Abstract
Adult body size in higher animals is dependent on the amount of growth that occurs during the juvenile stage. The duration of juvenile development, therefore, must be flexible and responsive to environmental conditions. When immature animals experience environmental stresses such as malnutrition or disease, maturation can be delayed until conditions improve and normal growth can resume. In contrast, when animals are raised under ideal conditions that promote rapid growth, internal checkpoints ensure that maturation does not occur until juvenile development is complete. Although the mechanisms that regulate growth and gate the onset of maturation have been investigated for decades, the emerging links between childhood obesity, early onset puberty, and adult metabolic disease have placed a new emphasis on this field. Remarkably, genetic studies in the fruit fly Drosophila melanogaster have shown that the central regulatory pathways that control growth and the timing of sexual maturation are conserved through evolution, and suggest that this aspect of animal life history is regulated by a common genetic architecture. This review focuses on these conserved mechanisms and highlights recent studies that explore how Drosophila coordinates developmental growth with environmental conditions.
Introduction
The life history of insects is similar to that of other animals, with discrete stages representing embryonic development, a juvenile growth phase, sexual maturation, and reproductive adulthood. In Drosophila, these stages correspond to four morphologically distinct developmental states: embryo, larva (three instar stages), pupa, and adult. Embryogenesis, along with the first and second larval instars (L1 and L2), each last one day, followed by two days of third instar larval development (L3). The larval growth stage is terminated by puparium formation and four days of metamorphosis, during which the sexually active adult fly is formed [1]. Progression through all of these stages is dictated by pulses of the steroid hormone 20-hydroxyecdysone (20E) [2,3]. A series of enzymatic steps within the endocrine organ of the insect, the prothoracic gland (PG), converts cholesterol into ecdysone, which is released into the circulatory system and modified by peripheral tissues into the active form of the hormone, 20E [4,5]. This steroid acts through a heterodimer of the ecdysone receptor (EcR) and Ultraspiracle (USP) nuclear receptors to trigger stage-specific transcriptional cascades that direct progression through each stage in the fly life cycle, determining the timing of developmental progression [2,3].
All growth in Drosophila normally occurs during the juvenile larval stages, resulting in a remarkable ~200-fold increase in body mass [6]. Thus, the 20E signaling events that determine the duration of larval development are critical for dictating final body size. The pulses of 20E during L1 and L2 trigger molting of the larval cuticle, accommodating the increase in animal size [2,3]. Additional low-titer hormone pulses during the L3 stage prepare the animal for metamorphosis [7], while a high-titer 20E pulse at the end of L3 terminates larval development, arrests growth, and signals the onset of adult maturation [2,3]. This critical role for 20E in determining the duration of larval development implies that key growth regulators must feed into the timing of these events. Recent genetic studies in Drosophila have identified these pathways, laying the groundwork for understanding how environmental factors can regulate growth and determine the timing of maturation.
Linking Body Size to the Timing of Maturation
Both developmental and nutritional signals feed into the timing of the 20E pulses that dictate the duration of larval growth. The manner in which animals respond to these signals, however, changes during larval development, and is centered on an important but poorly understood transition that occurs near the L2-to-L3 molt in laboratory strains of Drosophila [8,9]. At this time, larvae first surpass ‘minimal viable weight’, where they achieve sufficient body mass to successfully complete larval and pupal development in the absence of nutrients [10,11]. This event is followed by a key life history event that commits larvae to enter metamorphosis within a definite period of time — the attainment of ‘critical weight’ [9–14]. If animals encounter poor nutrient conditions prior to the onset of critical weight, larval development will stall and subsequent 20E pulses will be delayed until growth conditions improve (Figure 1). This link between nutrition and maturation makes sense, insofar as it provides a means of storing sufficient nutrients for survival during the non-feeding pupal stage of development as well as an opportunity for the animal to achieve an appropriate size for adult reproductive fitness. The time an animal spends developing as a pre-critical weight larva can vary greatly, as progression past the critical weight checkpoint is determined by body size and not time. Although physiological studies in other insects, such as the tobacco hookworm Manduca sexta, have demonstrated an important role for juvenile hormone in determining the attainment of critical weight, no studies have shown that this function is conserved in Drosophila [12,15].
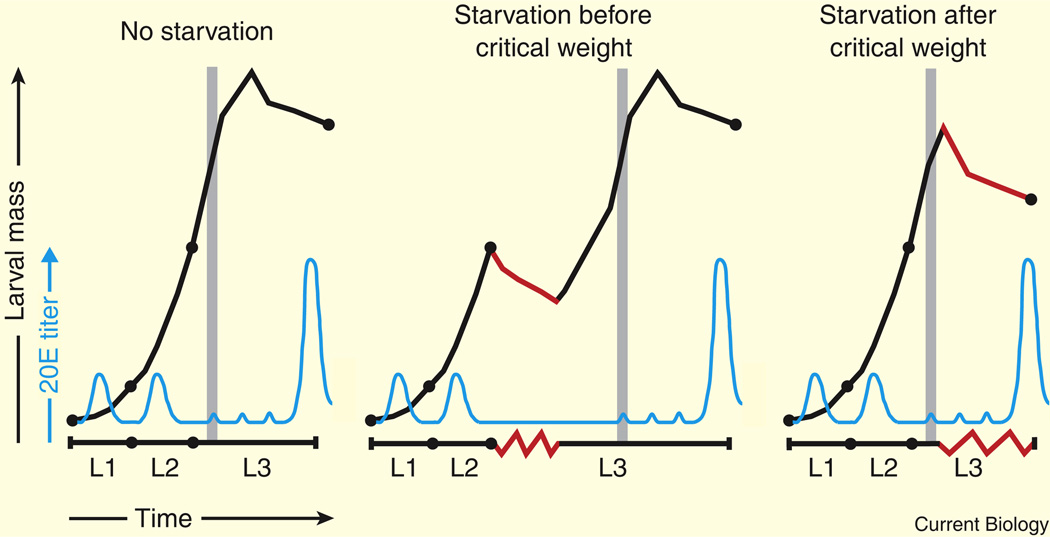
Figure 1. A schematic representation of Drosophila larval growth and development.
