Abstract
Zebrafish have become a widely used model organism to investigate the mechanisms that underlie developmental biology and to study human disease pathology due to their considerable degree of genetic conservation with humans. Chemical genetics entails testing the effect that small molecules have on a biological process and is becoming a popular translational research method to identify therapeutic compounds. Zebrafish are specifically appealing to use for chemical genetics because of their ability to produce large clutches of transparent embryos, which are externally fertilized. Furthermore, zebrafish embryos can be easily drug treated by the simple addition of a compound to the embryo media. Using whole-mount in situ hybridization (WISH), mRNA expression can be clearly visualized within zebrafish embryos. Together, using chemical genetics and WISH, the zebrafish becomes a potent whole organism context in which to determine the cellular and physiological effects of small molecules. Innovative advances have been made in technologies that utilize machine-based screening procedures, however for many labs such options are not accessible or remain cost-prohibitive. The protocol described here explains how to execute a manual high-throughput chemical genetic screen that requires basic resources and can be accomplished by a single individual or small team in an efficient period of time. Thus, this protocol provides a feasible strategy that can be implemented by research groups to perform chemical genetics in zebrafish, which can be useful for gaining fundamental insights into developmental processes, disease mechanisms, and to identify novel compounds and signaling pathways that have medically relevant applications.
Keywords: Developmental Biology, Issue 93, zebrafish, chemical genetics, chemical screen, in vivo small molecule screen, drug discovery, whole mount in situ hybridization (WISH), high-throughput screening (HTS), high-content screening (HCS)
Introduction
The identification of efficacious therapeutic drugs to treat human illness is the endgame for many biomedical researchers. While the identification and characterization of potential pharmacological agents for clinical applications has obvious value for healthcare, it has remained a significant challenge. Ultimately, the development of many therapeutic compounds comes from the knowledge of how specific cell types either develop or function. The zebrafish, Danio rerio, is a freshwater teleost of the Cyprinidae family that has become a mainstream model organism in developmental biology1. Zebrafish are genetically tractable vertebrates that are easy to maintain and manipulate for scientific studies2,3. The zebrafish embryo is transparent, externally fertilized, and can be produced by the hundreds from a single adult mating pair in only a week2,3. Furthermore, zebrafish share major organ systems and possess a large degree of genetic similarity with mammals, including humans4. Strikingly, 70% of human genes have a zebrafish orthologue4. Due to this similarity, zebrafish have been a highly relevant experimental system for studying the mechanisms of vertebrate development and physiology, and have been employed to recapitulate a multitude of human diseases5-7. These reasons, among others, have made the zebrafish an ideal candidate for continuing genetic interrogations and has resulted in a steadily increasing appreciation for the utility of zebrafish in biomedical research6,7.
Over the past few decades, an abundance of genetic tools have been developed in the zebrafish. The zebrafish genome is amenable to mutagenesis and transgenesis3. To date, a plethora of forward and reverse genetic studies have been performed, leading to the generation of over 9,000 mutants3. Furthermore, numerous transgenic lines have been created, which have enabled the labeling and isolation of specific cell types3. There have been significant ongoing efforts to centralize and distribute these resources to the research community, which are accessible for laboratories at a rather low cost through the Zebrafish International Resource Center (ZIRC)8. Further, an extensive repository of biological information and molecular tools, ranging from gene expression patterns to morpholino sequences and protocols, have been progressively annotated on the Zebrafish Information Network (ZFIN)9-11. These zebrafish research infrastructures have been made possible through significant NIH support and advocacy of this animal model8. This cache of zebrafish mutants, transgenic lines, and pertinent information continues to be of exceptional value to the biological research community at large. However, while zebrafish are now a well-established and pre-eminent model organism, they represent a largely untapped reservoir to study developmental biology and disease through the use of chemical genetic screens.
Chemical genetics is the use of small molecules, such as drugs or other compounds, to affect a biological process within a living system12. Chemical genetics can be implemented to understand the molecular mechanisms of gene function, such as to identify agents that rescue or exacerbate a specific genetic defect12. Further, chemical genetics represents one major avenue to conduct translational research12. While traditional forward genetic screens in animal models have been tremendously powerful in linking specific genes to diseases, they lack a temporal dimension, making it difficult or sometimes impossible to observe phenotypes that would arise after early embryogenesis. Additionally, gene redundancy can make the results observed from forward genetics difficult to interpret. Alternatively, chemical genetic screens allow for fine temporal control and simultaneously provide a valuable starting point for drug discovery12,13.
Chemical genetics can be performed using in vitro systems (e.g., cell lines, proteins) or using an in vivo system, such as a whole organism. Using an in vitro system for chemical genetics can be beneficial because it is simpler than a whole organism system, easier to work with on a large scale, less costly, and less time-intensive. As a result, in vitro systems allow for both high-throughput screening (HTS) and high-content screening (HCS). There are a number of advantages to using an in vitro system that should be considered when approaching chemical genetics, but also significant limitations. One major limitation to using a cell line is that small molecules can be efficacious and non-toxic in a specific cell line, yet become toxic when introduced into a whole organism. Thus, the crux of the issue is that in vitro systems do not capture the broad context of an organism and therefore are not always able to accurately mimic what happens in a whole organism. While compound testing in whole organisms can circumvent these issues, the use of mammalian whole organism models poses a number of physical and ethical limitations. For example, drug screening in the mouse is logistically hindered by the space and expense needed to test an adequate sample size. In addition, developmental questions can be difficult to study, due to the fact that mammals develop in utero. Finally, while the conduct of biomedical research has immense societal value, there are nevertheless substantial needs to protect animal welfare by limiting intense studies and alleviating the pain and suffering of animals used for research. Zebrafish provide a viable substitute to working with higher vertebrates like mammals, and furthermore, many topics can be studied using chemical genetics at the embryonic and larval stages when neurological processing is absent or operative at a low level12,13.
To date, a myriad of chemical genetic screens have been performed using zebrafish embryos in particular, as compound delivery can be accomplished by immersing embryos in media containing the drug of interest12-15. Further, there have been numerous scientific and technological advances to make chemical genetic screens possible and manageable16-20. Peterson and colleagues were the first to use zebrafish in a chemical genetic screen in 2000, and later in 2004 identified a compound that suppressed an aortic coarctation mutation in zebrafish21,22. Chemical screens have since been implemented to study several developing tissues (e.g., the heart, blood, vessels)23-25, physiological functions (e.g., neurological basis for behavior)26, adult regeneration27,28, and disease models (e.g., muscular dystrophy and cancer)29,30. From among these zebrafish chemical screens, the discovery that prostaglandins regulate hematopoietic stem cells has been taken to clinical trials in humans, demonstrating the power to move from “tank to bedside”23,31-33 (NIH, Clinical Trials.gov, NCT00890500; NCT01627314). More recently, promising candidate therapeutics have emerged for complex organs like the kidney, which remediate cellular pathology in polycystic kidney disease (PKD)34 and acute kidney injury27,35, although these have yet to move to clinical trials. These chemical genetic screens broadly demonstrate the utility of testing small compounds in the developing zebrafish and the ability to apply chemical genetics to interrogate adult tissue function. Furthermore, there is a growing list of commercially available pharmacology collections that are available for academic studies19.
