Abstract
The mechanism of toxicity of the leukemogen benzene is not entirely known. This pilot study used RNA-sequencing (RNA-seq) technology to examine the effect of benzene exposure on gene expression in peripheral blood mononuclear cells obtained from 10 workers occupationally exposed to high levels of benzene (≥5 ppm) in air and 10 matched unexposed control workers, from a large study (n = 125) in which gene expression was previously measured by microarray. RNA-seq is more sensitive and has a wider dynamic range for the quantification of gene expression. Further, it has the ability to detect novel transcripts and alternative splice variants. The main conclusions from our analysis of the 20 workers by RNA-seq are as follows: The Pearson correlation between the two technical replicates for the RNA-seq experiments was 0.98 and the correlation between RNA-seq and microarray signals for the 20 subjects was around 0.6. 60% of the transcripts with detected reads from the RNA-seq experiments did not have corresponding probes on the microarrays. Fifty-three percent of the transcripts detected by RNA-seq and 99% of those with probes on the microarray were protein-coding. There was a significant overlap (P < 0.05) in transcripts declared differentially expressed due to benzene exposure using the two technologies. About 20% of the transcripts declared differentially expressed using the RNA-seq data were non-coding transcripts. Six transcripts were determined (false-discovery rate < 0.05) to be alternatively spliced as a result of benzene exposure. Overall, this pilot study shows that RNA-seq can complement the information obtained by microarray in the analysis of changes in transcript expression from chemical exposures.
Keywords: benzene, RNA sequencing, gene expression, peripheral blood
INTRODUCTION
Benzene, a component of gasoline, is associated with various hematological cancers [Steinmaus et al., 2008; Vlaanderen et al., 2010]. Multiple possible mechanisms of action are thought to be involved in benzene toxicity [Rappaport et al., 2009; Zhang et al., 2010; Smith et al., 2011; McHale et al., 2012]. Previously, using microarrays, we reported dose-dependent changes in gene expression in peripheral blood mononuclear cells (PBMCs) associated with exposures to benzene ranging from less than 1 ppm to greater than 10 ppm in 83 workers in shoe manufacturing plants in China, compared to unexposed controls [McHale et al., 2011]. We observed significant effects on the expression of a relatively large number of genes across the range of exposures. The acute myeloid leukemia pathway and immune response related pathways were among the biochemical pathways that were found to be significantly affected in a dose-dependent manner.
Next generation sequencing [Wang et al., 2009] of expressed transcripts (RNA-seq) is a newer alternate technology to the hybridization and probe-specific-based microarray technology for quantification of transcript expression. This technology has been shown to be more sensitive than microarrays in detecting transcripts expressed at low levels [Marioni et al., 2008; Su et al., 2011] and differentially expressed transcripts [Liu et al., 2011] and also to have a greater dynamic range than microarrays [Raghavachari et al., 2012]. More importantly, RNA-seq is not restricted to the measurement of expression for a prespecified set of transcripts but is able to quantify all expressed transcripts including non-coding ones, for example, see [Beane et al., 2011] where the authors found pseudogenes, processed transcripts, long intergenic non-coding RNAs (lincRNAs) among those transcripts that were differentially expressed between smokers and non-smokers in the epithelial cells of their bronchial airways. Further, there is potential for discovery of novel transcripts and also for the detection of alternative splicing events resulting from a perturbation to control conditions (e.g., alternative splicing was detected in transcripts obtained from kidney and liver tissue samples [Marioni et al., 2008] and in the peripheral blood of patients with sickle cell disease [Raghavachari et al., 2012]). The comparison of RNA-seq technology with microarrays is described in greater detail in an accompanying paper in this issue [McHale et al., submitted].
This manuscript describes a pilot study designed to test the application of RNA-seq to estimate changes in transcript expression of PBMCs in workers exposed to benzene. The samples were chosen from a subset of 10 highly exposed workers and a corresponding set of 10 matched controls in the study described in our previous study [McHale et al., 2011]. The objectives of our current study were as follows: (1) Estimate the technical replicability of the RNA-seq approach. (2) Estimate the correlation between transcript expression levels as quantified by RNA-seq and the corresponding levels as determined by microarray. (3) Estimate the overlap of transcripts declared differentially expressed as a result of high exposure to benzene by the two technologies. (4) Determine if there are transcripts that are alternatively spliced as a result of benzene exposure.
MATERIAL AND METHODS
Study Subjects and Exposure Assessment
All subjects were from a molecular epidemiology study of occupational exposure to benzene that comprised 250 benzene-exposed shoe manufacturing workers and 140 unexposed age- and sex-matched controls who worked in three clothes-manufacturing factories in the same region near Tianjin, China [Vermeulen et al., 2004; Lan et al., 2006]. This study complied with all applicable requirements of U.S. and Chinese regulations, including institutional review board approval. Participation was voluntary, and written informed consent was obtained.