Drosophila larvae experience exponential growth (black line) as they develop through three distinct larval instars (L1, L2, and L3). Pulses of 20E (blue line) direct progression through the larval molts. The critical weight checkpoint (grey vertical line) occurs near the L2–L3 molt. A series of low-titer 20E pulses occur at ~8, 20, and 28 h after the L2–L3 molt, followed by a high-titer 20E pulse at the end of L3 that triggers puparium formation [7]. If an animal is starved (red lines) prior to the attainment of critical weight, development stalls until the larva finds a new food source, but final body size is unaffected. After critical weight is achieved, starvation inhibits growth but no longer affects developmental progression, resulting in a significantly smaller final body size.
Larvae that have achieved critical weight have sufficient stored energy to successfully complete metamorphosis, and the high-titer, late-L3 pulse of 20E will occur after a definite period of time, without regard to nutrient availability (Figure 1). With this strategy in effect, environmental conditions will dictate final body size. Larvae that develop in a favorable environment will continue to grow and can significantly increase their body size before entering metamorphosis [9,14,16]. In contrast, post-critical weight larvae that experience starvation will stop growing in size but will continue to mature into smaller fertile adults [9,14,16] (Figure 1). Ultimately, both fed and starved post-critical weight animals enter metamorphosis within a similar time frame, but animals that continue to feed are larger than animals that experience starvation. Larval development, therefore, is regulated by genetic mechanisms that coordinate developmental progression and growth with nutrient availability, uptake, and utilization. While many of these pathways control cell-intrinsic processes, successful development requires that growth is coordinated among all tissues within the larva. This level of systemic control is achieved by secreted factors that regulate cellular physiology. Intriguingly, these factors not only arise from the brain and endocrine organs, but also are produced and secreted by the main source of stored energy within the animal: the fat body.
Nutrient Signaling and the Fat Body
The larval fat body, which functions as a hybrid of the mammalian liver and white adipose tissue, plays a central role in sensing nutritional signals and allowing diverse tissues to coordinately respond to changes in metabolic status. The central role of the fat body in regulating organismal growth was first described nearly 35 years ago, when it was found to promote nonautonomous growth in cultured larval imaginal discs [17]. Similarly, quiescent neuroblasts will reenter the cell cycle in vitro when co-cultured with fat body tissue from fed larvae [18]. A key to understanding how the fat body regulates peripheral tissue growth was identified in genetic screens for growth modifiers. Mutations in the putative amino-acid transporter minidiscs result in developmental arrest and imaginal disc growth defects even though this gene is expressed primarily within the fat body [19]. Similarly, decreased expression of the gene slimfast (slif), which encodes a cationic amino-acid transporter and is highly expressed in the fat body, delays growth and produces abnormally small animals [20]. Furthermore, tissue-specific depletion of slif in the fat body elicits a whole-body growth defect, demonstrating that the fat body can retard organismal growth in response to decreased amino-acid availability [20] (Figure 2). Further studies revealed that this effect is mediated by the TOR signaling pathway, which is a critical regulator of nutrient signaling in Drosophila [21,22]. Mutations that disrupt the TOR pathway phenocopy the loss of slif function. These phenotypes can also be seen with fat body specific TOR inactivation, while overexpression of the TOR downstream target S6 kinase can partially rescue the growth defects caused by slif depletion [20]. The fat body, therefore, can monitor amino-acid levels via the TOR signaling pathway and can remotely coordinate growth and developmental progression (Figure 2).
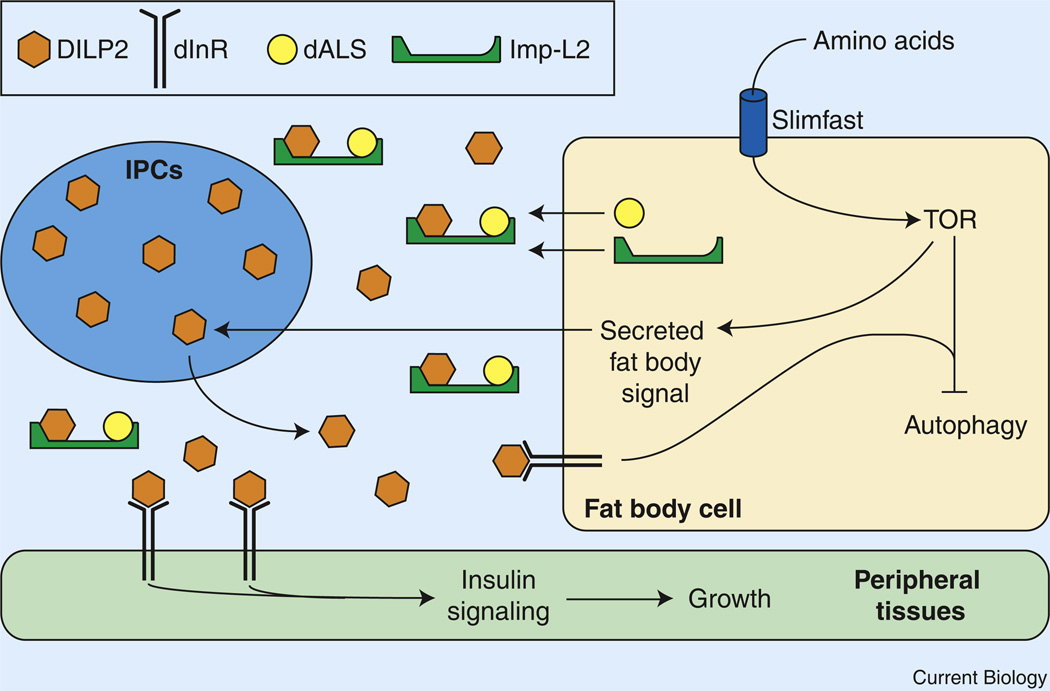
Figure 2. The larval fat body regulates systemic growth.
Pre-critical weight larval growth is regulated by nutrient-dependent signals that emanate from the fat body. Ingested amino acids are sensed by fat body cells and activate TOR kinase, which promotes the release of an unknown factor that stimulates DILP2 secretion from the insulin-producing cells (IPCs). DILP2, in turn, promotes growth and development in peripheral tissues by binding to the insulin receptor (dInR) and activating the insulin signaling pathway. Additionally, the fat body releasesd ALS and Imp-L2, which form a stable complex with DILP2 and dampen insulin signaling.