In the last several years, there have been prominent advances in chemical genetic screening technology, including the use of automated systems of robots for drug dispensing and handling, along with automated imaging systems36-38. Such systems allow for the possibility of testing many thousands of compounds to achieve both HTS and HCS36-38. Unfortunately, the undertaking of chemical genetic screens with these innovative robotic enterprises typically require considerable resources. These types of resources are a significant impediment for many institutions that lack the capital to acquire, operate, and maintain such instrumentation. While establishing the infrastructure for robotic handling of chemical libraries, including the transfer of embryos to arrayed plates, remains cost prohibitive for many labs, automated imaging and analysis is generally more accessible12. Automated image screening enables quantification of fluorescent transgenic reporters and facilitates specific phenotypic analysis12,15.
Although automated systems represent useful technological advances, many questions can be addressed by more focused manual screens, such as the query of several thousand biologically active compounds on a developmental process with a whole-mount in situ hybridization (WISH) readout39. Further, while an increasing number of labs have invoked zebrafish chemical screens to investigate a research question, few (if any) efforts have reached saturation and many topics have yet to be breached using chemical genetics. Chemical screens can be executed using a small zebrafish colony consisting of only a few wild type or transgenic tanks. Therefore, this form of genetic interrogation should be attainable by most zebrafish research groups interested in expanding/diversifying into this arena. Chemical libraries of varying size can be procured from a commercial distributor and/or collaborators. For these reasons, chemical genetics can be economically feasible even in harsh funding climates. However, there are hurdles to beginning a chemical genetics screen, such as how to plan logistical aspects of the screen: e.g., the number of personnel needed for a successful screen, the allotment of dedicated time and piecing together a physical protocol to manually process samples that number from the hundreds to several thousand. Here, we detail a manual high-throughput protocol to perform chemical genetics in the zebrafish embryo with WISH as a read-out. This method involves drug treatment of zebrafish embryos, followed by riboprobe detection of mRNA transcripts. Using this protocol, a small team or even one individual can reasonably screen and analyze a modest chemical library approximately 600 compounds in the span of 9 weeks.
Protocol
The procedures for working with zebrafish described here were approved by the Institutional Animal Care and Use Committee at the University of Notre Dame. A guide to the research design for this manual small molecule screen in the zebrafish is provided (Figure 1). An overview of the methodologies described in this protocol is provided as a flow chart (Figure 2). Please refer to the example labeling system (Figure 3) to plan the embryo processing steps described in Parts 4-10. For the staging of embryos, please refer to the zebrafish staging series40, and for preparation of WISH antisense riboprobes used in Parts 9-10, refer to recently published methods41.
1. Preparation of Zebrafish Mating Chambers
Place an adult male zebrafish and an adult female zebrafish into a mating chamber, separated by the chamber divider O/N. NOTE: Mass matings, such as those using groups of 2-3 females and 2-3 males can generate more embryos compared to single paired matings, and can be performed with appropriately sized mating chambers.
Repeat Step 1.1 such that a total of 25 pairs of zebrafish are set-up for every 96 well plate of small molecules that will be tested.
2. Collection of Zebrafish Embryos
The next day, remove the divider that separates each fish pair and after 1 hr, collect zebrafish embryos using a fine-mesh strainer and place them in a Petri dish containing E3 embryo medium (see material list for recipe). NOTE: Collecting embryos after 1 hr ensures that embryos are at similar developmental stages.
Use a plastic transfer pipette to remove debris (e.g., food and solid waste). Next, use a stereomicroscope to observe each clutch of embryos and remove any unfertilized eggs.
Decant the E3 embryo media and replace with fresh E3, then place each dish of embryos in a 28.5 °C incubator. After 2 hr observe each clutch of embryos using a stereomicroscope and remove any unfertilized eggs. NOTE: Unfertilized eggs can poison other embryos and will appear to be at the single cell stage or opaque.
Continue to monitor the developmental progress of the embryos using a stereomicroscope until they reach 40% epiboly. Observe the sphere shape of the embryo that looks to be composed of two separate layers at the 40% epiboly stage40.
3. Chemical Dilution Preparation
Remove the chemical library plate from storage at -80 °C, cover it with aluminum foil and thaw it at RT. NOTE: Different libraries will thaw at different rates depending primarily on the solvent being used. Typically, a chemical library frozen at -80 °C will be completely thawed in approximately 3 hr at RT. Store thawing plate(s) in a safe location with appropriate hazmat labeling. Wear appropriate eyewear, a laboratory coat, and two sets of gloves when handling chemicals.
Use an 8 channel multichannel pipette to add 99 µl of E3 into columns 2-12 of a 96 well plate. NOTE: From our experience, an appropriate beginning drug range is between 10 µM - 50 µM, and also as described in current protocols39. Here we demonstrate a 10 µM treatment if using a 1 mM chemical library stock plate.
Use an 8 channel multichannel pipette to add 1 µl of each compound to its respective well with E3.
Use a P10 to pipette 1 µl of DMSO (or the appropriate solvent) into the control column (here, column 12).
Cover the chemical dilution plate with aluminum foil, as the aluminum foil will protect any light-sensitive compounds in the library, and return the stock chemical library plate to the -80 °C for storage.
4. Arraying Zebrafish Embryos
Select a Petri dish containing zebrafish embryos at the 40% epiboly stage. Using a transfer pipette, carefully place approximately 10 embryos inside each well of a (1.1 ml) deep 96 well plate. Dispense embryos into columns 2-12 of the deep 96 well plate. NOTE: The researcher should select the timepoint of drug addition that is best tailored to the developmental process they wish to interrogate. Use of a plastic transfer pipet can minimize embryo damage. Since drug treatment prior to 40% epiboly can result in increased embryo lethality, develop the embryos for longer prior to drug exposure, especially when later developmental processes are being examined. Placing more than 15-20 embryos in each well can lead to developmental delay due to crowding and should be avoided. It is important to note that the deep 96 well plate containing the embryos is different from the chemical dilution 96 well plate.
Use a glass transfer pipette to remove excess E3 from each well, and expel the excess E3 into a liquid waste container. NOTE: This process typically takes 30 min to 1 hr depending on the researcher’s speed, so the embryos are between 50% and 60% epiboly by completion, which is the target developmental stage for addition of the chemical dilution described here. The plate can be tilted at a 45° angle to cause the embryos to fall to the bottom of the well so that the tip of the pipette can be pressed against the bottom of the well to draw off the excess E3.
5. Addition of Chemical Treatments
Use an 8 channel multichannel pipette to transfer the 100 µl chemical dilutions to their respective wells in the deep 96 well plate containing arrayed zebrafish embryos. NOTE: Ensure that the original well location of the compound matches with the well location in the corresponding deep 96 well plate (e.g., the B1 well location in the chemical dilution plate is pipetted into the B1 well location in the deep 96 well plate).
Wet a paper towel and place it into a light-sensitive container large enough to hold a 96 well plate. NOTE: The wet paper towel will help prevent evaporation of the chemical dilution.