Exposure assessment to benzene was performed as described previously [Vermeulen et al., 2004]. For this study, we categorized exposure groups using mean individual air benzene measurements obtained during the 3 months preceding phlebotomy. McHale et al. analyzed global gene expression in 125 subjects in five exposure categories—11 workers with very high exposure (>10 ppm), 13 with high exposure (5–10 ppm), 30 with low exposure (<1 ppm), 29 with very low exposure (<<1 ppm), and a set of 42 matched controls, using Illumina HumanRef-8 V2 BeadChips [McHale et al., 2011]. In this pilot study, we analyzed global transcript expression by RNA-seq in subsets of five subjects from each of the two highest exposure categories (5–10 ppm and >10 ppm) and in 10 unexposed controls, chosen and matched by age, sex, and smoking status. Mean age (± SD) for the exposed workers were 29.9 ± 11.1 and 29.2 ± 9 for the control workers. There were eight women and eight non-smokers in each group. In this article, we have compared differential expression by microarray and RNA-seq data in the 10 control versus 10 highly exposed subjects, analyzed by both methods. In addition, we have compared the RNA-seq data with the microarray data from all the 42 controls and 24 highly exposed subjects.
Biological sample collection was described in McHale et al. [2011]. RNA-Seq libraries were prepared from the PBMC RNA (1 µg) using Illumina’s mRNA TruSeq protocol, and sequenced on an Illumina HiSeq 2000. The first step in the TruSeq protocol workflow involves purifying the poly-A containing mRNA molecules. In the same experiment, using the same protocol, we also sequenced two replicates of Stratagene Universal Human Reference RNA (Stratagene), which is composed of 10 different human cell lines (Agilent Technologies, Santa Clara), to assess the technical replicability of the RNA-seq experiments.
Data Preprocessing
Paired-end Illumina reads (in .fastq files) were mapped to hg19 (Genome Reference Consortium GRCh37) using Tophat [Trapnell et al., 2009] followed by Cufflinks [Trapnell et al., 2010] for transcript assembly and estimation of transcript and transcript isoform expression levels. The fragment bias correction option [Roberts et al., 2011] was used in the Cufflinks program. The expression levels are provided in terms of FPKM (fragments per kilobase of exon model per million fragments mapped) reads. The resulting reads were quantile normalized across the 20 study subjects.
Identification of Coding and Non-Coding Transcripts
The Ensembl IDs of the transcripts identified using the RNA-seq data were used to query the BioMart database [Haider et al., 2009; Smedley et al., 2009; Guberman et al., 2011] for coding/non-coding information. The queries were made using the getBM function in the biomaRt package [Durinck et al., 2005] in R[Ihaka and Gentleman, 1996]. The relevant attribute in these queries was “gene_biotype.” Non-coding transcripts on the Illumina HumanRef-8 V2 BeadChips platform were identified as probes in the associated annotation file with RefSeq IDs [Pruitt et al., 2007] starting with “NR_” or “XR_.”
Differential Expression Analyses
To identify transcripts differentially expressed between the controls and the exposed subjects, we fit negative binomial generalized linear models given in Eq. (1) using the glm.nb function in R[Ihaka and Gentleman, 1996].
| (1) |
Where λij is the FPKM reads of the jth transcript on the ith subject and Xij = 0 or 1 according to whether the ith subject is a control or exposed subject. These models are used to estimate exp(b), the change in mean FPKM reads of the jth transcript from the control to the exposed workers. The resulting fits of the two models are subject to goodness of fit tests [McCullagh and Nelder, 1989]. The first one is a chi-squared test using the estimated residual degrees of freedom on the deviance of the model fit. The second goodness of fit tests the normality of the standardized deviance residuals of the model fit using the Anderson-Darling test. A model fit is deemed to pass the goodness of fit tests if the P-values from the two tests are each greater than 0.1. In addition to the above parametric model, we also use the non-parametric Wilcoxon rank sum test [Wilcoxon et al., 1970] to estimate the significance of the difference in median FPKM reads of the jth transcript in the control and exposed works. From the above analyzes, differentially expressed transcripts are identified as those with a false-discovery rate (FDR) [Benjamini and Hochberg, 1995] less than 0.05.
Alternative Splicing Analyses
To identify transcripts that are differentially spliced with benzene exposure, we fit two negative binomial generalized linear models given in Eqs. (2) and (3) using the glm.nb function in R.
| (2) |
| (3) |
where λij is the FPKM reads of the jth transcript isoform on the ith subject and Xij = 0 or 1 according to whether the ith subject is one of the controls or one of the exposed. Zij gives the index of the particular transcript isoform being considered for the jth transcript and lies in ․ {1, 2, ⋯, nj} I[k = = z] is equal to 1 if the indices k and z are equal else it is equal to 0. The model implied by Eq. (3) is nested inside the one implied by Eq. (2). The additional 3 nj terms in Eq. (2) attempts to capture the effect on the expression level reads due to the interaction between exposure to benzene and a particular isoform of transcript j. A likelihood ratio test of the model fits using Eqs. (2) and (3) is used to assess the significance that a transcript is not differentially spliced as a result of benzene exposure. In other words, the test is used to assess the significance that the changes in mean expression levels due to benzene exposure of a given transcript are the same across all the identified isoforms. From the above analyses, transcripts that are alternatively spliced as a result of benzene exposure are identified as those with a FDR [Benjamini and Hochberg, 1995] less than 0.05.
