Abstract
Venoms are chemically complex secretions typically comprising numerous proteins and peptides with varied physiological activities. Functional characterization of venom proteins has important biomedical applications, including the identification of drug leads or probes for cellular receptors. Spiders are the most species rich clade of venomous organisms, but the venoms of only a few species are well-understood, in part due to the difficulty associated with collecting minute quantities of venom from small animals. This paper presents a protocol for the collection of venom from spiders using electrical stimulation, demonstrating the procedure on the Western black widow (Latrodectus hesperus). The collected venom is useful for varied downstream analyses including direct protein identification via mass spectrometry, functional assays, and stimulation of venom gene expression for transcriptomic studies. This technique has the advantage over protocols that isolate venom from whole gland homogenates, which do not separate genuine venom components from cellular proteins that are not secreted as part of the venom. Representative results demonstrate the detection of known venom peptides from the collected sample using mass spectrometry. The venom collection procedure is followed by a protocol for dissecting spider venom glands, with results demonstrating that this leads to the characterization of venom-expressed proteins and peptides at the sequence level.
Keywords: Genetics, Issue 93, spider, toxin, proteomics, transcriptomics, electrical stimulation, Latrodectus
Introduction
Venoms are animal secretions predominantly injected into another animal for the purposes of predation or defense, and also have important biological applications with biomedical relevance1–3. Only certain animals synthesize venom, but its production is taxonomically widespread across invertebrates (e.g., cnidarians, cone snails, scorpions, and spiders) and vertebrates (e.g., snakes, some fish and mammals), because it has independently evolved multiple times1,4. Biochemical characterization of venoms typically show they are composed of a wide variety of proteins and peptides, which largely act on the circulatory and nervous systems of injected animals to rapidly deliver constituent toxins1. A small fraction of venomous animals may pose a threat to humans, particularly species with synanthropic distributions5. The study of venom composition and functional activities has played an important role in the functional characterization of vertebrate cellular components (particularly neuronal ion channels)6 and in elucidating fundamental cellular processes (e.g. neurosecretion)7. Moreover, select venom peptides have useful properties in biomedical contexts, including their uses as treatments for cancer and pain8,9, and are mined for candidate drug leads and antivenom development. Venoms also play prominent roles in ecological and evolutionary research1,4,10,11, thus the collection of these hazardous secretions has many utilities.
There are >40,000 described species of spider (Order Araneae), and all but one of the more than 100 spider families possess paired venom glands that terminate in fangs12. The high species diversity of spiders suggests that they represent the largest clade of venomous organisms. However, biochemical characterization of spider venoms has largely concentrated on a small number of species most often associated with human envenomation. Recent proteomic and transcriptomic studies of spider venoms indicate they typically contain many unique proteins and peptides2,13,14. Advances in high-throughput cDNA sequencing and mass spectrometry peptide fingerprinting has greatly facilitated the discovery of these venom proteins15. Nevertheless, such work begins with the collection of sufficient venom and/or venom glands from the spiders, and detailed documentation for such techniques are few16.
This paper presents a protocol for the collection of venom and venom glands from black widow spiders, which can be applied to similarly sized spiders. The collection of venom separately from the glands enables identification of proteins that are secreted into the venom as opposed to proteins performing other cellular functions. Black widows and other Latrodectus species are widely recognized as being among the most hazardous spiders due to their highly neurotoxic venom, which causes severe pain in humans, which is sometimes accompanied by profuse sweating, muscle contractions, hypertension, difficulty breathing and patchy paralysis5. The venom collection protocol presented here uses electro-stimulation to deliver electrical current to anesthetized spiders to elicit muscular contractions and release of venom. Venom droplets are quickly collected with micro-capillaries and dispensed into tubes for freezer storage. Because the protocol involves hazardous procedures, it should only be performed by well-trained individuals and caution is urged at key steps. The collected venom has numerous uses, such as characterization and isolation of constituent molecules2, for physiological experiments or functional assays18, and to stimulate venom gene expression11. The protocol concludes with a description of venom gland dissection and preservation useful for the cloning of venom-specific genes, the expression of which has been shown to occur 2-3 days following venom depletion in different spiders10,11.