Next, use a sealing mat to seal the deep 96 well plate, place it into the light sensitive container, and incubate it at 28.5 °C. NOTE: Let the zebrafish embryos develop until the next morning. Alternatively, let the embryos develop until the appropriate developmental stage for the process being analyzed.
6. Removal of Chemicals
Use an 8 channel multichannel pipette to add 300 µl of E3 to each row of the drug treated wells. Draw off and discard the E3/chemical dilution from each row of wells. Wash the embryos 3 times with 300 µl of E3. NOTE: Discard drug-waste in a labeled container used only for this purpose. For ease of washes, use a glass basin filled with E3 wide enough to use the multichannel pipette.
7. Pronase and Fixing Zebrafish Embryos
Transfer zebrafish embryos using a plastic transfer pipette into four 24 well plates, then remove any dead embryos. Label the 24 well plates so that they correspond to the original drug well locations. Place the pipette in front of a dark background after each well transfer to visualize embryos in the pipette and to prevent embryo well crossover.
Use a plastic transfer pipette to draw off E3 so that the embryos are barely submerged in liquid.
Use a glass pipette to add 2 drops of pronase stock (50 mg/ml) to each well. Let embryos incubate in pronase for 5 min and then agitate the wells to cause the chorion to break apart. Alternatively, pronase the embryos before they are at the desired developmental stage for analysis.
Use a plastic transfer pipette to wash the embryos twice with E3.
Once the embryos are at the desired stage of analysis, completely draw off and discard the remaining E3.
Use a plastic transfer pipette to add fresh 4% PFA/PBS to each well. Incubate the embryos at 4 °C O/N to fix. Use freshly thawed PFA to initially fix embryos. NOTE: Proceed to perform your screen assay of interest. Detailed here in Sections 8-10 is a procedure to process screen plates with WISH. Alternative methods can be substituted as desired, then proceed to Section 11 and evaluate the results.
8. Embryo Permeabilization
Use a plastic transfer pipette to draw-off the 4% PFA/PBS from the embryos and wash the embryos twice with 1x (Phosphate-buffered saline with Tween) PBST. Transfer the embryos into 48 well plates, then draw off the 1x PBST from the embryos. Label the 48 well plates to match the original well locations of the deep 96 well plate.
Wash by adding 100% methanol to the wells using a plastic transfer pipette then draw it off and add 100% methanol again to the embryos.
Place the embryos at -20 °C, and incubate for at least 20 min. Alternatively, keep the embryos in 100% methanol at -20 °C for long-term storage.
Remove the 100% methanol and wash the embryos with 50% MeOH/1x PBST, then 30% MeOH/1x PBST, and finally twice with 1x PBST. NOTE: Embryos older than 24 hr post fertilization (hpf) can be bleached at this point to remove pigmentation39.
Add 15 µl of thawed proteinase K stock (10 mg/ml) to 50 ml of 1x PBST to prepare a proteinase K (5 µg/ml) working solution.
Remove the 1x PBST from the embryos and add the proteinase K working solution. Remove the proteinase K after 4 min. NOTE: The experimenter should work quickly during this step to prevent embryo degradation. Proteinase K concentration and incubation period need to be varied depending on the age of the embryos being treated41. The parameters described here pertain to embryos that are 24 hpf.
Wash the embryos with 1x PBST, then add ice-cold 4% PFA/PBS. Let the embryos incubate in 4% PFA/PBS for 20 min at RT.
9. Hybridization and Probe Removal
Using a glass transfer pipette remove the 4% PFA/PBS and wash the embryos twice with 1x PBST. Replace the 1x PBST with 500 µl of HYB+ and incubate the embryos for 4 hr at 70 °C. NOTE: Pre-hybridization can be performed for as few as 2 hr, but 4 hr is ideal. Also, different hybridization and wash temperatures may be ideal for certain probes, such as 65 °C, but should be tested on an individual basis.
Generate digoxygenin-labeled riboprobes corresponding to the gene(s) of interest41.
Add 16 µg of riboprobe to 16 ml of HYB+. Pre-warm the riboprobe/HYB+ solution at 70 °C for at least 20 min before using. If using multiple probes, add 16 µg of each probe to the same HYB+ volume (16 ml).
Using a glass transfer pipette, remove the HYB+ and use a P1000 to add on 200 µl of the riboprobe/HYB+ mixture. Invert the riboprobe/HYB+ cocktail just prior to use and pipette up and down once before adding the mixture into each well. Use barrier pipette tips to add the riboprobe/HYB+ solution, this prevents the mixture vapor from entering the pipette.
Incubate the embryos in the riboprobe/HYB+ cocktail at 70 °C O/N. Use an adhesive film to cover the plate wells for the O/N probe incubation and all of the following hot washes.
Prepare 50% formamide/2x SSCT, 2x SSCT, and 0.2x SSCT and pre-warm at 70 °C O/N.
Using a P1000, remove the riboprobe/HYB+ and store the mixture at -20 °C. Optionally, re-use the riboprobe/HYB+ mixture up to three times. Prior to each reuse, add in 3 µg of riboprobe to the original riboprobe/HYB+ mixture.
Using a glass transfer pipette, wash embryos twice with 50% formamide/2x SSCT for 30 min at 70 °C. Wash the embryos once with 2x SSCT for 15 min at 70 °C. Wash the embryos twice with 0.2x SSCT for 30 min at 70 °C. Remove the 0.2x SSCT and wash the embryos with MABT twice at RT.
10. Anti-digoxygenin Antibody Incubation and Detection
Use a glass transfer pipette to remove the MABT from each well. Using a plastic transfer pipette, add fresh block solution to each well. Let the embryos incubate in block for 4 hr at RT. Add 8 µl of anti-digoxygenin antibody to 40 ml of block solution (5,000-fold dilution).
Using a plastic transfer pipette, replace the block solution with the antibody/block solution. Add enough antibody/block solution to completely cover the embryos.
Cover the 48 well plate with aluminum foil, so that no light can penetrate through to the embryos. Incubate the embryos O/N at 4 °C. Using a glass pipette, remove the antibody/block solution and wash the embryos 10 times with MABT for 5-10 min each. NOTE: The MABT washes can be performed continuously, as it will take the researcher 5-10 min to remove MABT from each plate and dispense fresh MABT into the wells.
Generate two tubes of 40 ml (80 ml total) of pre-staining buffer. Add 180 µl of NBT (50 mg/ml) and 140 µl of BCIP (50 mg/ml) to one of the prepared tubes of pre-staining buffer to create the staining solution. NOTE: This staining solution will generate a purple colored substrate.
Replace the MABT with pre-staining buffer and let the embryos incubate at RT for 5 min.
Replace the pre-staining buffer with the staining solution and leave at RT. Monitor the colorimetric reaction every 15 min using a stereomicroscope. NOTE: Staining reactions can take from a few minutes to many hours depending on the probe.
Replace the staining solution with 1x PBST after the staining reaction is complete and then wash the embryos twice with 1x PBST for 5 min.
