Abstract
Endoglin (ENG) is a TGF-β coreceptor and essential for vascular development and angiogenesis. A chimeric antihuman ENG (hENG) monoclonal antibody (mAb) c-SN6j (also known as TRC105) shows promising safety and clinical efficacy features in multiple clinical trials of patients with various advanced solid tumors. Here we developed a novel genetically engineered mouse model to optimize the ENG-targeting clinical trials. We designed a new targeting vector that contains exons 4–8 of hENG gene to generate novel genetically engineered mice (GEMs) expressing functional human/mouse chimeric (humanized) ENG with desired epitopes. Genotyping of the generated mice confirmed that we generated the desired GEMs. Immunohistochemical analysis demonstrated that humanized ENG protein of the GEMs expresses epitopes defined by 7 of our 8 anti-hENG mAbs tested. Surprisingly the homozygous GEMs develop normally and are healthy. Established breast and colon tumors as well as metastasis and tumor microvessels in the GEMs were effectively suppressed by systemic administration of anti-hENG mAbs. Additionally, test result indicates that synergistic potentiation of antitumor efficacy can be induced by simultaneous targeting of two distinct epitopes by anti-hENG mAbs. Sorafenib and capecitabine also showed antitumor efficacy in the GEMs. The presented novel GEMs are the first GEMs that express the targetable humanized ENG. Test results indicate utility of the GEMs for the clinically relevant studies. Additionally, we generated GEMs expressing a different humanized ENG containing exons 5–6 of hENG gene, and the homozygous GEMs develop normally and are healthy.
Keywords: genetically engineered mouse model, humanized endoglin, antiendoglin monoclonal antibodies, synergy, vascular targeting cancer therapy
Introduction
Although antiangiogenic agents showed varying degrees of antitumor efficacy to different tumors, clinical efficacy of these agents is less than anticipated.1–3 Many of the currently available antiangiogenic agents target/modulate the VEGF pathway. One approach to improving the antiangiogenic therapy will be to explore new antiangiogenic agents that target/modulate non-VEGF signaling pathways. A relevant approach will be to target the VEGF pathway and a non-VEGF pathway simultaneously or sequentially. We have been studying to develop a new vascular-targeting antiangiogenic agent targeting ENG4–8 that modulates the TGF-β pathway9 and also the BMP9 pathway.10 In the present study, we hypothesized that mice expressing targetable epitopes of hENG in the tumors will be highly valuable for optimizing the hENG-targeted therapy in cancer patients and that we may be able to develop such mice by partial humanization of the appropriate regions of mENG. To test these hypotheses, we developed novel GEMs expressing desired epitopes of hENG and performed therapeutic experiments relevant to the ongoing clinical trials of a chimeric anti-hENG mAb c-SN6j (also known as TRC105).
ENG is a proliferation-associated cell membrane antigen and strongly expressed on the tumor-associated angiogenic vascular endothelium4,5,11–13 and on lymphatic vessels in tumors.14 In addition, ENG is essential for vascular development and angiogenesis.15,16 ENG expression was stimulated in the presence of hypoxia and TGF-β.17 Anti-VEGF therapy may increase ENG expression.18 ENG promotes endothelial proliferation and TGF-β/ALK1 signal transduction.19
We hypothesized that ENG is a useful target for vascular-targeting antiangiogenic therapy based on the in vitro and animal model studies.4–8,13,20 The majority of anti-hENG mAbs do not crossreact with murine endothelial cells while a few anti-hENG mAbs showed weak cross-reactivity with murine endothelial cells.4,5,21 Anti-hENG mAbs suppress angiogenesis and tumor growth by multiple mechanisms that include antibody-dependent cell-mediated cytotoxicity, induction of apoptosis, direct suppression of cell proliferation, T cell-mediated mechanisms,7,13,22 and BMP9 signaling inhibition.10 To facilitate clinical application of anti-hENG mAbs, we generated a human/mouse chimeric anti-hENG mAb c-SN6j (TRC105) from one (i.e., SN6j) of our anti-hENG mAbs13,23 and performed studies of pharmacokinetics, toxicology and immunogenicity of c-SN6j in nonhuman primates.23 Subsequently, clinical trials of TRC105/c-SN6j were initiated in patients with advanced solid tumors. Results of a recently completed phase 1 clinical trial24 and several ongoing clinical trials of TRC105 (e.g., ClinicalTrials.gov Identifier: NCT01090765, NCT01306058, NCT01326481 and NCT01332721) are consistent with our hypothesis. TRC105 is the first and only anti-hENG mAb that was tested for therapy of cancer patients.
To optimize the anti-hENG mAb-based cancer therapy, we developed two types of GEMs; one by replacing a smaller size (exons 5–6) of mENG gene and the other by replacing a larger size (exons 4–8) of mENG gene with the corresponding gene segments of hENG. We found that hemizygous (heterozygous) and homozygous mice of both types of GEMs develop normally and are healthy without signs of telangiectasia; the results indicate that the generated humanized ENGs containing the two different sizes of hENG are functional in mice. In this report, we place our emphasis on the humanized ENG with the larger hENG because it will have wider utility than that with the smaller hENG.
Materials and Methods
Reagents, mAbs, cells and animal studies
Sorafenib that is an orally active multikinase inhibitor25 was kindly provided by Bayer HealthCare Pharmaceuticals (Wayne, NJ). Capecitabine was purchased from LC Laboratories (Woburn, MA). Various restriction enzymes were obtained from New England Biolabs (Beverly, MA). Twelve anti-hENG mAbs were generated in our laboratory. Details of the generation and characterization were previously reported for six of these mAbs, i.e., SN6, SN6a, SN6f, SN6h, SN6j and SN6k.4,5,20,26 Among the remaining six mAbs, three relevant mAbs (SN6c, SN6d and SN6i) for this report are generated as described in the Results section. BALB/c-l ES cells27 were obtained from Dr. Frank Koentgen of Ozgene (Bentley DC, Australia). Rat anti-mENG mAb MJ7/18 was purchased from BD Biosciences (San Diego, CA). 4T1 murine mammary carcinoma cell line and Col 26 murine colon adenocarcinoma cell line were described previously.7,8 All animal studies were performed at the Roswell Park Laboratory Animal Resources Facilities following the IACUC-approved protocols.