Protocol
1. Preparation of Electro-stimulator Apparatus
Connect electro-stimulator power cord to outlet and attach external foot pedal to the external signal input. Note: Electrical wires must be properly insulated and exposed metal delivering current should not be touched. Wear latex or nitrile gloves for protection. Electro-stimulator power switch should be in OFF position when attaching power cord and attaching wires.
Connect positive electrode to red output terminal on electro-stimulator (when polarity switch is in normal mode) and connect the negative electrode to the black output terminal. Note: Each electrode wire should terminate in an alligator clip.
Set stimulator to the following recommended initial settings: voltage=7 V, Frequency=1 pulses per second, delay=0 milliseconds, duration=200 milliseconds, twin pulses switch=regular, and mode switch=off.
Test output of electrodes by attaching alligator clips to voltmeter positive and negative test leads. Turn on stimulator power switch and deliver current with foot pedal.
2. Preparation of Immobilizing Forceps and Other Materials
Dip one prong of featherweight forceps into liquid plastic coating and wait four hours to let coating dry. Tightly wrap 15 mm of the tip of the opposing prong with a single layer of cotton sewing thread, and secure thread by tying the end in a knot. Note: The thread is used to absorb an applied saline solution that promotes electrical conductivity, while the plastic coating provides insulation to retard current.
Cut tip of blunt hypodermic needle with sharp scissors and smooth the opening with sand paper. Attach the needle to a vacuum waste trap (e.g., vacuum filter flask) via tubing. Note: The needle will suction away water and regurgitate, and will serve to complete the circuit created by the stimulator electrodes. The needle opening must be small enough to not harm the spider, yet large enough to vacuum solution without clogging (e.g., 25 G x 1 ½ in gauge, see Materials List).
Position featherweight forceps horizontally in a tightly closed position using a vinyl-covered two-pronged clamp secured to a magnetic base or fixed stand apparatus under a dissecting microscope’s field of view, with plastic coated prong of forceps on top and thread coated prong on bottom. Note: The two-pronged clamp is maintained in a stationary position to the magnetic base by a clamp holder (see Materials List). A rubber band may be tightly secured around prongs of the featherweight forceps, next to the clamp holder, to add additional pressure to keep the featherweight forceps in a closed position around the spider once introduced.
Attach positive electrode alligator clip to forceps’ bottom prong upstream of thread and attach negative alligator clip to blunt metal vacuum needle.
Test circuit current with voltmeter by placing positive lead on forceps prong between thread and alligator clip and placing negative lead on blunt vacuum needle. Press foot pedal to ensure sufficient current is detected and remove leads if sufficient current is detected.
Prepare several microcapillary tubes for venom collection. Hold a single microcapillary tube at one end with a gloved hand and at the other end with sterile metal forceps, placing the forceps-held end horizontally over a Bunsen burner flame. Pull on the microcapillary with the metal forceps away from the flame to create an elongated tip, while holding the other end in a stationary position. Note: wear protective eyewear as microcapillaries can easily fracture.
Rest prepared microcapillaries on a mounting putty strip in a petri dish. Examine microcapillary ends under a dissecting microscope and create a small, beveled opening at the pulled end with sterilized metal forceps.
Label variable number of sterile 0.5 ml tubes. Add sterile, deionized water to sterile tubes (e.g., 5 μl of autoclaved deionized water). Close tubes and place tubes on ice.
Prepare saline solution in a beaker and fill a second beaker with autoclaved water.
3. Immobilization of Spider in Forceps
Turn on CO2 tank and transfer spider from closed plastic collecting vial (see Materials List) into CO2 chamber using long metal forceps. Note: black widow spiders have hazardous venom; always wear gloves during this protocol.
Keep spider in chamber for 5-10 min or until anesthetized and no longer moving when gently prodded with long forceps. Use transfer pipette to wet thread on forceps with saline solution. Note: For guidelines on CO2 anesthesia parameters for black widows, refer to Spagna and Moore19.
Retrieve anesthetized spider from CO2 chamber by picking it up by its anterior legs using blunt dissecting forceps and gloved hands. Move anesthetized spider to featherweight forceps apparatus. Separate prongs of closed featherweight forceps and place the spider’s cephalothorax in between the prongs and allow the prongs to close shut to ensure spider is held tightly in place but is not being crushed. Ensure that the spider is placed carapace dorsum side downwards (see Foelix20 for guide to spider anatomy), resting on thread coated prong, and the carapace’s ventral side facing the plastic coating, while the positive electrode remains attached to bottom prong. Work quickly when handling spider.