Remove the 1x PBST and add 4% PFA/PBS to fix the embryos. NOTE: Alternatively, perform a double WISH to detect riboprobes labeled with an anti-fluorescein antibody. If performing a double in situ, skip adding 4% PFA/PBS (Step 10.9) after the 1x PBST wash (Step 10.8) and wash the embryos twice for 15 min with 0.1 M glycerol, twice for 15 min with MABT washes, and then incubate the embryos for 30 min in block. Then add on the correct antibody O/N and perform Steps 10.3-10.9 using the correct staining solution. To detect the fluoroscein-labeled riboprobe, prepare a staining solution with BCIP and INT to observe a red substrate (refer to Step 10.1).
11. Scoring the Chemical Screen Embryo Plates
Perform visual scoring of the results using a stereomicroscope to evaluate the embryo samples in the phenotype assay.
Using a plastic transfer pipette, transfer the embryos from the 48 well plates to 12 well plates for optimal visualization. Label the 12 well plates so that they correspond to the well locations from the original deep 96 well plate. When the transfer is complete, view under the stereomicroscope. NOTE: Representative embryos can be selected from 12 well plates and imaged on glass slides under a cover slip41.
Representative Results
The manual approach described here allows for a single researcher to efficiently and successfully conduct a high-throughput small molecule chemical genetics screen using the zebrafish embryo model (Figure 1). Using this method, it is practical for a single person to perform a chemical genetics screen, such that over the course of 9 weeks one person (committed to a 40 hr work week) can carry out a survey of ~600 compounds (Figure 1). Thus, the bench time of one person can bypass the need for high-resource demanding equipment, like automated systems. The trade-off is that a manual screen requires several weeks of dedicated effort. In contrast, the use of automated systems can compress the time involved into 1-2 weeks of robotic time, and may entail relatively minimal researcher effort overall. One way to increase the efficiency and/or scope of a manual screen is to enlist 2-3 researchers in total, which would enable screening of more compounds, varying compound concentration, and/or screening over a shorter period of time.
The primary steps involved in the protocol to complete a chemical genetics screen in zebrafish embryos with a WISH readout are illustrated as a flowchart (Figure 2). The experimental tasks can be divided into two types of work weeks: (1) chemical exposure and embryo labeling, and (2) analysis and imaging. In the chemical exposure work week, these steps include setting up zebrafish adults (Day 1), embryo collection, arraying embryos and drug treatment (Day 2), preparing embryos for in situ hybridization (Day 3), and in situ hybridization (Day 4-6). In the analysis/imaging week, the researcher analyzes mRNA expression to score embryos and image hits (Day 7-11).
A critical part of performing a chemical genetics screen is to consistently and accurately label well locations. As the chemical genetics screen process detailed here involves transferring sets of embryos from a well in one plate to a well in a different plate multiple times, the need for precise and clear labeling is paramount, so that when a chemical hit is scored it can be linearly and easily traced back to its identity within the chemical library (Figure 3). An easy way to ensure this is to label the plates in such a way that at any time the well location for a set of embryos treated by a compound directly matches the chemical library drug plate well location of the compound used to treat those embryos (Figure 3). Here, a dual system of color-coding and numerical labels is utilized to organize the samples, which were added to the screen plates using permanent markers (Figure 3).
While analyzing the results of a high-throughput screen, there is a critical need to develop a method to correctly organize, annotate, and analyze potential small molecule hits. Therefore, a chemical genetics screen must have a thorough and clear grading system that is carefully designed and strictly adhered to. Such a system should easily and completely convey the important dynamics of a compound and provide a concise way to present the results of the finished screen. One way to construct such a system is to grade embryos utilizing a class system, which would explain overall morphological severity and confidence of a hit. Further, the class categories could be coupled with a (+) or (-) to describe either an expansive or reductive effect on the region(s) of interest. Here, zebrafish embryos were subjected to individual compounds from a bioactive library (Enzo Chemicals) from 50% epiboly to 24 hpf, and then assayed by WISH using a cocktail of three riboprobes to detect nephron segments (Figure 4A). As an example grading system, a well that contains a group of embryos that had a dramatically enlarged proximal convoluted tubule (PCT) of the pronephros, the zebrafish embryonic kidney, would cause the compound used to treat that well being categorized as (Class I), which in this case would represent the class associated with the most severe defect and the highest level of confidence in the phenotype observed (Figure 4B). The compound would then be annotated with (PCT) and a (+), the complete notation being (Class I - PCT+) (Figure 4B). This concept of classification provides a simple and fast way to describe a trove of information concerning a small molecule of interest, including the severity of the compounds effect on the organism, the region that is affected by the drug and the manner in which the region is affected (Figure 4B).
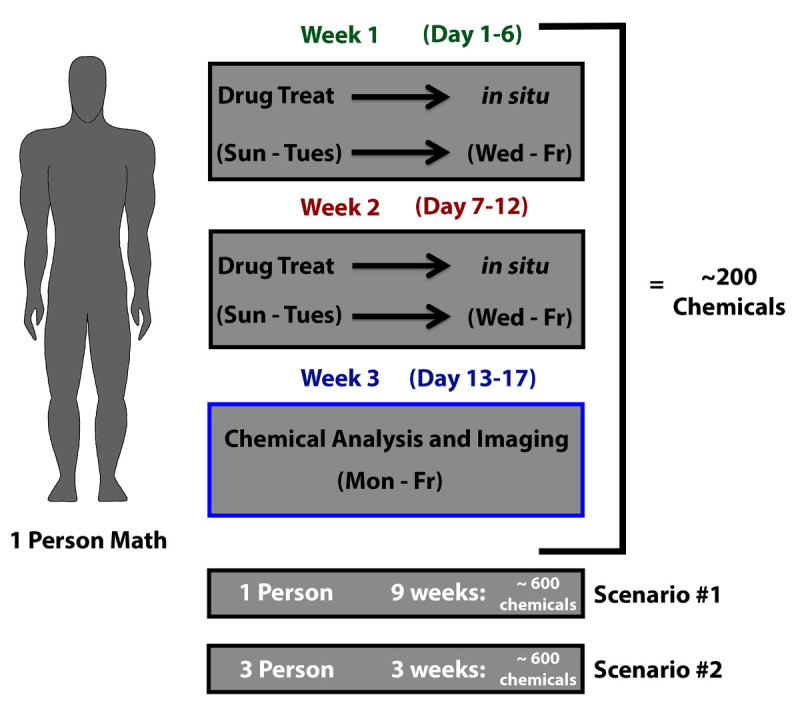 Figure 1. Manual zebrafish embryo chemical screen strategy. A researcher can feasibly test a 96 well plate of chemicals on zebrafish embryos and perform subsequent WISH processing during a single week. Performing two weeks of chemical testing (Week 1 (Day 1-6) and Week 2 (Day 7-12)) can be paired with a third week dedicated to sample analysis. Approximately two 96 well plates worth of screen samples can be scored and imaged Week 3 (Day 13-17). This scenario amounts to the screening of approximately 200 compounds in a 3 week time period, equating to a small library of approximately 600 compounds completely screened by 9 weeks. This manual strategy can be modified depending on the resources available. For instance, three researchers can reduce the time to complete a screen of approximately 600 compounds to 3 weeks compared to the 9 weeks it would take with one researcher. Screening strategies can be adapted by assigning researchers to specific aspects of the screen, such as drug treatment and in situ hybridization or directing researchers to independently perform the entire screening process on separate parts of the chemical library.