A primary focus of fat body regulated growth is the insulin signaling pathway [16,23] (Figure 2). The Drosophila genome encodes seven insulin-like peptides (DILP1–7) [24,25], three of which, DILP2, 3, and 5, are expressed in two clusters of neurosecretory cells within the larval brain. These insulin-producing cells (IPCs) are functionally similar to pancreatic β-cells, and can secrete DILPs into the hemolymph [26]. Circulating DILPs bind to the Drosophila insulin receptor (dInR) on target cells and activate a highly conserved phosphoinositide 3 kinase (PI3K) signaling cascade that inhibits the dFOXO transcription factor, thereby promoting cell-autonomous growth [27]. When a larva experiences nutrient deprivation there is a decrease in dInR-dependent PI3K activity [28]. As a result, dFOXO translocates from the cytoplasm into the nucleus and inhibits cell growth [29–31].
Since larvae possess an open circulatory system, nutrient-deprived animals can rapidly slow development by regulating DILP activity. The expression of both dilp3 and dilp5 are transcriptionally downregulated under low nutrient conditions [24]. In contrast, dilp2 transcript levels are insensitive to nutrient deprivation, but DILP2 protein secretion and signaling activity are heavily influenced by metabolic status. This regulation can be readily visualized in the IPCs, where DILP2 protein is present at a basal level in well-fed animals but accumulates to relatively high concentrations upon starvation or amino-acid deprivation [32].
DILP2 secretion is a primary target of nutrient-dependent fat body signaling (Figure 2). When slif expression or TOR activity is specifically disrupted in the fat body, DILP2 is not secreted and accumulates in the IPCs [32]. Furthermore, in a series of elegant experiments, co-culturing brains isolated from starved L3 larvae with fat body tissue or hemolymph from fed animals was shown to promote DILP2 secretion from the starved animals’ IPCs [32]. This response, however, did not occur when the brain co-culture was conducted using tissue from starved larvae. Similarly, quiescent larval neuroblasts re-enter the cell cycle in response to TOR activation within the fat body, promoting brain growth during larval stages [18,33,34]. The fat body of fed animals, therefore, must secrete an as yet unidentified factor that promotes DILP secretion. This approach paves the way for a clearer understanding of the mechanisms by which the fat body can sense nutritional status and relay that information to control developmental growth.
The fat body not only controls DILP secretion, but also releases two proteins, Imp-L2 and dALS (acid labile subunit), that interact with circulating DILP2 [35,36] (Figure 2). These two proteins, however, do not promote insulin-signaling, but rather appear to sequester and inactivate DILP2 in a stable trimeric complex, as fat-body-specific depletion of either Imp-L2 or dALS results in an overgrowth phenotype [20,35,36]. These studies provide a new context for understanding the mechanisms by which ALS contributes to mammalian insulin signaling as well as insights into how the larval fat body can control systemic insulin signaling and coordinate organismal growth.
The fat body also releases stored nutrients to ensure the survival of peripheral tissues during periods of starvation. Under normal growth conditions, nutrient-dependent TOR signaling functions cell autonomously to suppress autophagy — a process by which cells can non-specifically degrade bulk cytoplasm for energy production (Figure 2). When animals become nutrient-deprived, decreased TOR signaling results in autophagic degradation of the fat body, thereby releasing nutrients that help sustain the starving animal [37,38]. Intriguingly, decreased insulin signaling within fat body cells can also promote autophagy [37,38]. The fat body, therefore, both coordinates systemic growth and provides an essential source of energy in response to unfavorable environmental conditions.
An Evolutionarily Conserved Program to Coordinate Growth and Maturation
Once the larva achieves critical weight, the PG begins to release low-titer pulses of 20E, preparing the animal for the cessation of larval development [7] (Figure 1). This ‘mid-third instar transition’ consists of key behavioral and developmental changes, including a cessation of feeding, the onset of wandering behavior, glue protein synthesis in the salivary glands, and the initiation of fat body autophagy [37–39]. The regulation of steroid hormone activity to promote maturation appears to define an ancient regulatory pathway by which many animals control this key life history event. This common genetic architecture was first described in the context of the Caenorhabditis elegans life cycle, when the animal makes a decision, based on environmental factors, to either continue development to form a reproductive adult or enter a larval diapause state [40,41]. When juvenile worms encounter poor growth conditions during the first larval stage, they indefinitely arrest development by forming a dauer larva, an alternative to the L3 stage that is ideally suited for survival [42].
The genetic mechanisms that regulate developmental growth were discovered through an elegant and unbiased genetic screen for mutations that affect dauer formation [43,44]. The identification and subsequent characterization of these abnormal dauer formation (daf) mutants defined three key signaling pathways: insulin, TGFβ, and the steroid hormone dafachronic acid (DA) (Figure 3). Mutations that eliminate signaling through either the insulin or TGFβ pathways cause animals to become dauers independent of culture conditions [43,45,46]. In contrast, daf mutations that disrupt negative regulators of either pathway, such as the FOXO homolog [47,48], render animals incapable of forming dauers [43]. These genetic studies demonstrate that both pathways converge on the regulation of DA signaling. DA is a steroid hormone that binds to and modulates the activity of the nuclear receptor DAF-12 [49]. When DA is present, DAF-12 promotes continuous development, while in the absence of DA production DAF-12 induces dauer formation. Similarly, loss-of-function mutations that reduce DA synthesis or prevent DA from binding to the DAF-12 ligand-binding domain lead to dauer formation [50–52]. A commitment to adult maturation, therefore, is dependent on the coordinate activity of these three signaling pathways. Favorable growth conditions stimulate insulin and TGFβ signaling, which, in turn, promotes DA production, DAF-12 activation, and continued progression through larval development [53,54]. Conversely, poor growth conditions reduce insulin and TGFβ signaling, resulting in decreased DAF-12 signaling and dauer formation [53,54] (Figure 3).