Plasmids, primers and Southern blot probes for generation of GEMs
The 2.8 kbp gene fragment containing hENG exons 4–8 was subcloned from a human male BAC clone, RPCI11-228b15,28 into plasmid pUC19 using a recombineering system (gap repair; 29). The 2.0 kbp 5′- and 8.0 kbp 3′-homologous arms were also subcloned into pUC19 from a mouse BALB/cByJ BAC clone, CHORI28-21a20 (BACPAC Resources Center at Children's Hospital Oakland Research Institute, Oakland, CA) using a recombineering system. Fragments for gap repair were prepared by PCR with primers containing 5′ regions homologous to the target sequence and 3′ regions homologous to pUC19. In these primers, restriction enzyme sites, required for targeting vector construction and Southern blot analysis, were inserted between the target sequence and pUC19 sequence. PCR products were digested with DpnI (New England Biolabs, Ipswich, MA) to remove template (pUC19) and purified using QIAEX II gel extraction kit (QIAGEN, Valencia, CA). Electroporation of E. coli was performed as described by others.29 To obtain the targeting vector, the fragment containing human exons 4–8 and both homologous arms were sequentially assembled into pTKneoF vector (generous gift from Dr. Peter Aplan, NCI, Bethesda, MD), which contains loxP flanked neomycin resistant cassette for positive selection and a thymidine kinase gene for negative selection (Supporting Information Fig. 1).
Probes for Southern blot were amplified by PCR using a mouse C57BL/6J BAC clone, RPCI23-17p1230 as a template and cloned into EcoRI site of pUC19. Approximate sizes of 5′ and 3′ probes are 400 and 700 bp, respectively. All primers used in this study are listed in Supporting Information Table 1.
Generation of GEMs
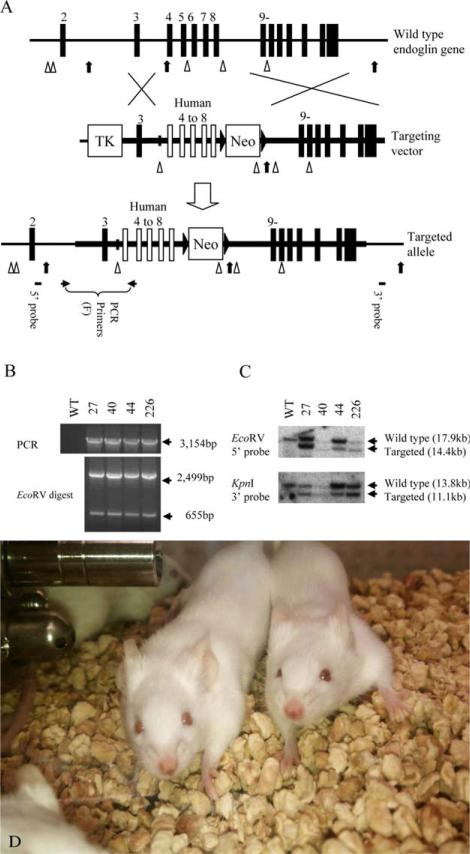
The targeting vector was linearized by restriction enzyme digestion and electroporated into mouse BALB/c-I ES cells.31 G418-resistant clones were first screened by PCR which amplifies a 3.2 kbp product specific to the targeted recombinant allele. PCR-positive clones were further analyzed by Southern blot using 5′ and 3′ external probes. Four out of 232 (1.7%) G418-resistant clones were found to be homologous recombinants. Two clones (No. 27 and 226) whose chromo-some karyotypes were verified to be normal by spectral karyotyping (SKY) imaging were microinjected into C57BL6/J blastocysts at the Roswell Park Gene Targeting Facility to obtain chimera mice. To flox out neomysin cassette, the resulting chimera mice were bred to Cre-deleter mouse line, BALB/c-Tg(CMV-cre)1Cgn/J (The Jackson Laboratory, Bar Harbor, ME), which is expressing Cre recombinase in all tissues including germ cells.32 Then we selected albino mice with BALB/c background for further studies. Specific deletion of neomycin marker was confirmed by PCR and Southern blot. The cre transgene was eliminated by backcrossing male mutant offspring to wild type BALB/cJ female (The Jackson Laboratory) and retrieving a male as a founder for establishing a mutant line (note that the cre transgene is X-linked). Genotyping of mice was performed by PCR and/or Southern blot analysis of DNA from the tail.
IHC
Tissue samples from mice were embedded in Tissue-Tek OCT compound (Sakura Fintek USA, Torrance, CA) and frozen in isopentane chilled with liquid nitrogen. Tissue sections (7–8 μm) were sliced with a Shandon Cryotome Cryostat (Thermo Fisher Scientific, Waltham, MA) and stained with DAKO LSAB1 Kit (Carpinteria, CA) using biotinylated anti-hENG mAb, control mAb or control IgG. The stained tissue sections were counterstained with hematoxylin.
Matrigel plug assay in mice
This assay was performed to determine microvessel density in tumors as described previously.7
Suppression of metastasis
The lungs were retrieved from the sacrificed GEMs 10 days after the last administration of an anti-hENG mAb or an isotype-matched control IgG. The lung metastasis was measured by staining the lung with India ink followed by destaining of the lung with Feket's solution. The surface area of the metastatic colonies was measure by use of ImageJ.33
Therapy of GEMs bearing established tumors
Col 26 colon cancer cells or 4T1 breast cancer cells (1.25 × 105 cells in 0.1 ml PBS for both cells) were inoculated s.c. into the left flank of GEMs. Mice were left untreated until palpable tumors appeared. Mice bearing established s.c. tumors of a similar size were distributed nearly evenly into different groups at the onset of therapy. Consequently, average size of the tumors in each group of mice became similar at the onset of therapy. Then, mice were treated by i.v. administration (via tail vein) of anti-hENG mAb (e.g., 50 or 100 μg/mouse) or the equal amount of an isotype-matched control IgG (MOPC 195v or CCL130; both IgG1-κ). In selected tests, mice received capecitabine (100 mg/kg) or sorafenib (20 mg/kg) by oral administration. Capecitabine was suspended in 40 mM citrate buffer (pH 6.0) containing 5% gum arabic. A stock solution of sorfenib was prepared in Cremophor EL/ethanol (50:50) and was diluted with three-volumes of distilled water before use following the manufacturer's manual. Tumor size and body weight of mice were measured as described previously.6 The measured tumor diameters were converted to tumor volume using the Excel program and the following formula: V = length × width × height × π/6. Statistical analysis of the data was performed using the Student's t-test, the log-rank test or the Gehan-Breslow-Wilcoxon test.