Figure 1. Manual zebrafish embryo chemical screen strategy. A researcher can feasibly test a 96 well plate of chemicals on zebrafish embryos and perform subsequent WISH processing during a single week. Performing two weeks of chemical testing (Week 1 (Day 1-6) and Week 2 (Day 7-12)) can be paired with a third week dedicated to sample analysis. Approximately two 96 well plates worth of screen samples can be scored and imaged Week 3 (Day 13-17). This scenario amounts to the screening of approximately 200 compounds in a 3 week time period, equating to a small library of approximately 600 compounds completely screened by 9 weeks. This manual strategy can be modified depending on the resources available. For instance, three researchers can reduce the time to complete a screen of approximately 600 compounds to 3 weeks compared to the 9 weeks it would take with one researcher. Screening strategies can be adapted by assigning researchers to specific aspects of the screen, such as drug treatment and in situ hybridization or directing researchers to independently perform the entire screening process on separate parts of the chemical library.
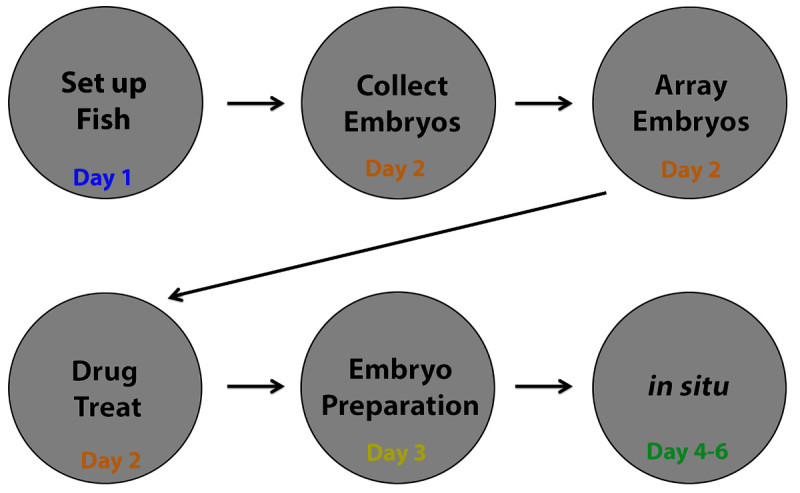 Figure 2. Flowchart: breakdown of 6 day screen workflow. The chemical screening process can be discretely broken down into 6 days. Day 1 consists of setting up zebrafish mating pairs (=/> 25) to supply embryos for each 96 well plate to be screened. Day 2 involves collecting embryos from the mating chambers and arraying them in a deep 96 well plate, followed by drug treatment. On Day 3, embryos are prepared for WISH, including dechorionation, tissue fixation, and embryo dehydration. Finally, WISH is performed on the embryos through Day 4-6. Please note that alternative staining procedures on fixed specimens can be readily substituted for WISH.
Figure 2. Flowchart: breakdown of 6 day screen workflow. The chemical screening process can be discretely broken down into 6 days. Day 1 consists of setting up zebrafish mating pairs (=/> 25) to supply embryos for each 96 well plate to be screened. Day 2 involves collecting embryos from the mating chambers and arraying them in a deep 96 well plate, followed by drug treatment. On Day 3, embryos are prepared for WISH, including dechorionation, tissue fixation, and embryo dehydration. Finally, WISH is performed on the embryos through Day 4-6. Please note that alternative staining procedures on fixed specimens can be readily substituted for WISH.
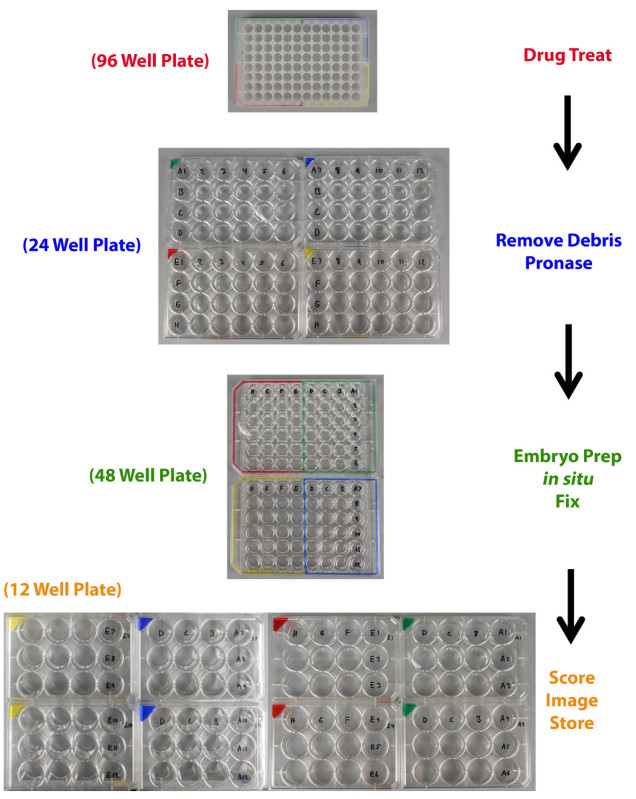 Figure 3. Plate transfer labeling schematic. Zebrafish embryos are arrayed and drug treated in a 96 well plate then moved to a 24 well plate for debris removal and pronase exposure. The embryos are then moved to 48 well plates for embryo preparation, WISH, and tissue fixation. Next, embryos are transferred to 12 well plates for scoring, imaging, and storage. Precise arraying of embryos is accomplished by the use of a color-coding system that enables the researcher to keep track of the chemical of origin for each test sample. Please click here to view a larger version of this figure.
Figure 3. Plate transfer labeling schematic. Zebrafish embryos are arrayed and drug treated in a 96 well plate then moved to a 24 well plate for debris removal and pronase exposure. The embryos are then moved to 48 well plates for embryo preparation, WISH, and tissue fixation. Next, embryos are transferred to 12 well plates for scoring, imaging, and storage. Precise arraying of embryos is accomplished by the use of a color-coding system that enables the researcher to keep track of the chemical of origin for each test sample. Please click here to view a larger version of this figure.