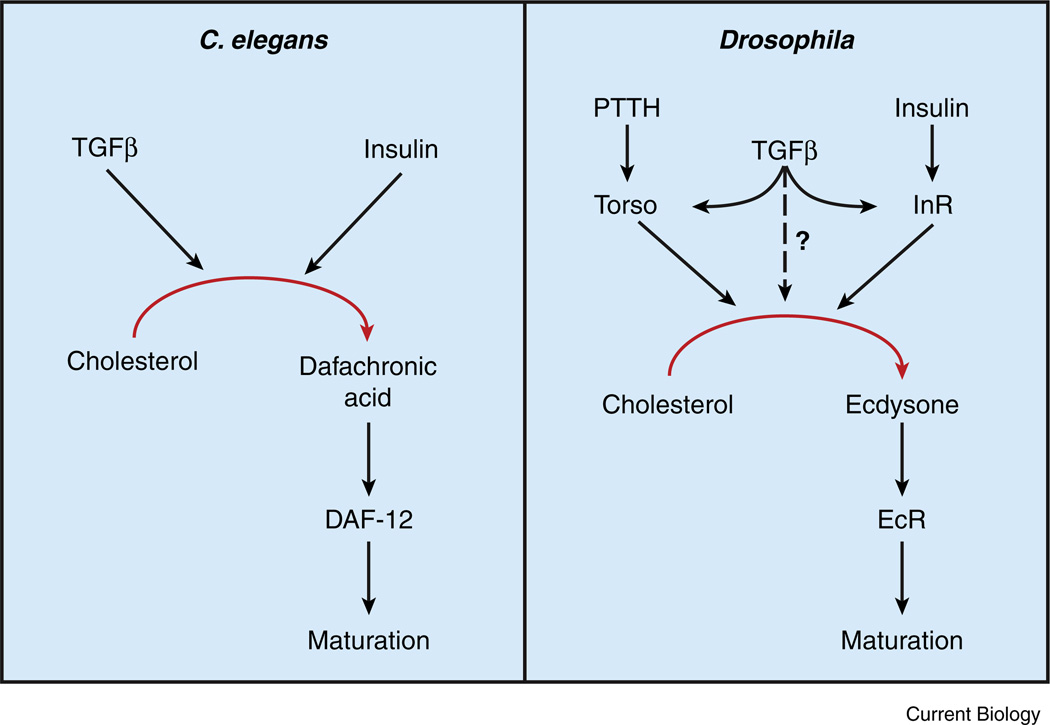
Figure 3. A conserved genetic hierarchy regulates animal maturation.
A combination of insulin and TGFβ signaling regulates steroid hormone production and maturation in both C. elegans and Drosophila. In worms, dietary nutrients and favorable growth conditions increase TGFβ and insulin signaling in endocrine tissues and stimulate dafachronic acid (DA) synthesis. DA systemically activates the nuclear receptor DAF-12, thereby preventing dauer formation and promoting maturation. In Drosophila, TGFβ signaling in the prothoracic gland (PG) upregulates expression of Torso and the insulin receptor (dInR), which promote ecdysone synthesis in response to PTTH and insulin, respectively. Ecdysone is then released from the PG, converted into 20E, and promotes maturation by systemically activating the ecdysone receptor (EcR).
Remarkably, recent studies suggest that a similar genetic framework controls the assessment of critical weight and maturation in Drosophila (Figure 3). These pathways exert their effect, in part, by sensitizing the PG to the activity of prothoracicotropic hormone (PTTH), a brain-derived neuropeptide that promotes proper ecdysone release [55]. The ptth gene is cyclically expressed throughout L3 development with a periodicity of approximately 8 hours [55]. Once the animal achieves critical weight, PTTH binds to the receptor tyrosine kinase Torso and activates a canonical mitogen-activated protein kinase (MAPK) signaling cascade [56]. This signaling event is essential for monitoring critical weight and for determining the timing of the onset of metamorphosis, as PTTH-dependent activation of MAPK signaling upregulates the expression of ecdysone biosynthetic genes [56]. If constitutively active forms of Torso, Ras, and Raf are expressed in the PG, animals precociously secrete ecdysone and pupariate early [56,57]. Similarly, both ecdysone release and pupariation are delayed when Torso, Ras, Raf, and Erk function are disrupted in the PG or if PTTH signaling is eliminated [56,57].
Interestingly, as in C. elegans, both TGFβ and insulin signaling feed into this hormone signaling pathway (Figure 3). When TGFβ signaling is reduced within the PG, Torso expression is significantly reduced and MAPK activity is downregulated [58]. As a result, the ecdysone biosynthetic genes are not properly expressed and the animals arrest development as L3. The observation that this phenotype is more severe than the elimination of PTTH signaling suggests that TGFβ can regulate ecdysone release through other pathways as well. At least one of these pathways appears to be insulin signaling [58]. Reduction of TGFβ signaling in the PG leads to reduced levels of dInR and reduced insulin signaling in this tissue. Moreover, expressing dInR or dAkt specifically in the PG is sufficient to overcome the block in larval development caused by reduced TGFβ signaling in this tissue [58]. This observation is consistent with several earlier studies that showed that activation of the insulin signaling pathway in the PG results in elevated ecdysone signaling and precocious initiation of metamorphosis [10,57,59]. Conversely, inhibiting insulin signaling within this organ dampens ecdysone signaling and extends larval growth. These results demonstrate that insulin and TGFβ signaling play a central role in coordinating growth with developmental progression and suggest that Dilp and TGFβ production or activity is sensitive to changes in body size. A key direction for future research will be to determine how the mechanisms that assess larval growth are linked to growth factor signaling.
It is interesting to note that a few studies in humans suggest that this pathway is conserved through evolution. Juvenile females diagnosed with type I diabetes mellitus exhibit a significant delay in menarche [60], while individuals with Marfan syndrome, which is likely caused by excessive TGFβ signaling, experience early onset puberty [61–63]. Thus, TGFβ and insulin signaling appear to control the timing of maturation in many higher organisms, defining a conserved genetic architecture that modulates steroid hormone signaling and the commitment to adult reproductive growth.