Epitope comparison
Three different experiments were performed to compare the epitope defined by SN6j (SN6j epitope) with the SN6c epitope. First, the molecular location of the epitopes in the linear sequence of the ENG molecule was determined by synthesizing recombinant polypeptide fragments corresponding to the different parts of ENG molecule and analyzing the fragments that bind to SN6j and/or SN6c as described previously.34 Second, the steric relationship among different epitopes including the SN6j and SN6c epitopes was determined by a competitive binding assay as described previously.26 Third, the degree of susceptibility of the two epitopes to enzymatic digestions with multiple enzymes was compared.
Results
Generation of three new anti-hENG mAbs
Anti-hENG mAbs SN6c, SN6d and SN6i were derived from hybridoma clones L4-1C8, D3-1A2, and K2-1C7, respectively that were generated by immunizing mice with an isolated cell membrane ENG preparation following a procedure reported previously.4 All three mAbs are of IgG1-κ. These mAbs along with a prototype anti-hENG mAb SN64,20 were characterized by testing against 25 hematopoietic cell lines in a cellular RIA followed by immunoprecipitation of the target antigen (i.e., ENG) as described previously.4 Briefly, SN6c, SN6d, SN6i and SN6 reacted with all of the five immature B-lineage leukemia cell lines tested (KM-3, NALM-16, REH, NALM-1 and NALM-6) and all of the three myelomonocytic leukemia cell lines tested (ML-2, HL-60 and U937). However, they did not react with any of the seven mature B-lineage leukemia- lymphoma cell lines (BALL-1, BALM-2, Daudi, Ramos, U698M, BALM-3 and SU-DHL-4), any of the seven T leukemia cell lines (MOLT-4, JM, CCRF-CEM, CCRF-HSB2, Ichikawa, HPB-MLT and HUT-78), or any of the three EBV-transformed B cell lines (CCRF-SB, RPMI 1788 and RPMI 8057). This pattern of reactivity is typical for anti-ENG mAbs.4,5,20 From the 125I-labeled antigen preparation, all of the four anti-ENG mAbs immunoprecipitated a single major component of M, 170,000 under unreduced conditions while an isotype-matched control IgG (MOPC 195v) did not immunoprecipitate any significant component. Under reduced conditions, a single major component of M, 90,000 was detected for the immunoprecipitates of SN6c, SN6d, SN6i and SN6. The results indicate that all three of SN6c, SN6d and SN6i define ENG. Epitopes defined by these mAbs were mapped as described above.
Selection of the target region
We selected the region of exons 4–8 for the targeted replacement because the ENG polypeptide segment encoded by these exons contains the majority (8 of the 12) of epitopes defined by our anti-hENG mAbs as described below. However, the major concern is whether the generated humanized ENG functions adequately and the generated GEMs survive (see Discussion section).
Generation of GEMs
We constructed a targeting vector for the targeted replacement of exons 4–8 in the mENG gene (Supporting Information Fig. 1). The scheme for generation of the GEMs is illustrated in Fig. 1. ES clones (Balb/c background) containing the human exons 4–8 recombinant allele were generated by targeted replacement of murine exons 4–8 (Fig. 1A) and identified by PCR (Fig. 1B) and Southern blot (Fig. 1C). ES clones 27 and 226 (Fig. 1C) whose chromosome karyotypes were verified to be normal were individually microinjected into blastocysts (C57BL/6J background), which were then transferred to pseudopregnant female mice to generate chimera mice. The Neo marker was deleted by breeding the chimera mice derived from the ES clone 226 with Cre-deleter mice and the cre transgene was eliminated as described in the Materials and Methods section. The generated homozygous GEMs develop normally and are healthy without signs of telangiectasia (Fig. 1D). We have visually inspected the homozygous GEMs (both male and female mice) for 14 months before they were euthanized. Both female and male mice were healthy and did not show signs of telangiectasia-like symptom. In addition, breeding between the hemizygous male and female GEMs produced wild-type, hemizygous and homozygous progeny in the ratio of 1:2:1. The result indicates that humanized ENG does not exert deteriorating effect on the embryonic development. We designate the generated mouse strain as the hue4-8ge (human endoglin exons 4-8 genetically engineered) mouse strain that was used in the subsequent tests of the targeted therapy of tumors.
Figure 1.
Targeted modification of the endoglin gene in ES cells. (A) Targeted replacement of mouse ENG (mENG) gene exons 4–8 with the corresponding human ENG (hENG) gene exons by homologous recombination in ES cells. The region drawn as a thick horizontal line represents the DNA used in the targeting vector. Mouse and human exons are shown as vertical closed and open boxes, respectively. EcoRV and KpnI restriction enzyme sites are indicated by arrow heads and arrows, respectively. Approximate location of PCR primers and Southern probes are also indicated. Primers used in these studies are shown in Supporting Information Table 1. (B) Four successfully targeted ES cell clones (i.e., 27, 40, 44 and 226) are identified by genomic PCR. Complete replacement events are confirmed by restriction enzyme digestion of the PCR product. (C) Genomic DNA from the ES cell clones are analyzed by Southern blot to confirm the result from genomic PCR. The chromosome karyotypes of clone 27 and 226 were examine by SKY imaging and found to be normal. These clones were individually microinjected into C57BL6/J blastocysts to obtain chimera mice. (D) Homozygous genetically engineered mice (GEMs) with novel human/mouse chimeric (humanized) ENG. The mice develop normally and are healthy without signs of telangiectasia.
As a plan B, we also generated GEMs bearing human exons 5–6 (data not shown). The generated homozygous GEMs (the hue5-6ge mouse strain) mice also develop normally and are healthy.