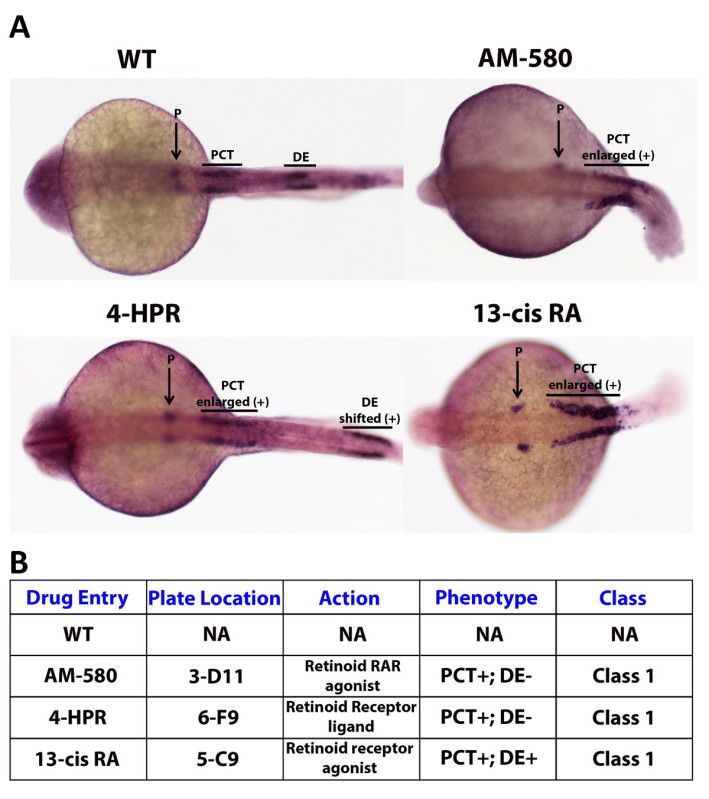 Figure 4. Example grading system: kidney segmentation assay. A WISH experiment enables the researcher to stain specific cell types based on the identity of the probes that are selected, allowing the evaluation of changes in embryo phenotypes according to changes in mRNA transcription location and/or levels. As an example, three segments of the zebrafish embryonic kidney, the pronephros, were labeled using Screen Mix 1. Screen Mix 1 is a cocktail of probes consisting of wt1b, which specifically localizes in the podocytes (P), slc20a1a which marks the proximal convoluted tubule (PCT), and slc12a1 which labels the distal early (DE) segment47. A (+) symbol next to a region label is used to annotate the expansion or positive shift of that region, while a (-) symbol is used to dictate the diminishment of a region. Here, embryos drug treated with 13-cis RA have an enlarged proximal convoluted tubule and a complete loss of the distal early segment with no apparent effect on the podocytes. Please click here to view a larger version of this figure.
Figure 4. Example grading system: kidney segmentation assay. A WISH experiment enables the researcher to stain specific cell types based on the identity of the probes that are selected, allowing the evaluation of changes in embryo phenotypes according to changes in mRNA transcription location and/or levels. As an example, three segments of the zebrafish embryonic kidney, the pronephros, were labeled using Screen Mix 1. Screen Mix 1 is a cocktail of probes consisting of wt1b, which specifically localizes in the podocytes (P), slc20a1a which marks the proximal convoluted tubule (PCT), and slc12a1 which labels the distal early (DE) segment47. A (+) symbol next to a region label is used to annotate the expansion or positive shift of that region, while a (-) symbol is used to dictate the diminishment of a region. Here, embryos drug treated with 13-cis RA have an enlarged proximal convoluted tubule and a complete loss of the distal early segment with no apparent effect on the podocytes. Please click here to view a larger version of this figure.
Discussion
Chemical genetics in the zebrafish can make a critical impact on the progression of drug discovery. This protocol provides a manual high-throughput method to perform chemical genetics in zebrafish embryos with a WISH read-out, effectively enabling an individual or small team to conduct a small molecule screen. This minimal-resource method extends the feasibility of chemical genetics to many research groups. Specifically, this screen strategy provides an important alternative to the use of robotic instrumentation, as such systems are not broadly accessible across research departments. This manual screen enables a single individual to test one 96 well plate of compounds with a WISH readout in a 6 day work week. Since zebrafish are oviparous, with the embryos developing outside the parent, and the embryos are large enough to see with the naked eye (between 1-3 mm), they are amenable for manual handling with simple instruments and the researcher can array samples in multi-well plates without the use of a microscope. Further, since zebrafish develop rapidly, small molecule exposure can be effective with a single treatment of a compound, rather than repeated doses, which minimizes further manual processing. Zebrafish embryos are easily cultured because their yolk provides a source of nutrition up to 5-6 days post fertilization, which eliminates the complication of fry feeding. Additionally, the majority of the larval organs have developed and become functional during this period, which provides the opportunity to study a wide array of research questions. The simple addition of small molecules to the embryo incubation media is non-invasive and allows the researcher(s) to select for drug dose, time of addition, and duration of exposure—thus providing tight control over many variables and allowing for specific developmental processes of interest to be queried. As described here, significant research productivity can be achieved in a short timeframe using this manual screen methodology. In our experience, there is a high return on the invested effort, which can be accentuated by using a known bioactive chemical library to analyze a process of interest, such as nephron segmentation during renal development (Figure 4; Poureetezadi and Wingert, unpublished).
The manual chemical screen described here is exceptionally versatile because it involves an assay on fixed zebrafish specimens. For example, this strategy can be revised to test several plates of compounds in one week and then to score embryos the following week. Alternative assays on fixed zebrafish samples, such as immunohistochemistry or other tissue staining preparations, could also be substituted for the WISH read-out. Experimental strategies to perform cellular assays in zebrafish are adaptable due to the optical clarity and transparency of early developmental stages. Further, researchers can inhibit pigmentation at later stages using chemicals or avoid the complication of pigment development altogether through the use of pigment-less genetic lines like casper or sheer zebrafish13. It is feasible to manually administer small molecules, process the stain(s) of interest, and to manually score embryos using a simple stereomicroscope. However, it is important to keep in mind that automated image analysis could be performed, and may in fact be required for high resolution phenotype analysis. For instance, it may be preferable to partner an automated image analysis system with a direct assay, such as a transgenic reporter, as this streamlines the manual processing of samples, quantifies differences not easily discernible to the naked eye, and eliminates researcher bias15. Fortunately, some automated imaging systems are both user-friendly and economical in terms of software costs15. The use and capabilities of other automated systems have rapidly evolved and can be used to orient organisms, administer drug treatments, and even to relocate organisms12,15. These automated systems have heralded a new technological age of research and have provided innovative solutions to manipulate large quantities of test subjects and thus perform HTS and HCS approaches in whole organisms12,15. While such machines are undeniably useful tools, they are also very costly and are out of reach for many basic research laboratories. Without access to these automated capabilities it may seem that chemical screens are not possible. However, this protocol outlines a guide for how to conduct a manual screen that can be used to complete a chemical genetics screen in a practical way over a reasonable period of time.
Drawbacks for both manual screening and chemical genetics in general exist, and should be kept in mind when considering a chemical screen. In particular, the method demonstrated here necessitates a moderate degree of physical dexterity. This concern is diminished or entirely obviated through repetition, as dexterity will improve with practice. Additionally, there is minimal room for gross error, as there are only 10 embryos per well for analysis. Small molecules can vary in the penetrance of the phenotype that is induced, so a reasonable scoring regimen to identify compounds of interest must be adhered to. It is vital that the researchers utilizing this protocol devise suitably rigorous criteria for what they will consider a hit. It is also important to keep in mind that the nature of this form of screen will most likely yield a certain portion of false positives, which can be delineated through further chemical work-up. Further, it is critical to stage embryos precisely and to collect clutches for screening shortly after fertilization, as asynchronous development can lead to complications in accurately evaluating the effect of drug treatments. Further, in regards to chemical screening as an experimental strategy, there are a host of limitations. Compound solubility and toxicity of carrier molecules (e.g., dimethyl sulfoxide (DMSO)) can also present an issue and may be prohibitive in testing many molecules. The chorion can also act as barrier to drug exposure. Finally, even though zebrafish are a powerful and translational model for chemical genetics, care should always be taken in determining whether an alternative chemical screen strategy, e.g., using an in vitro system such as a cell line, can be substituted for the use of whole animals. Nevertheless, chemical treatment in the whole zebrafish system is considered preferable to conducting similar approaches in mammals, as it provides the advantage of gathering the systemic effects of the compounds and can enable researchers to triage the list of compounds to be tested in a mammalian model.