Pathway Analyses
SEPEA, a network-based pathway enrichment method described in detail in Thomas et al. [2009], was used to evaluate the linkage between the benzene exposure and the KEGG human pathways [Kanehisa and Goto, 2000; Kanehisa et al., 2006, 2008] using the estimated expression levels for the control and exposed workers. Three analytic methods were described in Thomas et al. [2009]; we used SEPEA_NT3 in this study.
RESULTS AND DISCUSSION
Alignment to the Genome
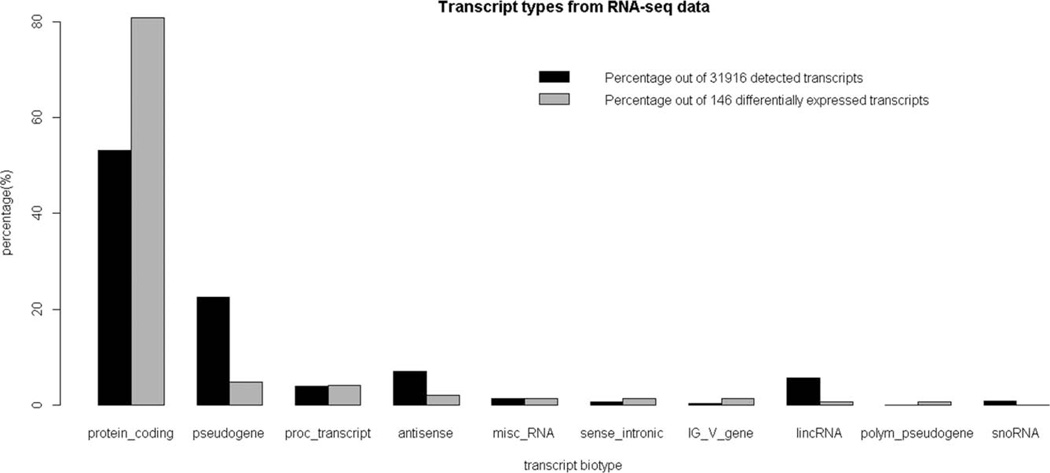
111 ± 20 million nt (100 bp paired-end reads) were generated per sample. 79 ± 6% of the reads was mapped to the human genome (hg19). This corresponds well with the experiments performed by Toung et al. who in part estimated the percent reads mapped using data derived at varying sequencing depths [Toung et al., 2011]. At about 100 million reads they estimated a 79% total alignment to the genome. Reads were detected in at least one of the 20 worker samples for 31,916 transcripts, in this study. This represents an overlap of 12,957 transcripts with the 18,190 transcripts that had probes on the microarray platform used in McHale et al. [2011]. Thus the RNA-seq experiments detected reads for 18,959 transcripts that did not have probes on the microarrays. Fifty-three percent of 31,916 transcripts were known to be protein coding while the remaining 47% were non-coding transcripts (Fig. 1). This contrasts with only 0.85% (155 out of 18,190) of the probes on the microarray being targeted to non-coding transcripts. It should be noted that the RNA-seq libraries were generated from poly-A enriched RNA samples. It is quite plausible that different non-coding transcripts may be identified using alternate library preparation protocols [Beane et al., 2011]. Genes have been typically associated with transcripts that have protein-coding capacity [Gerstein et al., 2007]. With the discovery that a significant part of transcription leads to non-coding transcripts, the definition of genes needs refinement [Gingeras, 2007]. Hence, we use the term “transcript” instead of “gene” (as suggested by Gingeras [2007]) in this article to refer to mapped reads from both the RNA-seq and microarray data.
Fig. 1.
Transcript types detected in the RNA-seq data. The results are given in terms of percentages of transcripts with detectable reads and also in terms of transcripts declared differentially expressed due to benzene exposure.