Expression of hENG epitopes on the tumor vasculature in the hue4-8ge GEMs
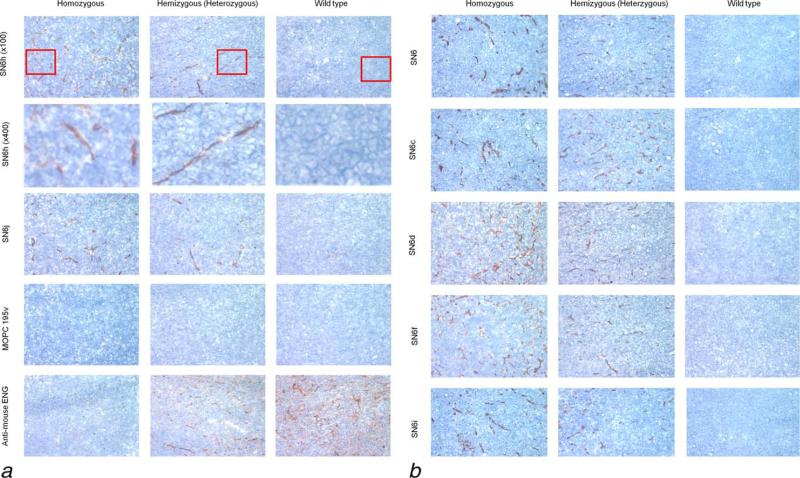
Homozygous and hemizygous GEMs as well as control wild-type mice were inoculated with 4T1 tumor cells. Tissues of the established tumors in these mice were tested by IHC staining with individual mAbs of the eight anti-hENG mAbs. An isotype-matched control IgG (MOPC195v, IgG1-κ) and rat anti-mENG mAb MJ7/18 were included in the test as controls (Fig. 2). MJ7/18 shows strong staining with tumor vasculature of a wild-type mouse and hemizygous GEM but not with tumor vasculature of homozygous GEM (Fig. 2a). The results indicate that MJ7/18 epitope is lost in the humanized ENG. Seven (i.e., SN6h, SN6j, SN6, SN6c, SN6d, SN6f and SN6i) of the eight anti-hENG mAbs whose defining epitopes are located in the hENG region encoded by exons 4–8 (34 and unpublished data) show strong staining with tumor vasculature from both homozygous and hemizygous GEM while these mAbs show weak or no significant staining with tumors from the wild-type mice. A portion of each panel of the top row was enlarged to show the larger images of the stained tumor vessels in the second row (Fig. 2a). Isotype-matched control IgG does not show any significant staining with tumors of any of the three types of mice. These findings with mAbs and control IgG were confirmed by repeating twice with two different sets of the three types of the mice. In each case the expression of ENG is restricted to the vascular endothelium and not detected in the tumor cells. SN6k does not show significant staining with tumor tissues from any of the three types of the mice. This lack of immunostaining of SN6k may be due to that SN6k epitope is a conformational epitope as suggested by Western blot analysis (data not shown). Anti-hENG mAbs (SN6, SN6h and SN6j) immunostained glomerulus of kidney tissue of the GEMs.
Figure 2.
IHC staining of humanized ENG of the blood vessels in the 4T1 tumor from GEMs. Seven (i.e., SN6h, SN6j, SN6, SN6c, SN6d, SN6f and SN6i) of the eight anti-hENG mAbs show strong staining with tumor vasculature from both homozygous and hemizygous GEMs, but show minimal or no significant staining of tumors from wild-type control mice. A portion (indicated by a red square) of each panel of the top row was enlarged to show the larger images of the stained tumor vessels in the second row (Fig. 2a). Rat anti-mENG mAb MJ7/18 shows strong staining with tumor vasculature from a wild-type mouse and hemizygous mouse but does not stain homozygous mouse tumor (Fig. 2a). The result indicates that the MJ7/18 epitope is lost in the humanized ENG. An isotype-matched control IgG (MOPC195v; IgG1-κ; Fig. 2a) does not show any staining with the tumors from the three types of mice.
Suppression of metastasis in GEMs
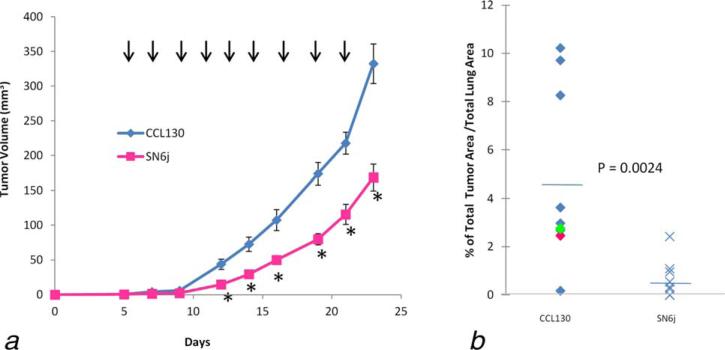
4T1 mammary tumor spontaneously metastasizes to the lung and liver from the primary tumor site. We inoculated 4T1 s.c. in the GEMs and evaluated activity of SN6j against the lung metastasis and the primary tumor. SN6j effectively suppresses growth of the established primary tumor of 4T1 (Fig. 3a). In addition, SN6j strongly suppresses lung metastasis of 4T1 (Fig. 3b).
Figure 3.
SN6j, the parental mAb of the chimeric anti-hENG mAb c-SN6j (also known as TRC105), is effective for suppressing metastasis and primary tumor of 4T1 breast cancer. (a) The GEMs with established tumors of 4T1 were treated by i.v. administration of SN6j (100 μg/mouse; n = 10) or an equal amount of an isotype-matched control IgG (CCL130; IgG1-κ; n = 10). The administration was performed at 2 day intervals as indicated by vertical arrows. Statistically significant (p < 0.05) experimental points are indicated by the star symbols (*). Abscissa indicates days after tumor inoculation. (b) The tumor-bearing GEMs were sacrificed 10 days after the last treatment and the lungs of individual GEMs were retrieved for measurement of metastasis.
Synergistic suppression of tumor growth by targeting two distinctive epitopes
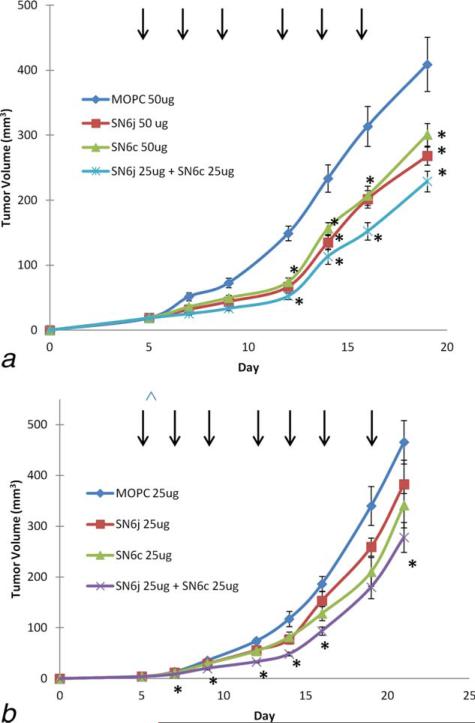
SN6j and SN6c define two distinctive epitopes as revealed by three different assays. First, SN6j epitope is located in the polypeptide region between Pro 231 and Glu 276 while SN6c epitope is located in the region between Tyr 277 and Cys 330 of human and the humanized ENG molecule. Second, these epitopes are sterically distinctive from each other as determined by a competitive binding assay. Third, SN6c epitope is more readily susceptible to enzymatic digestion with each of three proteases (thermolysin, trypsin and chymotrypsin) as compared with SN6j epitope; the results suggest that SN6c epitope is more readily accessible by surrounding aqueous media on the cell surface compared with SN6j epitope. On the basis of these findings, we investigated the tumor suppressive activity of the combination of SN6j and SN6c using a suboptimal dose (25 or 50 μg/mouse) of the mAbs (Fig. 4a). Combination of SN6j and SN6c improved antitumor efficacy compared with individual mAbs. In an additional study, we investigated the potential synergy between SN6j and SN6c in the tumor growth suppression following the previously described procedure.6,35 The test result indicates that combination of SN6j and SN6c is able to exert synergistic antitumor efficacy (Fig. 4b).