Adjust dissecting microscope magnification, focus, and light source until the spider’s chelicerae and fangs are clearly visible.
4. Venom Collection
Turn on vacuum and spray spider chelicerae with water solution using a second blunt tipped syringe needle. Suction away water with vacuum needle to remove impurities.
Touch the vacuum needle with attached negative electrode to spider rostrum and deliver pulse with foot pedal one or more times. Vacuum away regurgitate if necessary. Note: spider rostrum is inside mouth posterior to chelicerae on dorsal surface between chelicerae and endites (see Foelix20 for detailed spider anatomy). Note: With each pulse, the spider’s legs contract, and venom will be visible as clear droplets emerging from fangs.
Collect venom droplets with microcapillary tip and transfer capillary tip into 0.5 ml tube with water or buffer solution (solution will be sucked into capillary). Note: avoid puncturing spider with capillary which may lead to hemolymph contamination and death. Also avoid contamination of capillary with silk and glue from spinnerets.
Attach transfer syringe with tube adapter to microcapillary (at end opposite to collection tip) and dispense venom solution back into tube. Put tube on ice. Note: Dispose of waste capillaries in a nearby sharps container.
Place open plastic collecting vial, along with its cap, close to the spider. Carefully grasp the spider legs with blunt dissecting forceps held in one hand and carefully open featherweight forceps prongs with other hand, sliding spider into collecting vial with blunt forceps and quickly close cap. Continue with next spider and store venom-containing tubes in -80 °C freezer when finished. Note: continue to exercise caution when moving the spider from the forceps back into the vial. Make sure the collecting vial and cap is nearby to the spider for its rapid transfer and wear gloves.
5. Venom Gland Dissections
Two-to-three days after venom collection, anesthetize spider in CO2 chamber (see step 3.1). Fill liquid nitrogen carrier and place next to dissecting microscope. Note: when working with liquid nitrogen, use eye protection and cryogenic gloves.
Clean dissection area and dissecting forceps with solution that eliminates RNase and DNA contamination. Fill a small petri dish placed under dissecting microscope with dissecting buffer (e.g., 1x saline-sodium citrate (SSC) buffer).
Transfer spider from CO2 chamber to petri dish and use forceps to quickly separate the cephalothorax from the abdomen.
Hold the cephalothorax under the dissecting microscope with forceps and position the chelicerae into field of view. Use the sharp ends of a second pair of forceps to cut cuticle joining the carapace to the lateral aspects of the chelicerae. Laterally grasp the chelicerae using the second pair of forceps, and gently tug back and forth until venom glands are pulled out.
Separate glands from the chelicerae (or leave attached to chelicerae if desired) and transfer to a cyrosafe 0.5 ml tube. Place closed tube in liquid nitrogen. Note: each dissection should take no more than 15 min.
Repeat dissection with additional spiders until finished. Transfer tubes from liquid nitrogen into -80 °C freezer.
Representative Results
The collection of venom and venom gland dissections are frequently performed to characterize venom proteins and peptides at the sequence level10,11,15. Collected venom may also be used in physiological assays to determine their functional activities18. The collection of venom will stimulate venom gland gene expression, thereby facilitating the cloning of specific toxin transcripts via RT-PCR10,11. Identification of venom proteins can also be accomplished in a high-throughput manner by integrating standard mass spectrometry techniques with sequence databases generated from venom gland cDNA libraries15,21. An example of such work, starting from venom and glands collected using the protocol detailed above, demonstrating its effectiveness, is illustrated in Figure 1.