Replication Within and Across Platforms
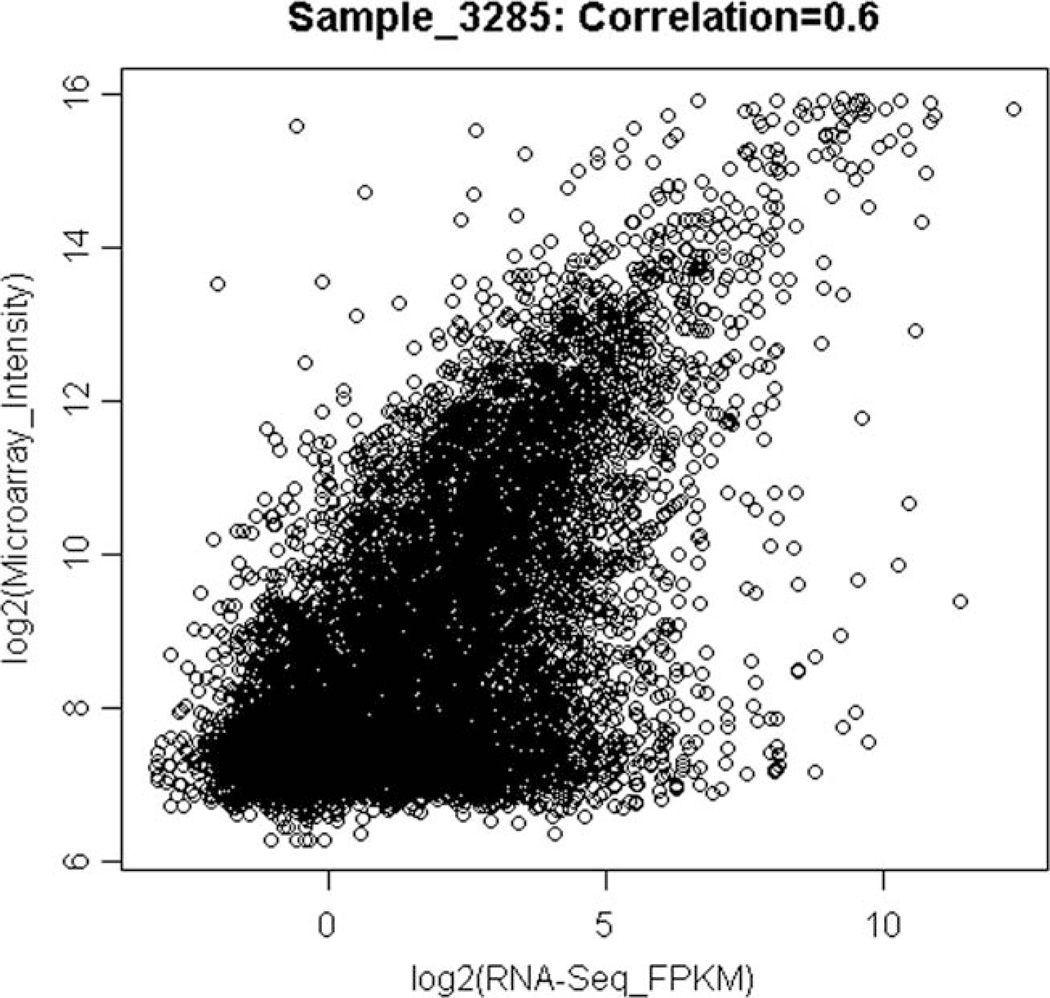
The correlation of the FPKM reads between the two technical replicates used was 0.98, suggesting a good technical replicability of the expression reads using RNA-seq. [Marioni et al., 2008] also showed high technical replicability of RNA-seq data. The correlation between the transcript expression signals as measured by Illumina microarrays and Illumina HiSeq 2000 was 0.596 on average (interquartile range: 0.592–0.601) across all the 20 samples. This value is within the range of correlation values between microarrays and RNA-seq data in the published literature [Marioni et al., 2008; Bradford et al., 2010]. Figure 2 displays the relationship between microarray and the RNA-seq expression values on the log scale for a given sample in our study. The flat cluster of points at the low intensity values for the microarray suggests that the RNA-seq-based approach is better able to detect transcripts expressed at lower levels than microarrays and/or that the RNA-seq approach could detect particular isoforms of transcripts that did not have matching probes on the microarray.
Fig. 2.
Comparison of log-2 transformed transcript expression signals in a sample using microarrays and RNA-seq. The signals from microarrays are in intensity units while those from RNA-seq are in FPKM units. The signals are from one of the exposed samples in the study. The correlation between the signals is 0.588.
Differentially Expressed Transcripts and Altered Biochemical Pathways
A total of 184 transcripts were identified as being differentially expressed between the control and exposed workers using the negative binomial generalized linear models. This number was reduced to 146 when only the transcripts that passed the goodness of fit tests were included. The top 10 differentially expressed transcripts and KEGG pathways identified by RNA-seq are given in Tables 1 and 2, respectively. The complete list of transcript expression changes and KEGG pathway changes are given in Supporting Information Tables S1 and S2. The non-parametric test did not identify any differentially expressed transcripts. Therefore, as we attempt to be more rigorous and make fewer assumptions about the distribution of reads from the RNA-seq experiments we identify fewer differentially expressed transcripts. The negative binomial probability distribution is commonly used to model reads from RNA-seq experiments [Anders and Huber, 2010; Robinson and Oshlack, 2010]. The results here suggest that one must be cautious in interpreting RNA-seq results that utilize either poisson or negative binomial distribution assumptions.