Figure 4.
Synergistic potentiation of antitumor activity of anti-hENG mAbs by simultaneous targeting of two distinct epitopes. (a) a suboptimal dose (25 μg or 50 μg/mouse) of anti-ENG mAb SN6j and/or SN6c or isotype-matched control IgG (MOPC 195v; IgG1-κ) was administered i.v., three times a week, into GEMs (n = 10) bearing established tumors of Col 26. Administration of mAb/control IgG is indicated by vertical arrows. Despite the suboptimal dose used, each mAb and the combination showed statistically significant tumor suppression as indicated by the star symbols (*). (b) The potential synergy between SN6j and SN6c was evaluated as described previously.6,35 The test indicates that the combination exerts synergistic antitumor efficacy as shown by the star symbols.
Suppression of tumor growth by SN6j, sorafenib and capecitabine, and suppressive effect of SN6j on tumor microvessels
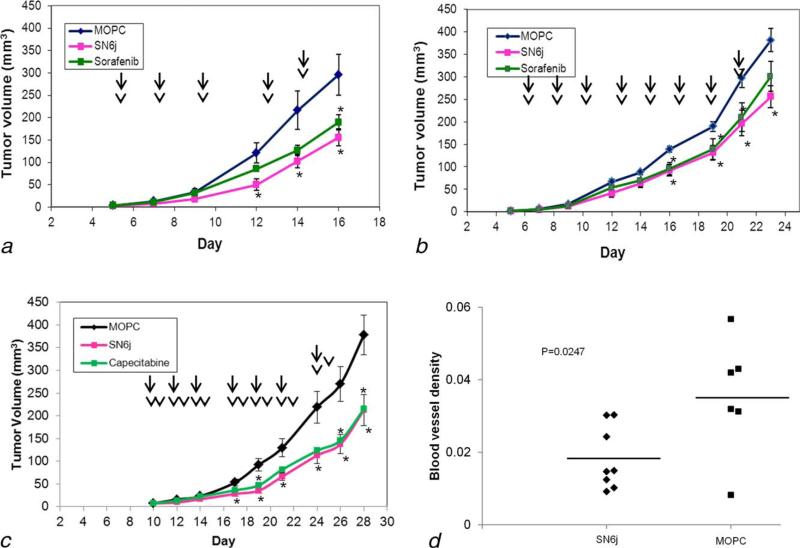
GEMs bearing established tumors of Col 26 (Fig. 5a) and 4T1 (Fig. 5b) were treated by i.v. administration of SN6j or an isotype-matched control IgG or by oral administration of sorafenib. Both SN6j and sorafenib show significant antitumor efficacy against Col 26 tumor (Fig. 5a) and 4T1 tumor (Fig. 5b). In a subsequent experiment, GEMs bearing established Col 26 tumor received SN6j, control IgG or capecitabine (Fig. 5c). Both SN6j and capecitabine showed significant antitumor activities. To gain information about the mechanisms by which SN6j suppresses tumor growth in the GEMs, we investigated the effect of SN6j on the tumor microvessels by use of Matrigel plug assay as described previously.7 SN6j suppressed microvessel density in the Col 26 tumor (p = 0.0247; Fig. 5d).
Figure 5.
Antitumor efficacy of SN6j, sorafenib and capecitabine, and suppressive effect of SN6j on the tumor microvessels in the GEMs. (a, b) Suppression of Col 26 tumor (a) and 4T1 tumor (b) by SN6j and sorafenib. GEMs bearing established tumors (n = 10) were given SN6j (50 μg for Col 26 and 100 μg for 4T1) i.v. three times a week while sorafenib (20 mg/kg) was given orally three times a week. Administration of mAb/control IgG and sorafenib is indicated by arrows and arrow heads, respectively. (c) Growth of established tumor of Col 26 was suppressed by SN6j or capecitabine. SN6j or an isotype-matched control IgG (MOPC 195v) was given i.v. three times a week as indicated by arrows, while capecitabine (100 mg/kg) was given orally six times a week as indicated by arrow heads. Statistically significant (p < 0.05) antitumor activity is indicated by the star symbols. (d) Matrigel plug assay to measure effect of the i.v administered SN6j on the microvessel density of Col 26 tumor in the GEMs. SN6j effectively suppressed the microvessel density (p = 0.0247).
Discussion
Metastasis is the primary cause of cancer-associated death of patients with solid tumors. Some patients may experience metastatic relapse within months whereas distant metastases emerge in others after years or even decades of latency of the disseminated tumor cells.36,37 A recent animal model study by Ghajar et al.38 shows that dormant disseminated tumor cells reside on microvasculature of distant tissues and that stable microvasculature constitutes a dormant niche whereas sprouting neovasculature sparks micrometastatic outgrowth. Therefore appropriate antiangiogenic agents may be able to prevent and/or suppress metastatic outgrowth of the dormant disseminated tumor cells. We hypothesize that ENG-targeted therapy may be useful for such purposes. This hypothesis is consistent with our finding that i.v. administered anti-ENG mAb SN6j strongly suppresses metastasis (Fig. 3).