To date, zebrafish chemical screens have provided a powerful experimental tool to delineate the mechanisms of development and to ascertain candidate pharmacological agents for use in medicine, such as the identification of molecules capable of preventing disease pathologies. For example, a study by Burns and colleagues developed an automated micro-well assay for heart rate using transgenic zebrafish embryos that expressed GFP in the myocardium42. They used this transgenic line to analyze heart rate in development and how it was affected upon drug treatment42. Another study, by testing the population density of hematopoietic stem cells in the zebrafish embryo, identified that prostaglandin E2 regulates the HSC niche23. This finding was validated in the mouse, and was later implemented for clinical testing in human umbilical cord transplants23,31-33 (NIH, Clinical Trials.gov, NCT00890500; NCT01627314). Through several recent studies, chemical genetics has proven to be useful in studying kidney disease using the zebrafish43. The zebrafish kidney provides an excellent paradigm to study nephrogenesis or regeneration of the nephron, the functional unit that comprise the kidney during development and acute kidney injury43. Nephron structure is highly conserved between the zebrafish embryonic kidney and the adult mammalian kidney44-50. Several studies examining the zebrafish embryonic kidney clearly illustrate its inherent translational potential when coupled with chemical genetics. A chemical genetics screen was performed on two zebrafish mutants that were mapped to the genes pkd2 and ift172, which are known to be key players in polycystic kidney disease (PKD)34. The screen resulted in identifying the pan-histone deacetylase (HDAC) inhibitor trichostatin A (TSA) as a small molecule that could nullify the morphological defects normally seen in the mutant embryos. A different approach taken by de Groh and colleagues screened for compounds that induced edema in wild-type zebrafish embryos, which would indicate a disruption in kidney function35. After identifying PTBA as a hit, they observed that in addition to causing edema, PTBA also increased the expression of pax2a, a marker for renal progenitors. Importantly, an optimized derivative of PTBA was then shown to decrease the percent fatality of adult zebrafish that underwent gentamicin-induced kidney damage and furthermore, accelerated the recovery of mice that suffered from renal damage27. Taken together, these contributions from zebrafish chemical genetics showcase the types of discoveries that can be made using the method described in this protocol.
While chemical genetics research bears substantial merit, it is not without certain challenges. Solely using a chemical genetics approach can make it complicated to determine the specific gene or genes that are being affected by a compound. Even if a target(s) is found, a significant gap can remain between compound identification and understanding the mechanism(s) of action in which the small molecule behaves. For example, it can be difficult to determine the mechanism in which a small molecule is acting because a single compound is capable of having a range of different effects within an organism. However, with the advent of new technology and more chemical genetic screens being completed in zebrafish, the problem of determining a mechanism is shrinking. One method to help determine the molecular effects of identified compounds is to select a drug library of known bioactives for screening, where mechanisms can be potentially deduced from previously annotated data. Additionally, chemoinformatic algorithms, like Discovery gate, are available for use that can compare structural similarities of a compound against a database of compounds with ascribed mechanisms14. Yet another way to elucidate the mechanism of a compound would be to generate a microarray profile after treatment and then query this profile in a compilation of microarray drug profiles, such as Connectivity Map, searching for mechanistically defined compounds that elicit a similar effect14. This approach can also be used to identify genetic mutants that have a similar effect as the compound of interest. Finding a connection between a compound and a mutant might suggest that the compound works molecularly upstream or downstream of the related gene, which could be delineated by treating the mutants with the compound and morpholinos. Another option would be mass spectrometry, which is continuing to become more optimized for zebrafish. This method can be used to delineate specific protein changes or post-translational modifications after drug treatment. Finally, small molecules or even entire libraries are sometimes capable of being tagged for pull-down of binding partners. It is also worth noting, that while the mechanism of how a small molecule functions is of significant import, there are FDA-approved drugs for which the mechanisms remain unknown.
Although zebrafish exhibit many similarities to mammals both genetically and physiologically, there is still an issue of drug crossover14. One study investigated the crossover capability of the hits from a screen for cell cycle inhibitors in zebrafish embryos50. The screen resulted in the identification of 14 hits that were previously unknown to possess cell cycle activity. Out of these 14 compounds, 6 were found to have cell cycle activity in both human and zebrafish cell culture assays, 3 were shown to be serum inactivated in cell culture assays, 1 was active only in zebrafish cells, and 4 had no activity in either zebrafish or human cell culture assays, but only in the zebrafish whole organism. Notably, the 4 compounds that only had activity in the zebrafish demonstrate that there are differences between zebrafish and mammals that may prohibit the translational potential of certain compounds. This is an important consideration to be mindful of, however there are examples of small molecules, like PGE2 which amplifies the ability of HSCs to engraft in human recipients and PTBA-derivatives that improve the rate of renal recovery in mammals, thus providing the proof of principle that crossover is possible23,31-33.
Regardless of these pitfalls, chemical genetics and this minimal-resource approach has much to offer the research community. Chemical genetics is particularly useful in the zebrafish and will continue to play a critical role in molecular pathway interrogation and drug discovery. Chemical screens based on discrete molecular targets have historically been subject to a high failure rate due to what is known as the constellation of adsorption, distribution, metabolism, excretion, and toxicological (ADMET) properties51. Chemical screens utilizing zebrafish to focus on a process, rather than a discrete target, can circumvent several ADMET problems and identify compounds that are efficacious in the context of the entire organism. With the increasing pool of resources available for zebrafish research, including current mutant strains and the ability to use genome editing to create targeted genetic mutants and transgenics, there are virtually a limitless number of potential chemical genetic screens using zebrafish. Many chemical libraries have been curated, and span collections with known bioactives, novel compounds, structurally diverse and structurally similar. These reagents provide a wealth of exciting opportunities for screen combinations that are currently available and are likely to further grow in the coming years. With these possibilities in hand, this manual and high-throughput protocol provides a practical and user-friendly way to increase the capability of research groups to undertake chemical genetic screens using zebrafish.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by funding to the Wingert research laboratory from the following: National Institutes of Health grants K01 DK083512, DP2 OD008470, and R01 DK100237; March of Dimes Basil O’Connor Starter Scholar grant award #5-FY12-75; start up funds from the University of Notre Dame College of Science and Department of Biological Sciences; and a generous gift to the University of Notre Dame from Elizabeth and Michael Gallagher on behalf of the Gallagher Family to foster stem cell research. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Paul T. Kroeger, Jr. for providing critical feedback on the manuscript. We thank the staff of the Department of Biological Sciences for their support, and the Center for Zebrafish Research at Notre Dame for their outstanding dedication in the care and welfare of our zebrafish colony. Finally, we thank the members of our research lab for their comments, discussions and insights about this work.