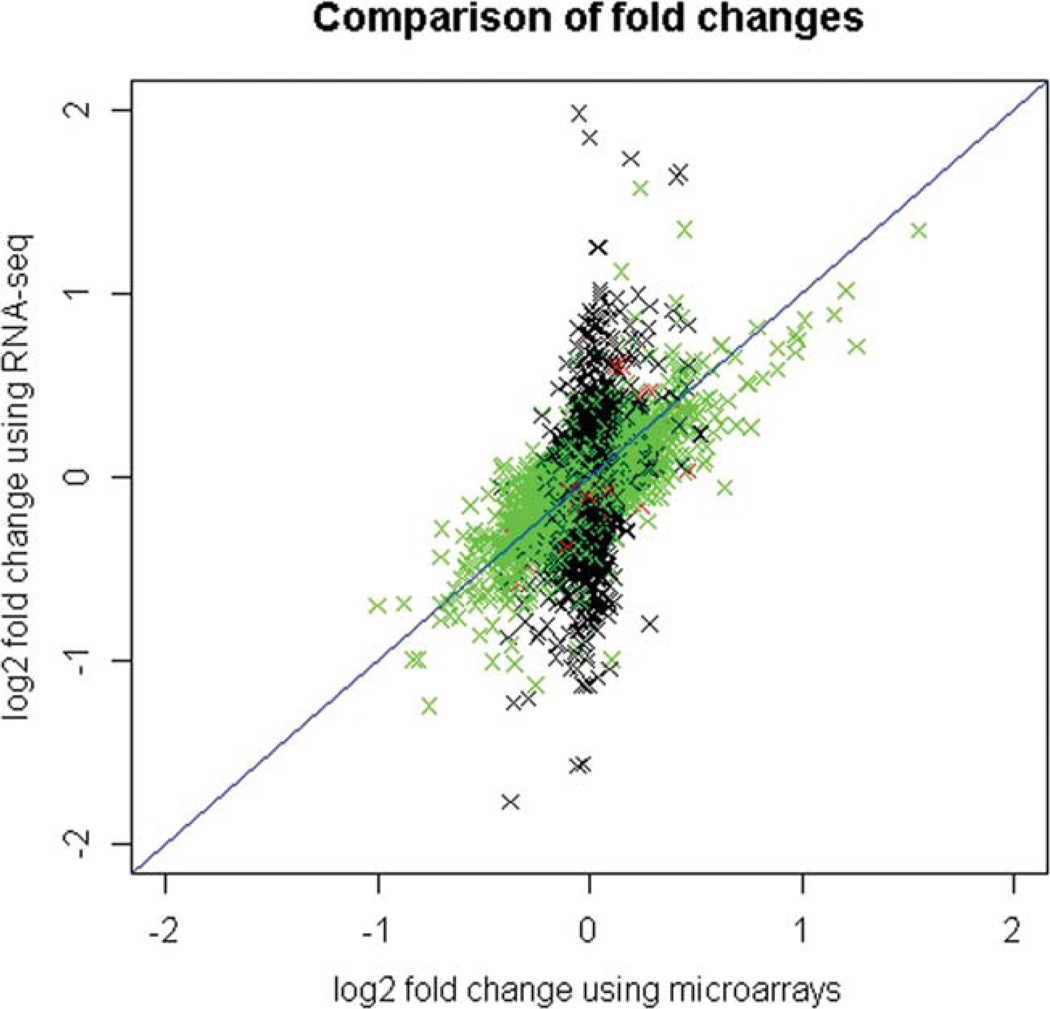
A comparison of mean log2 fold changes of expression as determined using data from corresponding transcripts on the microarray and RNA-seq technology is shown in Figure 3. There is a significant correlation (P < 2.2 × 10−16) of 0.53 between the corresponding fold changes. This correlation is of the same order as those detected by other authors in literature. For example, Marioni et al. estimated a correlation of 0.73 between fold changes by the two technologies of transcript expression in the kidney versus the liver of a single human male [Marioni et al., 2008], Liu et al. estimated a correlation of 0.61 of fold changes in transcript expression in the cerebellum of human, chimpanzee, and rhesus samples [Liu et al., 2011], Su et al. estimated a correlation of 0.49 of fold changes in transcript expression in the kidneys of rats treated with aristolochic acid [Su et al., 2011] and Beane et al. estimated a correlation of 0.36 of fold changes in transcript expression in the bronchial airway of smokers versus non-smokers [Beane et al., 2011]. The majority of changes in expression associated with benzene exposure determined by RNA-seq were subtle, with fold-changes of ≤2, in agreement with data from previous microarray analyses in McHale et al. [2011] The estimated fold-changes using the RNA-seq data was quite variable for transcripts with relatively low expression and also low fold-changes in the microarray data (see the black group of crosses in the center of Fig. 3). This may suggest that the RNA-seq data was more sensitive at detecting changes in low expressed transcripts.
Fig. 3.