Our hypothesis that ENG is an attractive target for cancer therapy was supported by several in vitro and animal model studies,4–8,13,20 which led us to generate a chimerized (partially humanized) anti-hENG mAb termed c-SN6j (also known as TRC105) to test it in nonhuman primates and cancer patients.23,24 Results of the ENG-targeting studies by other researchers39–41 are also consistent with our hypothesis. Recently, Nolan-Stevaux et al.10 synthesized TRC105 based on our published nucleotide and amino acid sequences of the VH and VL of TRC105.13. They confirmed the strong antiangiogenic activity of TRC105. They also found that TRC105 strongly interferes with the BMP9/pSMAD1 signaling. A recently completed phase 1 clinical trial24 and several ongoing clinical trials of TRC105 provide us with promising safety features and antitumor efficacy of TRC105 in advanced/metastatic cancer patients with various solid tumors.
Nevertheless, we still need to optimize the antitumor efficacy of TRC105 by various means one of which will be to combine TRC105 with other anticancer agents. To this end, novel GEMs expressing humanized ENG will be highly valuable for evaluating appropriate combinations that include selection of appropriate combination partners and determination of the optimal dosing schedule of individual agents. However, major obstacles exist in developing such GEMs because there is a substantial difference (>30% in the extracellular domain) in the amino acid sequence between mENG and hENG, mutations in the ENG gene are associated with HHT1; these HHT1-causing mutations are dispersed throughout the ENG gene.42 In addition, ENG is essential for vascular development/angiogenesis.
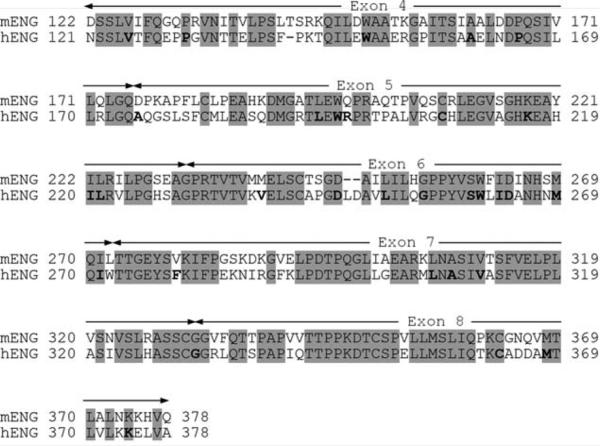
We considered the region of exons 4–8 for the targeted replacement because the ENG polypeptide segment encoded by these exons contains the majority (8 of 12) of epitopes defined by our anti-ENG mAbs. To gain information about the potential effects of the targeted replacement, we compared amino acid sequence between mENG and hENG in the region and identified amino acid residues of which mutations are associated with HHT1 (Fig. 6). The amino acid sequence encoded by exons 4–8 shows 66% identity between mENG and hENG. A total of 31 amino acid residues were identified in this region as the residues associated with the HHT1-causing mutations as indicated by bold letters in Fig. 6. It is assumed that these amino acid residues are important for maintaining functions of ENG and for vascular development in both humans and mice. Of these amino acid residues, 27 (87.1%) are conserved amino acid residues between mENG and hENG, and the remaining 4 are nonconserved residues. A major question is the effect of replacement of these four nonconserved amino acid residues on the functions of ENG in the GEMs; this replacement consists of Asp176Ala, Gln198Arg, Met240Val and Val279Phe. In addition, replacement of other nonconserved amino acids may still affect functions of chENG. These questions may be addressed only by generating the GEMs.
Figure 6.
Amino acid sequence comparison between mENG and hENG in the region encoded by exons 4–8. The conserved regions between mENG and hENG are indicated by shades. Amino acid residues whose mutations are associated with HHT1 by relevant mutations, i.e., missense mutations or single amino acid deletions, are indicated by bold letters. The data of these mutations are from the HHT Mutation Database (http://www.hhtmutation.org/).
We concluded that this and other relevant questions may be addressed only by actually generating such GEMs. To achieve this objective, we constructed a new targeting vector for the targeted replacement of exons 4–8 in mENG (Supporting Information Fig. 1) or of exons 5–6 in mENG (data not shown). Recently, Allinson et al.43 generated a floxed mENG allele in which loxP sites flank exons 5 and 6. In our study, the targeted replacement of exons 4–8 and exons 5–6 was carried out as illustrated for the exons 4–8 replacement (Fig. 1). To our great relief, the generated homozygous GEMs (for both exons 4–8 and 5–6) develop normally and are healthy without signs of telangiectasia (Fig. 1d).
Another important question is whether the generated humanized ENG protein expresses epitopes defined by the designated anti-hENG mAbs. This question was addressed by IHC analysis of tumors from wild-type mice (control) and the homozygous and hemizygous GEMs. IHC analysis of these tumors showed that the humanized ENG in the tumor vasculature strongly expresses epitopes defined by 7 of the eight anti-hENG mAbs whose epitopes are located in the hENG region encoded by exons 4–8 (Fig. 2). Next we evaluated antitumor efficacy of four anti-hENG mAbs SN6j, SN6c, SN6d and SN6i in the GEMs bearing established tumors. All four mAbs showed significant antitumor efficacy without significant side effects. Test results of SN6j and SN6c are shown in Figs. 3–5. Combination of SN6j and SN6c, which define distinctively different epitopes indicated synergistic antitumor efficacy (Fig. 4).
In additional studies, we evaluated antitumor efficacy and safety of other anticancer agents that are candidates for combination therapy with anti-hENG mAbs. One of such agents is sorafenib, a multikinase inhibitor.25 Another agent is cape-citabine, an oral prodrug of 5-fluorouracil (5-FU). Both sorafenib and SN6j show significant antitumor efficacy against established tumors of Col 26 (Fig. 5a) and 4T1 (Fig. 5b). Capecitabine is also effective for tumor suppression in the GEMs (Fig. 5c). Parameters for effective combination of SN6j with sorafenib or capecitabine for tumor therapy remain to be defined in the GEMs. In a clinical trial, the combination of TRC105 and sorafenib is tested to treat hepatocellular carcinoma patients who have not responded to other treatments (NCT01306058). In another trial, the combination of TRC105 and capecitabine is tested to treat Her2-negative metastatic breast cancer patients (NCT01326481, Ref. 44). Coordinated effort between the animal studies in the GEMs and the clinical trials will be valuable for the effective application of the combination therapies. We need to define the optimal doses and dosing schedules of individual components of these combinations in the GEMs.
A recent study of the ENG-heterozygous (Eng+/–) haploinsufficient mice by Anderberg et al.45 suggested that concurrent attenuation of ENG- and VEGF-induced signaling may provide effective control of metastatic dissemination of tumor. Ardelean et al.46 also studied the Eng+/– mice and found that anti-VEGF treatment reduced the hepatic micro-vesssel density and microvascular perfusion.