Regulation of Post-Critical Weight Growth
The mid-L3 pulses of 20E correlate with dramatic changes in the larval growth program that allow development to progress independent of nutrient availability. This developmental transition stems, in part, from a fundamental change in the role of insulin signaling. Prior to mid-L3, insulin controls the rate of developmental progression and, when young larvae that harbor temperature-sensitive dInR alleles are raised at a non-permissive temperature, development is significantly delayed [64]. In contrast, when these mutants are shifted to a non-permissive temperature after mid-L3, depletion of insulin signaling produces smaller adults but does not affect larval development [64]. Similarly, ectopic expression of dFOXO before mid-L3 elicits a developmental delay, but expression during mid- to late-L3 only affects body size [29].
These fundamental changes in insulin signaling are likely a result of increased 20E activity after the mid-L3 transition. This hormone inhibits larval growth, with body size being significantly reduced in animals that are fed exogenous 20E [59]. Elevated 20E signaling does not, however, affect growth in dFOXO mutants, indicating that 20E regulates growth by antagonizing insulin signaling [59] (Figure 4). Intriguingly, fat-body-specific depletion of EcR is sufficient to suppress the growth-inhibitory effects of 20E, while overexpression of the insulin signaling inhibitor dPTEN within the fat body has no effect on body size, demonstrating that ecdysone signaling in the fat body acts remotely to control organismal growth [59,65].
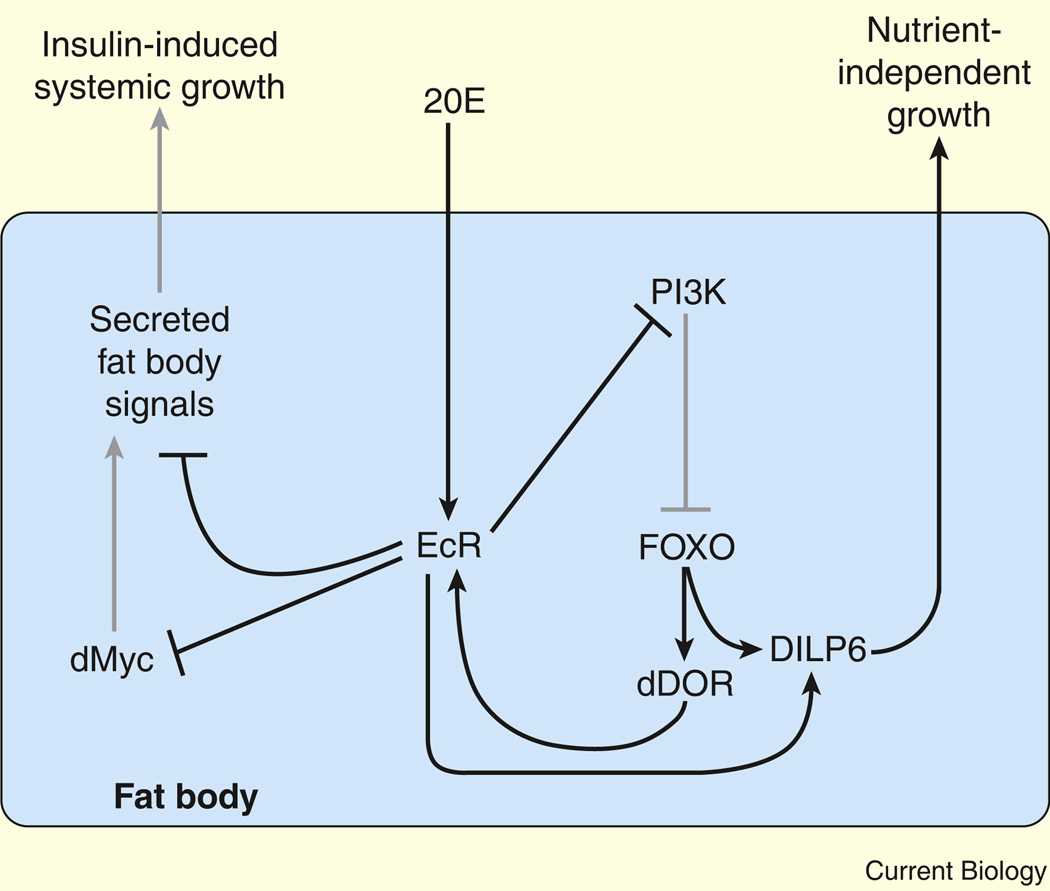
Figure 4. Ecdysone functions in the fat body to regulate systemic growth.
EcR activation by 20E in the fat body inhibits systemic insulin signaling and growth, in part by downregulating dMyc expression. EcR also inhibits PI3K signaling, which allows dFOXO to translocate to the nucleus and activate the expression of target genes. These include dDOR and dilp6, which is also upregulated by 20E–EcR. dDOR further activates EcR signaling, while dilp6 promotes nutrient-independent growth. Grey lines represent genetic interactions that are downregulated by 20E signaling.
Although the mechanism that links systemic growth and insulin signaling with EcR activity in the fat body remains unclear, this interaction is due, in part, to 20E-dependent regulation of the dMyc transcription factor, which plays a key role in promoting growth (Figure 4). EcR activation leads to a decrease in dMyc protein levels while, conversely, EcR depletion in the fat body results in cell-autonomous induction of dMyc expression [65]. The effects of 20E on growth are, at least in part, dependent on dMyc because a reduction in dMyc expression can suppress the increased growth that results from decreased EcR activity [65]. Intriguingly, TOR also regulates dMyc expression [66], hinting at a model whereby nutrient sensing and 20E signaling are integrated to coordinate systemic growth.
The interaction between 20E and insulin signaling is not limited to growth, but also influences maturation. Within the fat body, EcR activation antagonizes cell-autonomous insulin signaling by interfering with PI3K activity, thereby causing dFOXO to translocate into the nucleus [38,59] (Figure 4). While dFOXO controls the expression of many genes, recent studies have demonstrated that the stage-specific upregulation of two of these target genes helps larvae prepare for metamorphosis. One of these factors is dDOR, which encodes a transcriptional coactivator that physically interacts with EcR, and which is required for proper 20E signaling [67]. The 20E-induced translocation of dFOXO into the nucleus promotes dDOR expression, further activating EcR signaling and initiating a feed-forward loop in fat body cells (Figure 4). In addition, both EcR and dFOXO transcriptionally upregulate dilp6, an insulin-like peptide produced by the fat body [68,69]. Although this finding may seem paradoxical to the growth-inhibitory effects normally associated with these two transcription factors, successful development requires some growth after the cessation of larval feeding and the onset of metamorphosis. Hormone-induced expression of dilp6 allows animals to complete larval growth in the absence of external nutrients, likely by utilizing substrates that are released from the fat body and other larval tissues. In this manner, 20E and insulin act together to establish a transcriptional program that ensures successful completion of larval growth and development.