Comparison of log-2 transformed estimated fold changes using microarrays and RNA-seq. The points are colored according to the relative mean expression level of the transcript on the microarray platform. The mean log2 expression levels on the microarray ranged from 6.5 to 16. The transcripts with log2 expression levels less than 8 are colored black (low expressed), with expression levels between 8 and 14 are colored green and greater than 14 are colored red (high expressed).
We compared the differentially expressed transcripts identified by RNA-seq and microarray. Differential expression analyses using the microarray data from the 10 cases and 10 control subjects revealed only one differentially expressed transcript (LOC161931) at a FDR < 0.05, compared with 146 identified by RNA-seq. There were no reads for this transcript in the RNA-seq data. It has previously been reported that RNA-seq can detect a larger number of differentially expressed transcripts than microarrays can [Marioni et al., 2008; Beane et al., 2011; Liu et al., 2011; Su et al., 2011; Raghavachari et al., 2012].
Analysis of the microarray data from all of the controls (n = 42) and highly exposed subjects (n = 24) revealed 2,553 differentially expressed transcripts. Among the 146 transcripts declared differentially expressed by RNA-Seq, 89 had corresponding probes on the Illumina microarray platform [McHale et al., 2011]. Of these 89 probes, 32 were identified as commonly differentially expressed using data from the 10 controls and 10 exposed subjects analyzed by RNA-seq and from the 42 controls and 24 subjects analyzed by microarray. This overlap among the 12,957 transcripts that had corresponding probes on the microarrays and are detected in the RNA-seq experiments is statistically significant (P = 1.5 × 10−6 from Fisher’s exact test). The 57 transcripts not detected as differentially expressed by the microarrays represent potential novel findings of the newer RNA-seq technology. A large number of transcripts (n = 1,937) identified as differentially expressed by the microarrays were not detected by the RNA-seq. This could be a consequence of lower statistical power due to smaller sample size in the RNA-seq experiments. There are of course caveats to the comparison of the smaller RNA-seq dataset to the larger microarray dataset as opposed to comparing data only from the same subjects. First, there is the obvious sample size difference (42 vs. 24 subjects as compared with 10 vs. 10 subjects). Second, the subjects conditions are not matched as well on the potential confounders to changes in transcript expression (gender, smoking status).
Of the transcripts differentially expressed using the RNA-seq data, 80% (118 out of 146) were protein-coding while the rest were non-coding (Fig. 1). In contrast, 99.8% (2,548 out of 2,553) of transcripts identified as differentially expressed by microarray were protein coding transcripts. This reflects this microarray platform’s bias by design toward protein coding transcripts.
Transcripts Alternatively Spliced With Benzene Exposure
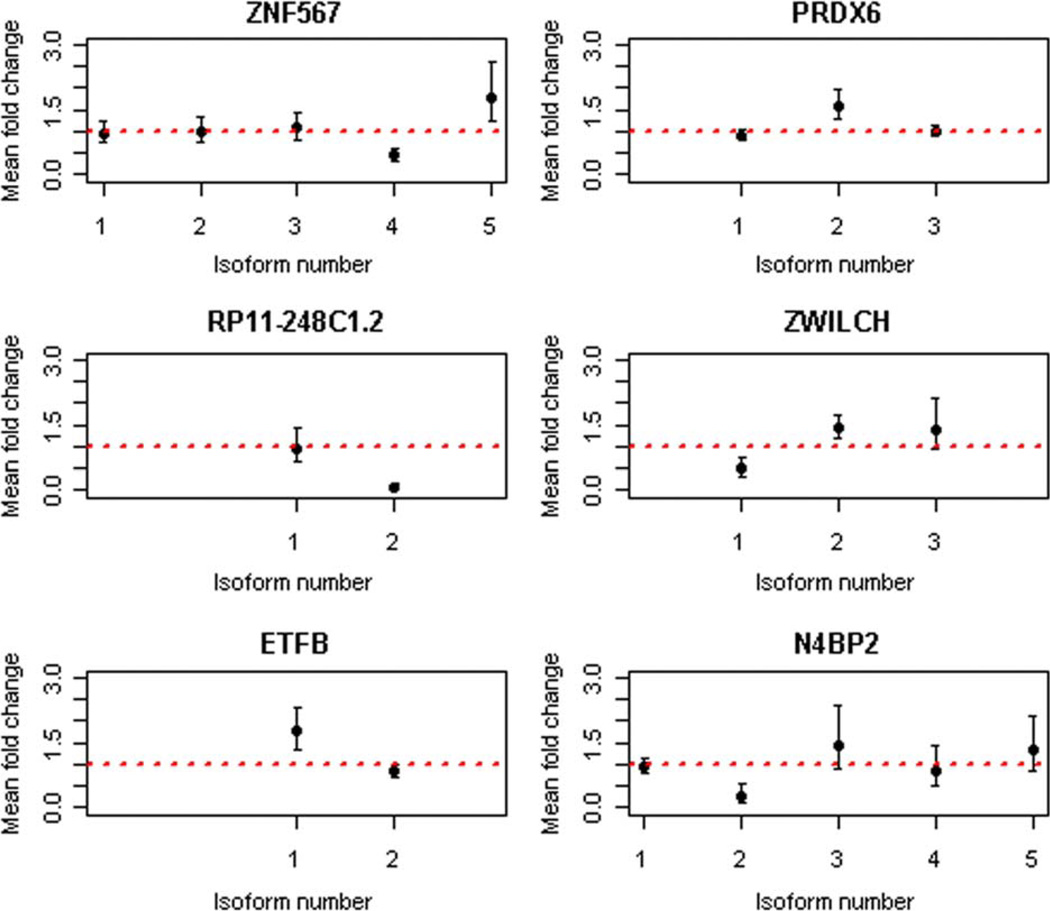
Six transcripts [ZNF567 zinc finger protein 567, PRDX6 peroxiredoxin 6, RP11-248C1.2, ZWILCH Zwilch kinetochore associated homolog (drosophila), ETFB electron transfer flavoprotein beta polypeptide, and N4BP2 NEDD4 binding protein 2] were identified as differentially spliced (FDR < 0.05) with exposure to benzene (Fig. 4). This provides evidence that alternative splicing of transcripts in PBMCs could be a response to benzene exposure.
