Abstract
Polybrominated diphenyl ethers (PBDEs) are known endocrine disrupting chemicals used commonly as flame retardants in everything from electronics to furniture. Exposure to PBDEs during early development has been linked to neurodevelopmental delays. Despite mounting evidence of neurological harm from PBDE exposure, the molecular mechanisms underlying these effects on brain function remain unknown. We examined the effects of perinatal exposure to BDE-47, the most biologically active and prevalent BDE congener in North America, on epigenetic patterns in the frontal lobe of Wistar rats. Dams were gavaged with BDE-47 (0.002 and 0.2 mg/kg body weight) at gestation days 9 and 16, and postnatal days 1, 8, and 15. Frontal lobes from offspring at postnatal day 41 were collected to measure 5-methylcytosine (5mC) in mitochondrial cytochrome c oxidase genes (Mt-co1, Mt-co2, and Mt-co3), global nuclear 5-hydroxymethylcytosine (5hmC) content, 5mC in repetitive elements L1Rn, and 5mC in nuclear genes (Bdnf, Crhr1, Mc2r, Nr3c1, and Snca) related to behavioral and brain functions in the nuclear genome. We observed a significant decrease in %5mC in Mt-co2 (difference from control= −0.68%, p=0.01 at the 0.2 mg/kg BDE-47). 5mC in repetitive elements L1Rn decreased at 0.002 mg/kg BDE-47 (difference= −1.23%, p=0.02). Decreased nuclear 5mC was observed in Bdnf and Nr3c1 in BDE-47 exposed rats. However, we did not observe significant effects of PBDE toxicity on DNA methylation patterns for the majority of genes in the brain.
Keywords: BDE-47, PBDE, Endocrine Disruptors, Epigenetics, DNA methylation, Brain
1.1 INTRODUCTION
Polybrominated diphenyl ethers (PBDEs) are organic chemicals used as flame retardants in a range of materials including textiles, plastics, wire insulation, and automobiles. PBDEs are toxic and accumulate in blood, breast milk, and fat tissue in humans, as well as in soil, sediment, and household dust (de Wit 2002).
PBDEs are highly lipophilic and therefore tend to accumulate in lipids or fats. They also easily cross the placenta and transfer to newborns through breastfeeding (Cui et al., 2012). Lipid-rich tissues, like the brain, have shown damage following PBDE exposure, possibly due to increased bioaccumulation (Mitchell et al., 2012). In rodents, gestational or postnatal exposure to the most prevalent PBDE congeners (i.e., PBDE 47, 99, 153, and 209) found in human blood (Fischer et al., 2006; Vizcaino et al., 2011) and in the environment (de Wit 2002) has been associated with significant postnatal hyperactivity (Branchi et al., 2002), decreased habituation capacity, and irreversible defects in the cholinergic system (e.g., marked hypoactivity after a nicotine challenge, decreased cholinergic binding sites in the hippocampus) (Eriksson et al., 2002; Goodman 2009). We previously observed substantial BDE-47-induced hyperactivity in offspring from rats exposed to low-dose BDE-47, which is considered the most biologically active PBDE congener and is found commonly in human blood samples across the U.S. and Canada (Suvorov et al., 2009). We have also reported significant changes in gene expression from the brain frontal lobe in the same animals (Suvorov and Takser 2011) occurring 3 weeks [postnatal day (PND) 40] after exposure. Consistent with animal experiments, three epidemiological studies have reported attention deficit and cognitive delay in relation to PBDE levels in blood (Gascon et al., 2012; Lengyel et al., 1985; Roze et al., 2009).
PBDE congeners have the potential to cross the cell membrane and accumulate in cells, particularly in mitochondria (Huang et al., 2010). Mitochondria produce the cellular energy that is critical to brain function. Mitochondrial cytotoxicity following PBDE exposure appears to occur by inhibition of mitochondrial respiration and dissipation of the mitochondrial membrane potential (Pereira et al., 2013). Even small amounts of PBDEs (e.g., 0.02 mM of BDE-49) inhibit electron transport at Complex V and Complex IV in neural mitochondria, as shown both in animal and in vitro studies (Napoli et al., 2013), which can affect the brain’s energy balance. Despite these findings, the root cause of PBDE-induced mitochondrial toxicity has remained elusive.