Generating an Adult Body
Progenitors for the structures of the adult fly are carried inside the larva in the form of diploid imaginal tissues (ITs), which are specified during embryogenesis and go through several rounds of cell division during larval development. Perhaps not surprisingly, IT growth is influenced by many of the same humoral factors and cell-intrinsic signaling pathways that regulate growth in larval tissues [16,70]. Moreover, communication is maintained between IT growth and maturation to ensure that these tissues are ready for their terminal differentiation during pupal stages. If these tissues are damaged with X-rays or apoptosis, critical weight is increased and maturation is delayed while the tissue regenerates [71,72]. This delay is a direct result of restricting PTTH production and 20E responses, due, at least in part, to a retinoid-dependent signal produced in response to damaged ITs [71]. Animals that are raised on a diet lacking the substrates required for retinoid synthesis or that harbor mutations that disrupt retinoid production are unable to delay larval development in response to IT damage [71]. This finding suggests that a retinoid-dependent mechanism helps synchronize IT growth with the onset of maturation.
The low-titer 20E pulses that correlate with the achievement of critical weight also produce changes in the ITs. Prior to this event, unliganded EcR, in complex with USP, represses gene expression in the ITs, reflecting a well-defined role for unliganded nuclear receptors to function as repressors [73,74]. The low-titer 20E pulses that follow the onset of critical weight relieve this repression. In the wing imaginal discs, for example, this switch is reflected by the expression patterns of the genes cut and senseless, which are repressed during early L3 stages, and are initially expressed in response to the 20E pulses during mid-L3 [75].
Metabolism and Maturation
Many of the genetic pathways that control developmental growth in higher organisms — insulin, TOR signaling, and nuclear receptors — are also essential metabolic regulators. This relationship is conserved in flies, as mutations that disrupt insulin, TOR, or 20E signaling not only affect growth, but also produce metabolic phenotypes [26,67,76,77]. Development and metabolism, therefore, are inseparably linked, and current studies are focused on better defining the mechanisms that underlie this interaction.
Recent studies have shown that larvae adopt a unique metabolic program that efficiently converts nutrients into biomass to support growth. This growth program is regulated by a nuclear receptor, the Drosophila ortholog of mammalian estrogen-related receptor (dERR), which coordinately upregulates the transcription of genes encoding enzymes involved in glycolysis, the pentose phosphate pathway, and lactate production during mid-embryogenesis [78]. The resultant metabolic program, which lasts throughout larval development, is a form of aerobic glycolysis that has been demonstrated in normal proliferating cells and cancer cells to support their remarkable growth [79–81]. Although only previously considered in the context of cell proliferation, it is interesting that this program has been adapted to facilitate the dramatic increase in body mass that occurs during Drosophila larval stages.
Intriguingly, many of the genes that are upregulated by dERR at the onset of larval development are coordinately downregulated as the animal prepares for metamorphosis [82]. Although the mechanism that controls this metabolic transition remains unclear, the downregulation of lactate dehydrogenase (ImpL3) correlates with the transcriptional changes induced by 20E during mid-L3 [39]. A similar phenomenon occurs in the silkworm Bombyx mori, where 20E signaling downregulates expression of many of the genes that encode glycolytic enzymes [83]. Thus, in addition to defining the end of larval growth, 20E arrests the metabolic program that supports this process.
A few studies also suggest that 20E initiates distinct metabolic programs at the onset of metamorphosis that are directed toward utilizing stored forms of energy to allow proper growth and development during the non-feeding pupal stages. A central aspect of this response is the discovery that 20E signals the onset of fat body autophagy during mid-L3 [37,38]. This is in parallel with the ability of 20E signaling to arrest growth through the fat body, mediated by insulin signaling [59], as well as an arrest of cell division in the ITs [84]. The developmentally programmed increase in fat body autophagy suggests that larvae utilize the nutrients that were stored earlier in development, which may explain how post-critical weight animals can develop independent of nutrient availability. This model is supported by observations in Manduca sexta, where the concentration of trehalose (the primary circulating sugar in insects) is depleted by starvation in pre-critical weight animals, but remains unaffected when animals are starved post-critical weight [85]. The metabolic transition that occurs at critical weight therefore allows the animal to develop independent of external nutrients and is due, in part, to the interplay between 20E and insulin.
Perspectives and Future Directions
The discovery that evolutionarily conserved signaling systems regulate larval growth and development establish Drosophila as an ideal platform for exploring the basic principles of animal maturation. These findings, combined with the observation that the fat body plays a central role in sensing nutrients and coordinating organismal growth, have emerged at a time when childhood obesity is increasing at an alarming rate. A number of studies have described correlations between obesity and insulin resistance in children and the premature onset of female puberty, linking nutritional status to sexual maturation [86,87]. The onset of puberty is influenced by leptin, a hormone produced by the adipose tissue in response to fat synthesis [88]. The importance of fat storage in regulating maturation makes the identity of fat-body-derived growth regulators of special interest. In addition, identifying the factor(s) that regulate insulin secretion would represent a significant advance. This signal is dependent on TOR and therefore reflects a point at which nutrient availability can be integrated into the systemic growth program.
The role of nutrient sensing in controlling growth and maturation is not limited to the fat body, however, because TOR functions within the PG to regulate nutrient-dependent ecdysone release, although its interactions with PTTH, TGFβ, and insulin remain poorly understood. When TOR activity is inhibited in the PG, decreased 20E signaling results in delayed pupariation [89]. This effect appears to be the result of TOR-dependent ecdysone production, as reduced TOR signaling causes delayed induction of key ecdysone biosynthetic genes. These results imply a role for TOR in contributing to the genetic hierarchy that regulates the onset of maturation. This hypothesis is supported in C. elegans, where loss-of-function mutations in let-363 and daf-15, the C. elegans homologs of TOR and the TOR complex component Raptor, respectively, lead to inappropriate dauer formation [90]. The phenotype of these C. elegans mutants, however, is morphologically distinct when compared with dauers produced by mutations in the insulin, TGFβ, or DA signaling pathways [44,90], suggesting that TOR regulation of dauer development is unique. Similarly, pharmacological manipulation of mTOR in the mouse brain suggests that TOR activity normally promotes maturation, although the mechanisms by which TOR regulates puberty remain largely unknown [88].