Fig. 4.
Alternatively spliced transcripts due to benzene exposure. The x-axes of the subplots for the six transcripts contain the indices of the all the isoforms of the corresponding transcripts with detectable reads from the RNA-seq experiment. The scale on the y-axes corresponds to the mean fold change. The error bars correspond to the 95% confidence intervals of these mean fold changes. The dotted line corresponds to a fold change of 1.
CONCLUSIONS
Data from RNA-seq and microarray analyses of benzene-exposed subjects and controls were well correlated at the transcript and fold-change levels though RNA-seq was more sensitive in detecting transcripts with low levels of expression. RNA-seq identified more transcripts as differentially expressed than microarrays and detected differential expression of non-coding transcripts. RNA-seq also identified alternative splicing as a potential mechanism of benzene toxicity. Thus, RNA-seq data can complement microarray data in toxicogenomic studies and provide novel information. We are currently expanding our benzene RNA-Seq study to analyze more samples and additional alterations in the transcriptome.
TABLE1.
Top 10 Differentially Expressed Transcripts Identified by the Negative Binomial Models That Passed the Goodness of Fit Tests
| Gene symbol | Gene name | Gene type | Fold change | FDR |
|---|---|---|---|---|
| CMYA5a | Cardiomyopathy associated 5 | Protein coding | 2.00 | 0.000 |
| ZNF703a | Zinc finger protein 703 | Protein coding | 0.51 | 0.000 |
| TTC9Ba | Tetratricopeptide repeat domain 9B | Protein coding | 2.39 | 0.001 |
| COG7a | Component of oligomeric golgi complex 7 | Protein coding | 1.18 | 0.001 |
| PLCL1a | Phospholipase C-like 1 | Protein coding | 17.27 | 0.002 |
| SIK2 | Salt-inducible kinase 2 | Protein coding | 1.22 | 0.002 |
| RBM23a | RNA binding motif protein 23 | Protein coding | 1.25 | 0.002 |
| RP5-837I24.1 | NA | Pseudogene | 0.87 | 0.003 |
| AC092135.1 | NA | Protein coding | 1.32 | 0.003 |
| GOT1a | Glutamic-Oxaloacetic Transaminase 1 | Protein coding | 1.26 | 0.003 |
This gene has a probe on Illumina HumanRef-8 V2 BeadChip.
TABLE 2.
Top 10 Pathways Altered by Benzene Exposure
| KEGG pathway name | P |
|---|---|
| Fc epsilon RI signaling pathwaya | 0.000 |
| MAPK signaling pathwaya | 0.000 |
| Adipocytokine signaling pathwaya | 0.001 |
| Leishmaniasis | 0.001 |
| Cysteine and methionine metabolism | 0.002 |
| Type II diabetes mellitus | 0.002 |
| Graft-versus-host disease | 0.003 |
| Pancreatic cancera | 0.003 |
| Insulin signaling pathwaya | 0.004 |
| Proteasome | 0.005 |
This pathway was detected as significant in either one of the two highest dose ranges of benzene exposure in the earlier microarray analyses [McHale et al., 2011].
Acknowledgments
Grant sponsor: National Institutes of Environmental Health Sciences; Grant number: P42ES004705 (to M.T.S); Grant sponsor: National Cancer Institute (intramural funds).
Footnotes
Additional Supporting Information may be found in the online version of this article.
AUTHOR CONTRIBUTIONS
Drs. Smith, Rothman, Zhang, and Lan designed the study. Dr. McHale performed the RNA-Seq sample preparation. Dr. Thomas analyzed the data and prepared the manuscript draft with important intellectual and editorial input from Drs. McHale, Zhang, Smith, Hubbard, Rothman, Lan, Vermeulen, Li, Yin, and Rappaport. Drs. Vermeulen and Rappaport performed the exposure assessment. All authors approved the final manuscript.