Venom collected from L. hesperus adult females was digested with trypsin and subjected to an in solution MuDPIT analysis, which links HPLC (high performance liquid chromatography) to tandem mass spectrometry (MS/MS). Here the MuDPIT analysis was performed by the University of Arizona Proteomics Consortium. Masses of detected peptides and their dissociated fragments (daughter ions, inferred from spectra) were compared to peptide sequences theoretically predicted from translations of cDNA sequences obtained from a gene expression library constructed from L. hesperus venom glands (glands were acquired by the dissection protocol described above)21. In this experiment, 36 proteins were detected from all collected spectra, at the 99.9% probability threshold, and contained at least one detected peptide. One of the detected proteins is supported by 43 total spectra (exemplar spectra shown in Figure 1A), corresponding to three exclusive peptides, covering 35% of the protein sequence (Figure 1B). The detected protein (translated from a collected cDNA) has a top BLASTp hit to latrodectin from L. tredecimguttatus (GenBank Accession P49125.1), with an e-score of 2e-46, and shares 80% identity at the amino acid sequence level with the protein detected in this experiment. Latrodectin, which is also known as alpha-latrotoxin low molecular weight protein, is a recognized venom component of black widow spiders22,23, verifying the effectiveness of the presented protocol. Some proteins identified from the venom correspond to sequences for which no BLAST hit is found. It is unclear whether such a result is due to the limited genomic resources available for spiders as well as the limited functional information for their proteins, or because the unknown protein represents a venom contaminant. Nevertheless, gene or protein expression analyses examining the abundance of such a novel protein in venom glands relative to other tissues should reveal whether it is a genuine venom component.
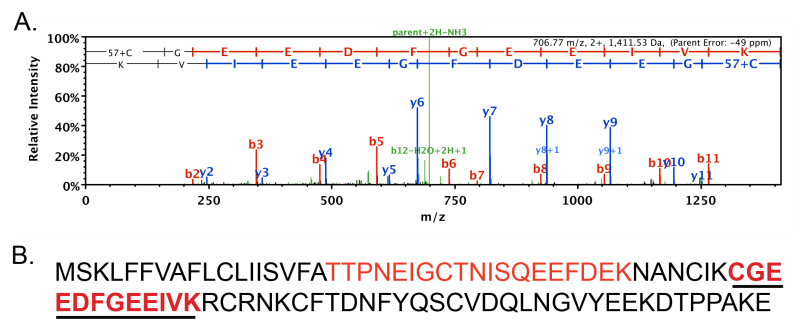 Figure 1: Latrodectin peptide identified from black widow venom using mass spectrometry.(A) Representative spectra (one of 43) detected via MS/MS portion of MuDPIT analysis of L. hesperus venom which was assigned to predicted translation of a collected cDNA sequence shown in part B. Spectra shows mass to charge ratio (m/z) of detected daughter ions on the horizontal axis produced by fragmenting parent peptide (CGEEDFGEEIVK), with letters at top indicating the peptide sequence based on daughter ion masses. (B) Protein sequence to which spectra in part A was assigned, showing in red, bold and underlined text the corresponding sequence from the spectra shown in part A, additional red text (not bold) represents another peptide in this protein detected with other collected spectra. The protein sequence is translated from a cDNA sequence obtained from dissected venom glands.
Figure 1: Latrodectin peptide identified from black widow venom using mass spectrometry.(A) Representative spectra (one of 43) detected via MS/MS portion of MuDPIT analysis of L. hesperus venom which was assigned to predicted translation of a collected cDNA sequence shown in part B. Spectra shows mass to charge ratio (m/z) of detected daughter ions on the horizontal axis produced by fragmenting parent peptide (CGEEDFGEEIVK), with letters at top indicating the peptide sequence based on daughter ion masses. (B) Protein sequence to which spectra in part A was assigned, showing in red, bold and underlined text the corresponding sequence from the spectra shown in part A, additional red text (not bold) represents another peptide in this protein detected with other collected spectra. The protein sequence is translated from a cDNA sequence obtained from dissected venom glands.
Discussion
Venoms represent an important source of physiologically reactive proteins, peptides and other molecules with applications for drug discovery, as well as for fundamental aspects of cellular and ecological research1–3. However, the collection of venom, particularly from dangerous or small animals, is a challenging task. This protocol demonstrates how venom and venom glands can be collected from black widow spiders, and confirms the success of this approach via a combination of MuDPIT analysis of the venom and a protein database derived from cDNAs cloned from venom glands21. While this protocol works well for black widows and medium sized spiders, other venom collection techniques have been employed for larger mygalomorph (tarantula-like) spiders, such as direct aspiration of venom from fangs into glass pipettes e.g., 24. This latter approach, however, will not work well for smaller-sized spiders that are not aggressive.
One especially critical aspect of the venom collection protocol described here is the initial phases of preparation and optimization of the collection process so that it becomes more routine, consistent and quicker. The protocol is initially challenging to master, but with repeated trials, it becomes easier and faster. Caution is also urged at all critical phases involving the handling of hazardous spiders, the use of electrical current, fine-point glass micro capillaries, and syringe needles. It is important to wear appropriate personal protective equipment such as nitrile gloves, a lab coat, long pants and closed shoes, as well as eyewear when shaping micro capillaries.
