R, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama K, Nakayama N, Jinawath N, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–2617. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 32.Courjal F, Louason G, Speiser P, et al. Cyclin gene amplification and overexpression in breast and ovarian cancers: evidence for the selection of cyclin D1 in breast and cyclin E in ovarian tumors. Int J Cancer. 1996;69:247–253. doi: 10.1002/(SICI)1097-0215(19960822)69:4<247::AID-IJC1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 33.Marone M, Scambia G, Giannitelli C, et al. Analysis of cyclin E and CDK2 in ovarian cancer: gene amplification and RNA overexpression. Int J Cancer. 1998;75:34–39. doi: 10.1002/(sici)1097-0215(19980105)75:1<34::aid-ijc6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Sehdev AS, Kurman RJ, Kuhn E, et al. Serous tubal intraepithelial carcinoma upregulates markers associated with high-grade serous carcinomas including Rsf-1 (HBXAP), cyclin E and fatty acid synthase. Mod Pathol. 2010;23:844–855. doi: 10.1038/modpathol.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolligs FT, Hu G, Dang CV, et al. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrix ND, Wu R, Kuick R, et al. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 37.Komiya T, Park Y, Modi S, et al. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene. 2006;25:6128–6132. doi: 10.1038/sj.onc.1209627. [DOI] [PubMed] [Google Scholar]
- 38.Bommer GT, Jager C, Durr EM, et al. DRO1, a gene down-regulated by oncogenes, mediates growth inhibition in colon and pancreatic cancer cells. J Biol Chem. 2005;280:7962–7975. doi: 10.1074/jbc.M412593200. [DOI] [PubMed] [Google Scholar]
- 39.Foster KW, Ren S, Louro ID, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 40.Kolligs FT, Nieman MT, Winer I, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 41.Hanai K, Furuhashi H, Yamamoto T, et al. RSF governs silent chromatin formation via histone H2Av replacement. PLoS Genet. 2008;4:e1000011. doi: 10.1371/journal.pgen.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jirawatnotai S, Hu Y, Michowski W, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connolly DC, Bao R, Nikitin AY, et al. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–1397. [PubMed] [Google Scholar]
- 44.Young T, Mei F, Liu J, et al. Proteomics analysis of H-RAS-mediated oncogenic transformation in a genetically defined human ovarian cancer model. Oncogene. 2005;24:6174–6184. doi: 10.1038/sj.onc.1208753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih Ie M, Nakayama K, Wu G, et al. Amplification of the ch19p13.2 NACC1 locus in ovarian high-grade serous carcinoma. Mod Pathol. 2011;24:638–645. doi: 10.1038/modpathol.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakayama N, Nakayama K, Shamima Y, et al. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 2010;116:2621–2634. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 47.Marchini S, Mariani P, Chiorino G, et al. Analysis of gene expression in early-stage ovarian cancer. Clin Cancer Res. 2008;14:7850–7860. doi: 10.1158/1078-0432.CCR-08-0523. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhn E, Kurman RJ, Vang R, et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma- evidence supporting the clonal relationship of the two lesions. J Pathol. 2012;226:421–426. doi: 10.1002/path.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.