Finally, environmental factors other than nutrient availability also influence growth and maturation through poorly understood mechanisms. In particular, day length is likely to play a critical role in this process, as PTTH expression is influenced by circadian rhythms [55], and activation of insulin signaling in the PG renders the onset of metamorphosis sensitive to changes in light signals [10]. Furthermore, the neurons that comprise the central circadian clock in the brain directly innervate the PG and are located within close proximity to the PTTH-producing neurons, providing a way to achieve this control [55,91]. Intriguingly, secretion of mammalian gonadotropin-releasing hormone, a neuropeptide that is synthesized in the hypothalamus and is critical during puberty, appears to be regulated by circadian rhythms [92]. A similar relationship was also recently described in C. elegans, where LIN-42, the worm homolog of the circadian rhythm protein Period, genetically interacts with DAF-12 and DA signaling to regulate dauer formation [93]. The intersection between growth factor signaling, circadian rhythms, and maturation therefore appears to be conserved throughout animals. Further studies in simple systems such as Drosophila and C. elegans provide an opportunity to better define this poorly understood regulatory pathway.
Conclusion
Even though Beadle and his colleagues [9] described Drosophila critical weight nearly 75 years ago, only now are we beginning to understand the genetic pathways that coordinate larval growth and maturation. The discovery of these developmental mechanisms appears to define a conserved genetic architecture that regulates juvenile growth and maturation throughout the animal kingdom and provides a new direction for understanding how human puberty and maturation are controlled.
Acknowledgements
We apologize for our inability to cite other papers that made important contributions to this area of research due to length restrictions.
We thank M.B. O’Connor at the University of Minnesota for sharing unpublished results. Research in the Thummel lab is supported by NIH 1RC1DK086426 and 5R01DK075607. J.M.T. is supported by NIH National Research Service Award F32DK083864 from the NIDDK.
References
- 1.Robertson CW. The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. J. Morphol. 1936;59:351–399. [Google Scholar]
- 2.Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- 3.Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam. Horm. 2001;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert LI, Warren JT. A molecular genetic approach to the biosynthesis of the insect steroid molting hormone. Vitam. Horm. 2005;73:31–57. doi: 10.1016/S0083-6729(05)73002-8. [DOI] [PubMed] [Google Scholar]
- 5.Huang X, Warren JT, Gilbert LI. New players in the regulation of ecdysone biosynthesis. J. Genet. Genomics. 2008;35:1–10. doi: 10.1016/S1673-8527(08)60001-6. [DOI] [PubMed] [Google Scholar]
- 6.Church RB, Robertson FW. A biochemical study of the growth of Drosophila melanogaster. J. Exp. Zool. 1966;162:337–351. [Google Scholar]
- 7.Warren JT, Yerushalmi Y, Shimell MJ, O’Connor MB, Restifo LL, Gilbert LI. Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Dev. Dyn. 2006;235:315–326. doi: 10.1002/dvdy.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Moed GH, Kruitwagen CLJJ, De Jong G, Scharloo W. Critical weight for the induction of pupariation in Drosophila melanogaster: genetic and environmental variation. J. Evol. Biol. 1999;12:852–858. [Google Scholar]
- 9.Beadle GW, Tatum EL, Clancy CW. Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol. Bull. 1938;75:447–462. [Google Scholar]
- 10.Mirth C, Truman JW, Riddiford LM. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr. Biol. 2005;15:1796–1807. doi: 10.1016/j.cub.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Bakker K. Feeding period, growth, and pupation in larvae of Drosophila melanogaster. Ent. Exp. Appl. 1959;2:171–186. [Google Scholar]
- 12.Nijhout HF. The control of body size in insects. Dev. Biol. 2003;261:1–9. doi: 10.1016/s0012-1606(03)00276-8. [DOI] [PubMed] [Google Scholar]
- 13.Robertson F. The ecological genetics of growth in Drosophila. 6. The genetic correlation between the duration of the larval period and body size in relation to larval diet. Genet. Res. 1963;4:74–92. [Google Scholar]
- 14.Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays. 2007;29:344–355. doi: 10.1002/bies.20552. [DOI] [PubMed] [Google Scholar]
- 15.Stern DL, Emlen DJ. The developmental basis for allometry in insects. Development. 1999;126:1091–1101. doi: 10.1242/dev.126.6.1091. [DOI] [PubMed] [Google Scholar]
- 16.Edgar BA. How flies get their size: genetics meets physiology. Nat. Rev. Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- 17.Davis KT, Shearn A. In vitro growth of imaginal disks from Drosophila melanogaster. Science. 1977;196:438–440. doi: 10.1126/science.403606. [DOI] [PubMed] [Google Scholar]
- 18.Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- 19.Martin JF, Hersperger E, Simcox A, Shearn A. minidiscs encodes a putative amino acid transporter subunit required non-autonomously for imaginal cell proliferation. Mech. Dev. 2000;92:155–167. doi: 10.1016/s0925-4773(99)00338-x. [DOI] [PubMed] [Google Scholar]
- 20.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 21.Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu. Rev. Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- 24.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 25.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 26.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 27.Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 28.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 29.Kramer JM, Davidge JT, Lockyer JM, Staveley BE. Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev. Biol. 2003;3:5. doi: 10.1186/1471-213X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geminard C, Rulifson EJ, Leopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chell JM, Brand AH. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honegger B, Galic M, Kohler K, Wittwer F, Brogiolo W, Hafen E, Stocker H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 2008;7:10. doi: 10.1186/jbiol72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arquier N, Geminard C, Bourouis M, Jarretou G, Honegger B, Paix A, Leopold P. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 2008;7:333–338. doi: 10.1016/j.cmet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- 40.Hu PJ. Dauer. WormBook. 2007:1–19. doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cassada RC, Russell RL. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- 43.Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 44.Albert PS, Riddle DL. Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 1988;126:270–293. doi: 10.1016/0012-1606(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 45.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 46.Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 47.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 48.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 49.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 50.Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 51.Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- 52.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 53.Gerisch B, Antebi A. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development. 2004;131:1765–1776. doi: 10.1242/dev.01068. [DOI] [PubMed] [Google Scholar]