Our GEM model allows us to perform comprehensive tests for the combination of anti-hENG mAbs with other anticancer agents (one or multiple agents), which will be valuable for translating the test results to the designs of the TRC105 trials in patients with various advanced/metastatic solid tumors.
Supplementary Material
What's new?
Endoglin (ENG) is a TGF-β coreceptor that is essential for vascular development and angiogenesis. A chimeric anti-human ENG (hENG) monoclonal antibody has shown promising safety and clinical efficacy in clinical trials of patients with various advanced solid tumors. Here, the authors developed a novel genetically engineered mouse model expressing a functional humanized ENG to optimize ongoing and future hENG antibody-based cancer therapy in patients. The homozygous genetically engineered mice developed normally and were healthy. Growth of established breast and colon tumors as well as metastasis and microvessel density in the mice were effectively suppressed by systemic administration of hENG mAbs.
Acknowledgements
The authors thank Dr. Peter Aplan for providing us with pTKneoF vector, and thank Drs. Scott Wilhelm and Dennis Healy for providing sorafenib. They thank Dr. Kazutoyo Osoegawa, Ms. Aimee Stablewski, Dr. Kenneth Gross, Dr. Xinwei She and Dr. Kazunori Kanehira for technical advice and help. They also thank Dr. Seiichi Matsui for SKY imaging analysis and Ms. Hilda Tsai for technical assistance.
Grant sponsor: Clinical Translational Research grant, from the Department of Defense Breast Cancer Research Program; Grant number: BC020043; Grant sponsors: Tracon Pharma, a Roswell Park Alliance Foundation grant.
Abbreviations
- BMP9
bone morphogenetic protein 9
- ENG
endoglin
- GEMs
genetically engineered mice
- GEMM
genetically engineered mouse model
- hENG
human ENG
- HHT1
hereditary hemorrhagic telangiectasia type 1
- IHC
immunohistochemistry
- mENG
mouse ENG
- mAb
monoclonal antibody
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Ebos JM, Kerbel RS. Antiangiogenic therapy: impact on invasion, disease progression, and metastasis. Nat Rev Clin Oncol. 2011;8:210–21. doi: 10.1038/nrclinonc.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreno Garcia V, Basu B, Molife LR, et al. Combining antiangiogenics to overcome resistance: rationale and clinical experience. Clin Cancer Res. 2012;18:3750–61. doi: 10.1158/1078-0432.CCR-11-1275. [DOI] [PubMed] [Google Scholar]
- 3.Jayson GC, Hicklin DJ, Ellis LM. Antiangiogenic therapy—evolving view based on clinical trial results. Nat Rev Clin Oncol. 2012;9:297–303. doi: 10.1038/nrclinonc.2012.8. [DOI] [PubMed] [Google Scholar]
- 4.Seon BK, Matsuno F, Haruta Y, et al. Long-lasting complete inhibition of human solid tumors in SCID mice by targeting endothelial cells of tumor vasculature with antihuman endoglin immunotoxin. Clin Cancer Res. 1997;3:1031–44. [PubMed] [Google Scholar]
- 5.Matsuno F, Haruta Y, Kondo M, et al. Induction of lasting complete regression of preformed distinct solid tumors by targeting the tumor vasculature using two new anti-endoglin monoclonal antibodies. Clin Cancer Res. 1999;5:371–82. [PubMed] [Google Scholar]
- 6.Takahashi N, Haba A, Matsuno F, et al. Antiangiogenic therapy of established tumors in human skin/severe combined immunodeficiency mouse chimeras by anti-endoglin (CD105) monoclonal antibodies, and synergy between anti-endoglin antibody and cyclophosphamide. Cancer Res. 2001;61:7846–54. [PubMed] [Google Scholar]
- 7.Tsujie M, Tsujie T, Toi H, et al. Anti-tumor activity of an anti-endoglin monoclonal antibody is enhanced in immunocompetent mice. Int J Cancer. 2008;122:2266–73. doi: 10.1002/ijc.23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uneda S, Toi H, Tsujie T, et al. Anti-endoglin monoclonal antibodies are effective for suppressing metastasis and the primary tumors by targeting tumor vasculature. Int J Cancer. 2009;125:1446–53. doi: 10.1002/ijc.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheifetz S, Bellon T, Cales C, et al. Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J Biol Chem. 1992;267:19027–30. [PubMed] [Google Scholar]
- 10.Nolan-Stevaux O, Zhong W, Culp S, et al. Endoglin requirement for BMP9 signaling in endothelial cells reveals new mechanism of action for selective anti-endoglin antibodies. PLoS One. 2012;7:e50920. doi: 10.1371/journal.pone.0050920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrows FJ, Derbyshire EJ, Tazzari PL, et al. Up-regulation of endoglin on vascular endothelial cells in human solid tumors: implications for diagnosis and therapy. Clin Cancer Res. 1995;1:1623–34. [PubMed] [Google Scholar]
- 12.Miller DW, Graulich W, Karges B, et al. Elevated expression of endoglin, a component of the TGF-beta-receptor complex, correlates with prolifera tion of tumor endothelial cells. Int J Cancer. 1999;81:568–72. doi: 10.1002/(sici)1097-0215(19990517)81:4<568::aid-ijc11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Seon BK, Haba A, Matsuno F, et al. Endoglin-targeted cancer therapy. Curr Drug Deliv. 2011;8:135–43. doi: 10.2174/156720111793663570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clasper S, Royston D, Baban D, et al. A novel gene expression profile in lymphatics associated with tumor growth and nodal metastasis. Cancer Res. 2008;68:7293–303. doi: 10.1158/0008-5472.CAN-07-6506. [DOI] [PubMed] [Google Scholar]
- 15.Li DY, Sorensen LK, Brooke BS, et al. Defective angiogenesis in mice lacking endoglin. Science. 1999;284:1534–7. doi: 10.1126/science.284.5419.1534. [DOI] [PubMed] [Google Scholar]
- 16.Arthur HM, Ure J, Smith AJ, et al. Endoglin, an ancillary TGFbeta receptor, is required for extra-embryonic angiogenesis and plays a key role in heart development. Dev Biol. 2000;217:42–53. doi: 10.1006/dbio.1999.9534. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Elsner T, Botella LM, Velasco B, et al. Endoglin expression is regulated by transcriptional cooperation between the hypoxia and transforming growth factor-beta pathways. J Biol Chem. 2002;277:43799–808. doi: 10.1074/jbc.M207160200. [DOI] [PubMed] [Google Scholar]