References
- Grunwald DJ, Eisen JS. Headwaters of the zebrafish—emergence of a new model vertebrate. Nat. Rev. Genet. 2002;3(9):717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- Dahm R, Geisler R. Learning from small fry: the zebrafish as a genetic model organism for aquaculture fish species. Mar. Biotechnol. (NY) 2006;8(4):329–345. doi: 10.1007/s10126-006-5139-0. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view) Nat. Rev. Genet. 2007;8(5):353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Wen ZH, Lin CS, Chakraborty C. The zebrafish model: use in studying cellular mechanisms for a spectrum of clinical disease entities. Curr. Neurovasc. Res. 2007;4(2):111–120. doi: 10.2174/156720207780637234. [DOI] [PubMed] [Google Scholar]
- Goldsmith JR, Jobin C. Think small: zebrafish as a model system of human pathology. J. Biomed. Biotechnol. 2012. [DOI] [PMC free article] [PubMed]
- Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J. Clin. Invest. 2012;122(7):2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henken DB, Rasooly RS, Javois L, Hewitt AT. The National Institutes of Health and the growth of the zebrafish as an experimental model organism. Zebrafish. 2004;1(2):105–110. doi: 10.1089/zeb.2004.1.105. [DOI] [PubMed] [Google Scholar]
- Sprague J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DG, et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessman CA. The developing zebrafish (Danio rerio): A vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res. C Embryo Today. 2011;93(3):268–280. doi: 10.1002/bdrc.20212. [DOI] [PubMed] [Google Scholar]
- Pickart MA, Klee EW. Zebrafish approaches enhance the translational research tackle box. Transl. Res. 2014;163(2):65–78. doi: 10.1016/j.trsl.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Peal DS, Peterson RT, Milan D. Small molecule screening in zebrafish. J. Cardiovasc. Transl. Res. 2010;3(5):454–460. doi: 10.1007/s12265-010-9212-8. [DOI] [PubMed] [Google Scholar]
- Tsang M. Zebrafish: a tool for chemical screens. Birth Defects Res C Embryo Today. 2010;90(3):185–192. doi: 10.1002/bdrc.20183. [DOI] [PubMed] [Google Scholar]
- Mandrekar N, Thakur NL. Significance of the zebrafish model in the discovery of bioactive molecules from nature. Biotechnol. Lett. 2009;31(2):171–179. doi: 10.1007/s10529-008-9868-1. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Fishman MC. Designing zebrafish chemical screens. Methods Cell Biol. 2011;105:525–541. doi: 10.1016/B978-0-12-381320-6.00023-0. [DOI] [PubMed] [Google Scholar]
- Tan JL, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 2011;105:493–516. doi: 10.1016/B978-0-12-381320-6.00021-7. [DOI] [PubMed] [Google Scholar]
- Mathias JR, Saxena MT, Mumm JS. Advances in zebrafish chemical screening technologies. Future Med. Chem. 2012;4(14):1811–1822. doi: 10.4155/fmc.12.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin OJ, et al. Small molecule screening in zebrafish: swimming in potential drug therapies. Wiley Interdiscip. Rev. Dev. Biol. 2012;1(3):459–468. doi: 10.1002/wdev.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, et al. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl. Acad. Sci. USA. 2000;97(24):12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 2004;22(5):595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129(8):2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- Molina G, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat. Chem. Biol. 2009;5(9):680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokel D, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat. Chem. Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino CC, et al. Histone deacetylase inhibitor enhances recovery after AKI. J. Am. Soc. Nephrol. 2013;24(6):943–953. doi: 10.1681/ASN.2012111055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppendal D, Goldsmith MI. A chemical screen to identify novel inhibitors of fin regeneration in zebrafish. Zebrafish. 2010;7(1):53–60. doi: 10.1089/zeb.2009.0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara G, et al. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2011;108(13):5331–5336. doi: 10.1073/pnas.1102116108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471(7339):518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord AM, North TE, Zon LI. Prostaglandin E2: Making more of your marrow. Cell Cycle. 2007;6(24):3054–3057. doi: 10.4161/cc.6.24.5129. [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE. Hematopoietic stem cell development: Using the zebrafish to identify the signaling networks and physical forces regulating hematopoiesis. Methods Cell Biol. 2011;105:117–136. doi: 10.1016/B978-0-12-381320-6.00005-9. [DOI] [PubMed] [Google Scholar]
- Cutler C, et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood. 2013;122(17):3074–3081. doi: 10.1182/blood-2013-05-503177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, et al. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc. Natl. Acad. Sci. USA. 2009;106(51):21819–21824. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh ED, et al. Inhibition of histone deacetylase expands the renal progenitor population. J. Am. Soc. Nephrol. 2010;21(5):794–802. doi: 10.1681/ASN.2009080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, et al. Automated image-based phenotypic analysis in zebrafish embryos. Dev. Dyn. 2009;238(3):656–663. doi: 10.1002/dvdy.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Codore H, Day BW, Hukriede NA, Tsang M. Development of automated imaging and analysis for zebrafish chemical screens. J. Vis. Exp. 2010. [DOI] [PMC free article] [PubMed]
- Tran TC, et al. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res. 2007;67(23):11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- Jing L, Durand EM, Ezzio C, Pagliuca SM, Zon LI. In situ hybridization assay-based small molecule screening in zebrafish. Curr. Protoc. Chem. Biol. 2012;4 doi: 10.1002/9780470559277.ch110236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TB. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Cheng CN, Li Y, Marra A, Verdun V, Wingert RA. Flat mount preparation for observation and analysis of fixed zebrafish embryo specimens. J. Vis. Exp. 2014. [DOI] [PMC free article] [PubMed]
- Burns CG, et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 2005;1(5):263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Poureetezadi SJ, Wingert RA. Congenital and Acute Kidney Disease: Translational Research Insights from Zebrafish Chemical Genetics. General Med. 2013;1 doi: 10.4172/2327-5146.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: Insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2013;2(5):559–585. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wingert RA. Regenerative medicine for the kidney: stem cell prospect., & challenges. Clin. Transl. Med. 2013;2(1) doi: 10.1186/2001-1326-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCampbell KK, Wingert RA. Renal stem cells: fact or science fiction. Biochem. J. 2012;444(2):153–168. doi: 10.1042/BJ20120176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Davidson AJ. The zebrafish pronephros: a model to study nephron segmentation. Kidney Int. 2008;73(10):1120–1127. doi: 10.1038/ki.2008.37. [DOI] [PubMed] [Google Scholar]
- Wingert RA, Davidson AJ. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev. Dyn. 2011;240(8):2011–2027. doi: 10.1002/dvdy.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cheng CN, Verdun VA, Wingert RA. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and notch signaling. Dev. Biol. 2014;386(1):111–122. doi: 10.1016/j.ydbio.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem. Biol. Drug Des. 2006;68(4):213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- Williams CH, Hong CC. Multi-step usage of in vivo models during rational drug design and discovery. Int. J. Mol. Sci. 2011;12(4):2262–2274. doi: 10.3390/ijms12042262. [DOI] [PMC free article] [PubMed] [Google Scholar]