- 54.Mak HY, Ruvkun G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development. 2004;131:1777–1786. doi: 10.1242/dev.01069. [DOI] [PubMed] [Google Scholar]
- 55.McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O’Connor MB. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell. 2007;13:857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326:1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- 57.Caldwell PE, Walkiewicz M, Stern M. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Curr. Biol. 2005;15:1785–1795. doi: 10.1016/j.cub.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 58.Gibbens YY, Warren JT, Gilbert LI, O’Connor MB. Neuroendocrine regulation of Drosophila metamorphosis requires TGFβ/Activin signaling. Development. 2011;138:2693–2703. doi: 10.1242/dev.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- 60.Kjaer K, Hagen C, Sando SH, Eshoj O. Epidemiology of menarche and menstrual disturbances in an unselected group of women with insulin-dependent diabetes mellitus compared to controls. J. Clin. Endocrinol. Metab. 1992;75:524–529. doi: 10.1210/jcem.75.2.1639955. [DOI] [PubMed] [Google Scholar]
- 61.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 62.Lee B, Godfrey M, Vitale E, Hori H, Mattei MG, Sarfarazi M, Tsipouras P, Ramirez F, Hollister DW. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352:330–334. doi: 10.1038/352330a0. [DOI] [PubMed] [Google Scholar]
- 63.Erkula G, Jones KB, Sponseller PD, Dietz HC, Pyeritz RE. Growth and maturation in Marfan syndrome. Am. J. Med. Genet. 2002;109:100–115. doi: 10.1002/ajmg.10312. [DOI] [PubMed] [Google Scholar]
- 64.Shingleton AW, Das J, Vinicius L, Stern DL. The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. 2005;3:e289. doi: 10.1371/journal.pbio.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delanoue R, Slaidina M, Leopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev. Cell. 2010;18:1012–1021. doi: 10.1016/j.devcel.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 66.Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 67.Francis VA, Zorzano A, Teleman AA. dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr. Biol. 2010;20:1799–1808. doi: 10.1016/j.cub.2010.08.055. [DOI] [PubMed] [Google Scholar]
- 68.Slaidina M, Delanoue R, Gronke S, Partridge L, Leopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev. Cell. 2009;17:874–884. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okamoto N, Yamanaka N, Yagi Y, Nishida Y, Kataoka H, O’Connor MB, Mizoguchi A. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell. 2009;17:885–891. doi: 10.1016/j.devcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu. Rev. Genet. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- 71.Halme A, Cheng M, Hariharan IK. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr. Biol. 2010;20:458–463. doi: 10.1016/j.cub.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stieper BC, Kupershtok M, Driscoll MV, Shingleton AW. Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev. Biol. 2008;321:18–26. doi: 10.1016/j.ydbio.2008.05.556. [DOI] [PubMed] [Google Scholar]
- 73.Schubiger M, Carre C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–5248. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- 74.Schubiger M, Truman JW. The RXR ortholog USP suppresses early metamorphic processes in Drosophila in the absence of ecdysteroids. Development. 2000;127:1151–1159. doi: 10.1242/dev.127.6.1151. [DOI] [PubMed] [Google Scholar]
- 75.Mirth CK, Truman JW, Riddiford LM. The ecdysone receptor controls the post-critical weight switch to nutrition-independent differentiation in Drosophila wing imaginal discs. Development. 2009;136:2345–2353. doi: 10.1242/dev.032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang H, Liu J, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl. Acad. Sci. USA. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, Bodmer R, Oldham SM. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 78.Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 80.Warburg O, Posener K, Negelein E. Ueber den stoffwechsel der tumoren. Biochem. Z. 1928;152:319–344. [Google Scholar]
- 81.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 83.Tian L, Guo E, Wang S, Liu S, Jiang RJ, Cao Y, Ling E, Li S. Developmental regulation of glycolysis by 20-hydroxyecdysone and juvenile hormone in fat body tissues of the silkworm, Bombyx mori. J. Mol. Cell. Biol. 2010;2:255–263. doi: 10.1093/jmcb/mjq020. [DOI] [PubMed] [Google Scholar]
- 84.Graves BJ, Schubiger G. Cell cycle changes during growth and differentiation of imaginal leg discs in Drosophila melanogaster. Dev. Biol. 1982;93:104–110. doi: 10.1016/0012-1606(82)90243-3. [DOI] [PubMed] [Google Scholar]
- 85.Tobler A, Nijhout HF. A switch in the control of growth of the wing imaginal disks of Manduca sexta. PLoS One. 2010;5:e10723. doi: 10.1371/journal.pone.0010723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ahmed ML, Ong KK, Dunger DB. Childhood obesity and the timing of puberty. Trends Endocrinol. Metab. 2009;20:237–242. doi: 10.1016/j.tem.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 87.Burt Solorzano CM, McCartney CR. Obesity and the pubertal transition in girls and boys. Reproduction. 2010;140:399–410. doi: 10.1530/REP-10-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roa J, Tena-Sempere M. Energy balance and puberty onset: emerging role of central mTOR signaling. Trends Endocrinol. Metab. 2010;21:519–528. doi: 10.1016/j.tem.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 89.Layalle S, Arquier N, Leopold P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell. 2008;15:568–577. doi: 10.1016/j.devcel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 90.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 91.Siegmund T, Korge G. Innervation of the ring gland of Drosophila melanogaster. J. Comp. Neurol. 2001;431:481–491. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 92.Chappell PE. Clocks and the black box: circadian influences on gonadotropin-releasing hormone secretion. J. Neuroendocrinol. 2005;17:119–130. doi: 10.1111/j.1365-2826.2005.01270.x. [DOI] [PubMed] [Google Scholar]
- 93.Tennessen JM, Opperman KJ, Rougvie AE. The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development. 2010;137:3501–3511. doi: 10.1242/dev.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]