Another challenging aspect to venom collection is the small amount of venom produced by any one spider, particularly Latrodectus species, from which the amounts collected may be limited to 1-2 microliters per individual at best. Obtaining sufficient venom for downstream applications, such as protein gels or functional assays may require the combination of venom from multiple individuals into one tube. In such cases, venom should only be combined from individuals of the same sex, ontogenetic stage, and population given the recognition of intersexual, developmental and geographic variability in some venoms25,26. Spiders may also exhibit considerable variation in the amount of venom produced among individuals, where lesser amounts may reflect the recent depletion of the gland. Thus it may be advisable to collect venom several days after their last feeding. If little venom is released, excessive current should not applied to the spider, which may cause the cuticle to rupture, leading to contamination of the venom with hemolymph or death.
Contamination of venom samples with spider silk or human sources should also be avoided through the use of sterile or clean equipment. Despite these challenges, collection of pure venom, leaving the spider alive, is preferable to methods that obtain venom from gland homogenates (which do not separate venom components from other cellular proteins) and kill the spider. It is also critical to ensure that samples are quickly frozen to prevent protein degradation.
The extraction of venom promotes subsequent venom production, thereby stimulating venom gene expression in the venom gland. Thus, because this protocol allows for spiders to survive venom depletion, their glands may be dissected several days later (killing the spider) at a point where venom gene expression is expected to be sufficient for genetic studies, such as transcript cloning10,11. Several important precautions must also be taken in venom gland dissections. Emphasis should be placed on using lab equipment and reagents that are free of RNases that degrade RNA. Thus it is recommended to wipe forceps and other non-disposable equipment and surfaces with solutions that eliminate RNase and DNA contamination. The dissections should be performed as quickly as possible and directly frozen to further ensure RNA integrity of the tissue. Finally, dissections should only be performed on anesthetized spiders, after their cephalothorax and abdomen are quickly separated.
In conclusion, this article provides a verified protocol to obtain spider venom and venom glands. Venom and venom glands allow for the isolation and characterization of their protein and peptide components using proteomic and transcriptomic approaches. In addition, venom samples may represent the starting point of functional assays, which determine the biomedical and pharmacological potential of their constituent molecules. Nearly all spiders produce venom, and the wide diversity of venom components synthesized by individual species suggests a vast diversity of venom molecules are yet to be discovered13. Accordingly, this protocol provides tools to investigate the rich source of biologically active molecules present in spider venoms.
Disclosures
The author has nothing to disclose and no competing financial interests.
Acknowledgments
I thank the following individuals for their assistance in the development of this protocol: Chuck Kristensen, Greta Binford, Alex K. Lancaster, Konrad Zinsmaier, and Mays Imad. Mass spectrometry and proteomics data were acquired by the Arizona Proteomics Consortium supported by NIEHS grant ES06694 to the SWEHSC, NIH/NCI grant CA023074 to the AZCC and by the BIO5 Institute of the University of Arizona. Funding for this work was provided from the National Institutes of Health (National Institute of General Medicine) from grants 1F32GM83661-01 and 1R15GM097714-01 to Jessica E. Garb.