- 18.Bockhorn M, Tsuzuki Y, Xu L, et al. Differential vascular and transcriptional responses to anti-vascular endothelial growth factor antibody in orthotopic human pancreatic cancer xenografts. Clin Cancer Res. 2003;9:4221–6. [PubMed] [Google Scholar]
- 19.Lebrin F, Goumans MJ, Jonker L, et al. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 2004;23:4018–28. doi: 10.1038/sj.emboj.7600386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haruta Y, Seon BK. Distinct human leukemia-associated cell surface glycoprotein GP160 defined by monoclonal antibody SN6. Proc Natl Acad Sci U S A. 1986;83:7898–902. doi: 10.1073/pnas.83.20.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong H, Yang Y, Zhang Y, et al. Positron emission tomography imaging of CD105 expression during tumor angiogenesis. Eur J Nucl Med Mol Imaging. 2011;38:1335–43. doi: 10.1007/s00259-011-1765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She X, Matsuno F, Harada N, et al. Synergy between anti-endoglin (CD105) monoclonal antibodies and TGF-beta in suppression of growth of human endothelial cells. Int J Cancer. 2004;108:251–7. doi: 10.1002/ijc.11551. [DOI] [PubMed] [Google Scholar]
- 23.Shiozaki K, Harada N, Greco WR, et al. Antiangiogenic chimeric anti-endoglin (CD105) antibody: pharmacokinetics and immunogenicity in nonhuman primates and effects of doxorubicin. Cancer Immunol Immunother. 2006;55:140–50. doi: 10.1007/s00262-005-0691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen LS, Hurwitz HI, Wong MK, et al. A phase I first-in-human study of TRC105 (anti-endoglin antibody) in patients with advanced cancer. Clin Cancer Res. 2012;18:4820–9. doi: 10.1158/1078-0432.CCR-12-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilhelm S, Carter C, Lynch M, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–44. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi N, Kawanishi-Tabata R, Haba A, et al. Association of serum endoglin with metastasis in patients with colorectal, breast, and other solid tumors, and suppressive effect of chemotherapy on the serum endoglin. Clin Cancer Res. 2001;7:524–32. [PubMed] [Google Scholar]
- 27.Dinkel A, Aicher WK, Warnatz K, et al. Efficient generation of transgenic BALB/c mice using BALB/c embryonic stem cells. J Immunol Methods. 1999;223:255–60. doi: 10.1016/s0022-1759(99)00005-8. [DOI] [PubMed] [Google Scholar]
- 28.Osoegawa K, Mammoser AG, Wu C, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–96. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee EC, Yu D, Martinez de Velasco J, et al. A highly efficient Escherichia coli-based chromo-some engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 30.Osoegawa K, Tateno M, Woon PY, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–28. [PMC free article] [PubMed] [Google Scholar]
- 31.Noben-Trauth N, Kohler G, Burki K, et al. Efficient targeting of the IL-4 gene in a BALB/c embryonic stem cell line. Transgenic Res. 1996;5:487–91. doi: 10.1007/BF01980214. [DOI] [PubMed] [Google Scholar]
- 32.Schwenk F, Baron U, Rajewsky K. A cretransgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–1. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43(1 Suppl):25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 34.She X, Seon BK. Epitope mapping of endoglin, a TGF-b receptor, using recombinant fragments and twelve monoclonal antibodies. Proc Am Assoc Cancer Res. 2001;42:825. [Google Scholar]
- 35.Yokoyama Y, Dhanabal M, Griffioen AW, et al. Synergy between angiostatin and endostatin: inhibition of ovarian cancer growth. Cancer Res. 2000;60:2190–6. [PubMed] [Google Scholar]
- 36.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–46. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhr JW, Pantel K. Controversies in clinical cancer dormancy. Proc Natl Acad Sci U S A. 2011;108:12396–400. doi: 10.1073/pnas.1106613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghajar CM, Peinado H, Mori H, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–17. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkel T, Holig P, Merdan T, et al. Targeting of immunoliposomes to endothelial cells using a single-chain Fv fragment directed against human endoglin (CD105). Biochim Biophys Acta. 2004;1663:158–66. doi: 10.1016/j.bbamem.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Munoz R, Arias Y, Ferreras JM, et al. Targeting a marker of the tumour neovasculature using a novel anti-human CD105-immunotoxin containing the non-toxic type 2 ribosome-inactivating protein nigrin b. Cancer Lett. 2007;256:73–80. doi: 10.1016/j.canlet.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Dolinsek T, Markelc B, Sersa G, et al. Multiple delivery of siRNA against endoglin into murine mammary adenocarcinoma prevents angiogenesis and delays tumor growth. PLoS One. 2013;8:e58723. doi: 10.1371/journal.pone.0058723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald J, Bayrak-Toydemir P, Pyeritz RE. Hereditary hemorrhagic telangiectasia: an overview of diagnosis, management, and pathogenesis. Genet Med. 2011;13:607–16. doi: 10.1097/GIM.0b013e3182136d32. [DOI] [PubMed] [Google Scholar]
- 43.Allinson KR, Carvalho RL, van den Brink S, et al. Generation of a floxed allele of the mouse Endoglin gene. Genesis. 2007;45:391–5. doi: 10.1002/dvg.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine EG, Forero A, O’Connor T, et al. A phase 1b dose-escalation study of TRC105 (anti-endoglin antibody) in combination with capecitabine for advanced solid tumors (including patients with progressive or recurrent Her-2-negative metastatic breast cancer).. J Clin Oncol; 2013 ASCO Annual Meeting Proceedings.2013. p. 15S.p. 187s. [Google Scholar]
- 45.Anderberg C, Cunha SI, Zhai Z, et al. Deficiency for endoglin in tumor vasculature weakens the endothelial barrier to metastatic dissemination. J Exp Med. 2013;210:563–79. doi: 10.1084/jem.20120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ardelean DS, Jerkic M, Yin M, et al. Endoglin and activin receptor-like kinase 1 heterozygous mice have a distinct pulmonary and hepatic angiogenic profile and response to anti-VEGF treatment. Angiogenesis. 2014;17:129–46. doi: 10.1007/s10456-013-9383-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.