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res. 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995:289–300. [Google Scholar]
- Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, Miller CJ. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics. 2010;11:282. doi: 10.1186/1471-2164-11-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- Gingeras TR. Origin of phenotypes: Genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, Chelala C, Croft D, Cros A, Cutts RJ. BioMart Central Portal: An open database network for the biological community. Database: J Biol Databases Curation. 2011 doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal—Unified access to biological data. Nucleic Acids Res. 2009;37(suppl 2):W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graphical Stat. 1996;5:299–314. [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita K, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354. doi: 10.1093/nar/gkj102. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(suppl 1):D480. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Vermeulen R, Zhang L, Li G, Rosenberg PS, Alter BP, Shen M, Rappaport SM, Weinberg RS, Chanock S, Waidyanatha S, Rabkin C, Hayes RB, Linet M, Kim S, Yin S, Rothman N, Smith MT. Benzene exposure and hematotoxicity: Response. Science. 2006;312:998–998. [Google Scholar]
- Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. Boca Raton, Florida: Chapman & Hall/CRC; 1989. [Google Scholar]
- McHale CM, Zhang L, Lan Q, Vermeulen R, Li G, Hubbard AE, Porter KE, Thomas R, Portier CJ, Shen M, Rappaport SM, Yin S, Smith MT, Rothman N. Global gene expression profiling of a population exposed to a range of benzene levels. Environ Health Perspect. 2011;119:628–640. doi: 10.1289/ehp.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Smith MT. Current understanding of the mechanism of benzene-induced leukemia in humans: Implications for risk assessment. Carcinogenesis. 2012;33:240–252. doi: 10.1093/carcin/bgr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(suppl 1):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari N, Barb J, Yang Y, Liu P, Woodhouse K, Levy D, Christopher JD, Munson PJ, Kato GJ. A systematic comparison and evaluation of high density exon arrays and RNA-seq technology used to unravel the peripheral blood transcriptome of sickle cell disease. BMC Med Genomics. 2012;5:28. doi: 10.1186/1755-8794-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Kim S, Lan Q, Vermeulen R, Waidyanatha S, Zhang L, Li G, Yin S, Hayes RB, Rothman N. Evidence that humans metabolize benzene via two pathways. Environ Health Perspect. 2009;117:946. doi: 10.1289/ehp.0800510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart—Biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Zhang L, McHale CM, Skibola CF, Rappaport SM. Benzene, the exposome and future investigations of leukemia etiology. Chem Biol Interact. 2011;192:155–159. doi: 10.1016/j.cbi.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C, Smith AH, Jones RM, Smith MT. Meta-analysis of benzene exposure and non-Hodgkin lymphoma: Biases could mask an important association. Occup Environ Med. 2008;65:371. doi: 10.1136/oem.2007.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Li Z, Chen T, Li Q-Z, Fang H, Ding D, Ge W, Ning B, Hong H, Perkins RG. Comparing next-generation sequencing and microarray technologies in a toxicological study of the effects of aristolochic acid on rat kidneys. Chem Res Toxicol. 2011;24:1486–1493. doi: 10.1021/tx200103b. [DOI] [PubMed] [Google Scholar]
- Thomas R, Gohlke JM, Stopper GF, Parham FM, Portier CJ. Choosing the right path: Enhancement of biologically relevant sets of genes or proteins using pathway structure. Genome Biol. 2009;10:R44. doi: 10.1186/gb-2009-10-4-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toung JM, Morley M, Li M, Cheung VG. RNA-sequence analysis of human B-cells. Genome Res. 2011;21:991–998. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen R, Li G, Lan Q, Dosemeci M, Rappaport SM, Bohong X, Smith MT, Zhang L, Hayes RB, Linet M. Detailed exposure assessment for a molecular epidemiology study of benzene in two shoe factories in China. Ann Occup Hyg. 2004;48:105. doi: 10.1093/annhyg/meh005. [DOI] [PubMed] [Google Scholar]
- Vlaanderen J, Portengen L, Rothman N, Lan Q, Kromhout H, Vermeulen R. Flexible meta-regression to assess the shape of the benzene-leukemia exposure-response curve. Environ Health Perspect. 2010;118:526–532. doi: 10.1289/ehp.0901127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon F, Katti S, Wilcox RA. Critical values and probability levels for the Wilcoxon rank sum test and the Wilcoxon signed rank test. Selected Tables Math Stat. 1970;1:171–259. [Google Scholar]
- Zhang L, McHale CM, Rothman N, Li G, Ji Z, Vermeulen R, Hubbard AE, Ren X, Shen M, Rappaport SM. Systems biology of human benzene exposure. Chem Biol Interact. 2010;184:86–93. doi: 10.1016/j.cbi.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]