References
- Fry BG, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. G. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Quinton L, Nicholson GM. Venomics: unravelling the complexity of animal venoms with mass spectrometry. J. Mass. Spectrom. 2008;43(3):279–295. doi: 10.1002/jms.1389. [DOI] [PubMed] [Google Scholar]
- Twede VD, Miljanich G, Olivera BM, Bulaj G. Neuroprotective and cardioprotective conopeptides: An emerging class of drug leads. Curr. Opin. Drug Disc. 2009;12(2):231–239. [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013;28(4):219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Vetter RS, Isbister GK. Medical aspects of spider bites. Annu. Rev. Entomol. 2008;53:409–429. doi: 10.1146/annurev.ento.53.103106.093503. [DOI] [PubMed] [Google Scholar]
- Adams ME, Myers RA, Imperial JS, Olivera BM. Toxityping rat brain calcium channels with omega-toxins from spider and cone snail venoms. Biochemistry. 1993;32(47):12566–12570. doi: 10.1021/bi00210a003. [DOI] [PubMed] [Google Scholar]
- Silva J-P, et al. Latrophilin 1 and its endogenous ligand Lasso/Teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. PNAS. 2011;108(29):12113–12118. doi: 10.1073/pnas.1019434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Day M, Heavner JE. Ziconotide: an update and review. Expert Opin. Pharmacother. 2008;9(9):1575–1583. doi: 10.1517/14656566.9.9.1575. [DOI] [PubMed] [Google Scholar]
- Veiseh M, et al. Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007;67(14):6882–6888. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- Binford GJ, et al. Molecular evolution, functional variation, and proposed nomenclature of the gene family that includes sphingomyelinase D in sicariid spider venoms. Mol. Biol. Evol. 2009;26(3):547–566. doi: 10.1093/molbev/msn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garb JE, Hayashi CY. Molecular evolution of α-latrotoxin, the exceptionally potent vertebrate neurotoxin in black widow spider venom. Mol. Biol. Evol. 2013;30(5):999–1014. doi: 10.1093/molbev/mst011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnick NI. The World Spider Catalog., Version 14.0., DOI: 10.5531/db.iz.0001. 2013. Available online at: http://research.amnh.org/entomology/spiders/catalog/index.html.
- Sollod BL, Wilson D, Zhaxybayeva O, Gogarten JP, Drinkwater R, King GF. Were arachnids the first to use combinatorial peptide libraries. Peptides. 2005;26(1):131–139. doi: 10.1016/j.peptides.2004.07.016. [DOI] [PubMed] [Google Scholar]
- King GF, Hardy MC. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 2013;58(1):475–496. doi: 10.1146/annurev-ento-120811-153650. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Sollod B, King GF. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47(6):650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Kristensen C. Comments on the natural expression and artificial extraction of venom gland components from spiders. Toxin Rev. 2005;24(3-4):257–270. [Google Scholar]
- Ushkaryov YA, Volynski KE, Ashton AC. The multiple actions of black widow spider toxins and their selective use in neurosecretion studies. Toxicon. 2004;43(5):527–542. doi: 10.1016/j.toxicon.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Graudins A, et al. Cloning and activity of a novel α-latrotoxin from red-back spider venom. Biochem. Pharmacol. 2012;83(1):170–183. doi: 10.1016/j.bcp.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Spagna JC, Moore AMF. Safe immobilization by CO2 of Latrodectus hesperus (Arachnida: Theridiidae) Pan-Pac. Entomo. l. 1998;74(4):210–213. [Google Scholar]
- Foelix R. Biology of Spiders. Oxford University Press; 2010. [Google Scholar]
- Haney RA, Ayoub NA, Clarke TH, Hayashi CY, Garb JE. Dramatic expansion of the black widow toxin arsenal uncovered by multi-tissue transcriptomics and venom proteomics. BMC Genomics. 2014;15:366. doi: 10.1186/1471-2164-15-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescatori M, Bradbury A, Bouet F, Gargano N, Mastrogiacomo A, Grasso A. The cloning of a cDNA encoding a protein (latrodectin) which co-purifies with the α-latrotoxin from the black widow spider Latrodectus tredecimguttatus (Theridiidae) Eur. J. Biochem. 1995;230(1):322–328. doi: 10.1111/j.1432-1033.1995.tb20566.x. [DOI] [PubMed] [Google Scholar]
- Grasso A, Pescatori M. Structural and functional studies of latrodectin from the venom of black widow spider (Latrodectus tredecimguttatus) Adv. Exp. Med. Biol. 1996;391:237–243. doi: 10.1007/978-1-4613-0361-9_17. [DOI] [PubMed] [Google Scholar]
- Szeto TH, et al. Isolation of a funnel-web spider polypeptide with homology to mamba intestinal toxin 1 and the embryonic head inducer Dickkopf-1. Toxicon. 2000;38:429–442. doi: 10.1016/s0041-0101(99)00174-9. [DOI] [PubMed] [Google Scholar]
- Binford GJ. An analysis of geographic and intersexual chemical variation in venoms of the spider Tegenaria agrestis (Agelenidae) Toxicon. 2001;39:955–968. doi: 10.1016/s0041-0101(00)00234-8. [DOI] [PubMed] [Google Scholar]
- Alape-Girón A, et al. Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J. Proteome. Res. 2008;7(8):3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]