Recently, mitochondrial DNA methylation has been identified as a novel epigenetic mechanism with specific sensitivity to environmental exposures (Byun and Baccarelli 2014; Byun et al., 2013). Altered DNA methylation in mitochondria, as in the nuclear genome, leads to dysregulated gene expression (Feng et al., 2012). Given data that suggest that PBDE toxicity in neurodevelopment and brain activity may be mediated by mitochondrial dysfunction, we hypothesized that this dysfunction is driven by epigenetic changes caused by exposure. In this study, we examined the effects of BDE-47 exposure during perinatal neuronal development on DNA methylation in the frontal lobes of rats. The frontal lobes of the cerebral cortex are involved in complex behavior, cognition, and language (Fuster 2002). Specifically, we measured the methylation of mitochondrial genes involved in respiration, i.e., cytochrome c oxidase I (Mt-co1), cytochrome c oxidase II (Mt-co2), and cytochrome c oxidase III (Mt-co3), in exposed and control rats. Altered cytochrome c oxidase activity has been implicated in cognitive function (Gu et al., 2014), neurodegenerative disease (Griguer et al., 2013) and brain damage (Novgorodov et al., 2014).
The stimuli received by the mitochondria can be transmitted to the nucleus to induce changes in the regulation of nuclear genes (Woodson and Chory 2008), including by epigenetic mechanisms (Smiraglia et al., 2008). Therefore, we also examined DNA methylation in the nuclear genome. We measured nuclear 5-hydroxymethylation (5hmC), an emerging alternative methylation marker, as well as 5mC methylation at the left end of rat L1Rn (long interspersed repeated), a marker of methylation in retrotransposons, the ‘jumping’ DNA sequences that have been shown to have key roles in neuronal plasticity (Jakovcevski and Akbarian 2012). To further understand neuronal epigenetic PBDE toxicity, we measured changes in the DNA methylation of nuclear candidate genes related to behavioral and brain functions. These included the brain-derived neurotrophic factor (Bdnf), corticotropin releasing hormone receptor 1 (Crhr1), melanocortin 2 receptor (Mc2r), nuclear receptor subfamily 3 group C, member 1 (Nr3c1), and alpha-synuclein (Snca).
We observed that perinatal exposure to PBDE affects minor DNA methylation patterns in the brain in both the mitochondrial and nuclear genomes. However, the majority of DNA methylation patterns that we studied were not affected by PBDE exposure in the brain.
1.2 MATERIAL AND METHODS
1.2.1 Animals and Exposure
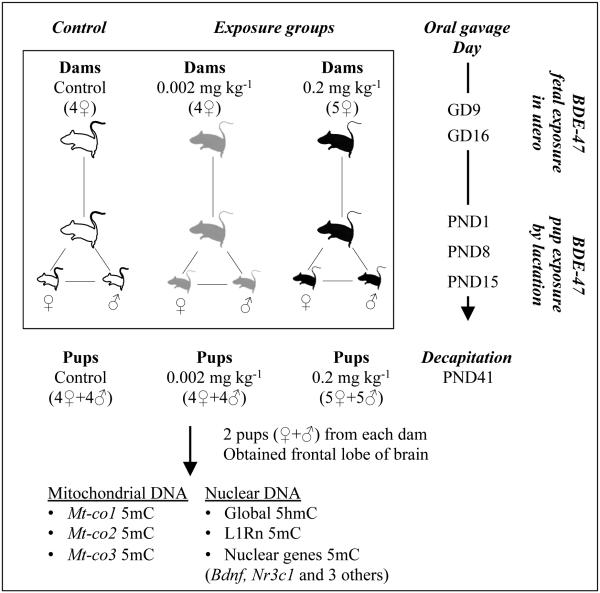
Seven-week-old Wistar female and male rats were received from Charles River Laboratories and then housed individually, except when breeding, in polypropylene cages (38×48×20) with sawdust bedding and under controlled temperature (21 ± 2°C), relative humidity (50 ± 10%), and 12-h light/dark cycle. Food (Rodent CHOW 5075, Charles River’s Laboratories, Wilmington, MA) and tap water were provided ad libitum. All animals received care in compliance with the Guide to the Care and Use of Experimental Animals from the Canadian Council of Animal Care, and all experimental protocols were approved by the institutional animal research ethics review board. Breeding pairs were established one week after acclimatization: each female rat was placed with a male overnight and copulation was confirmed in the morning by observation of vaginal plug. The day of detection was assigned as the first day of pregnancy [gestation day (GD) 0]. Dams were randomly assigned to three groups: 0.002 mg/kg body weight (4 dams) or 0.2 mg/kg body weight (5 dams) of neutral standard of 2,2’, 4,4’-tetrabromodiphenyl ether (BDE-47, Chromatographic Specialties Inc., Brockville, Canada), or no exposure (control group, 4 dams) (Figure 1). The vehicle was administered to the control group according to the same protocol. We note that the control group also had background BDE-47 level (Suvorov and Takser 2011). A preparation of BDE-47 solution was used for all experiments to provide a consistent vehicle solution (990 μL peanut oil with 10 μL toluene) and administered at 500 μL/body weight each morning via oral gavage at GD 9 and 16 and at PND 1, 8, and 15 through stainless steel needles (Harvard Apparatus Inc., 16×4”). Dams were allowed to deliver and the litter size was not artificially altered. Dams and pups were kept together until weaning on PND 21. At PND 41, one male pup and one female pup per litter were selected randomly and killed by decapitation after stunning as described in the Guide to the Care and Use of Experimental Animals from the Canadian Council of Animal Care (Canadian Council on Animal Care, 1993). Frontal lobe brain samples from control (4♀+4♂), 0.002 mg/kg (4♀+4♂), and 0.2 mg/kg (5♀+5♂) rats were isolated, snap-frozen in liquid nitrogen, and stored at −80°C until use (Figure 1).
FIGURE 1. Experimental model of BDE-47 exposure in Wistar rats and measurements of DNA methylation.
Oral gavage day: the day of BDE-47 exposure on dams by oral gavage, GD: gestational day, PND: postnatal day, ♀: female rat, and ♂: male rat.
1.2.2 DNA Extraction and Bisulfite Conversion
Genomic DNA was isolated using a Qiagen extraction kit (QIAGEN Inc., Valencia, CA) according to manufacturer’s instructions and kept at −80°C until use. Mitochondrial DNA was extracted from the frontal lobe of rat brains using the FOCUS™ Mitochondria prep kit (G-Biosciences, St. Louis, MO) according to the manufacturer’s instructions. One microgram of genomic DNA was bisulfite-converted using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. The final elution was performed with 30 μL of M-Elution Buffer. Bisulfite-treated DNA was aliquoted and stored at −80°C until use.
1.2.3 DNA Methylation Measurement
We developed bisulfite PCR pyrosequencing assays for rat mitochondrial genes (Mt-co1, Mt-co2, and Mt-co3), repetitive elements (L1Rn-ORF1 and L1Rn-5UTR), and nuclear genes (Bdnf, Crhr1, Mc2r, Nr3c1, and Snca). Specific regions were chosen for being promoter regions or known regulators of gene expression. Primer sequences, PCR conditions, and genomic locations can be found in Supplemental Table S1. Between one and six CpGs were evaluated in each assay. PCR amplification was performed at standard conditions using the GoTaq® Hot Start Polymerase (Promega, Madison, WI). We used a PSQ Q96 MD Pyrosequencing System (QIAGEN, Valencia, CA), as previously described (Byun et al., 2013). Pyrogram peak patterns from every sample were visually inspected to confirm the quality of the reactions. The Pearson correlation coefficient between technical replication was between 0.44 (Mt-co1) and 0.99 (Crhr1). Pearson correlation coefficient for L1Rn-5UTR is r=0.38, p=0.05.
5hmC content was quantified using a MethylFlash Hydroxymethylated DNA Quantification Kit (Epigentek, NY, USA) according to the manufacturer's instructions. Briefly, after subtracting the negative control from the sample and the standard, the relative fluorescence units (RFU) value of 5hmC for each sample was calculated as a ratio of sample OD relative to the standard OD as follows: 5hmC%=[(Sample OD-Negative control OD)/Slope × Input DNA amount] × 100 (Li and Liu 2011). The Pearson Correlation coefficient between technical replication was 0.87 (p<0.0001).
1.2.4 Statistical Analysis
The 5mC data were generated through pyrosequencing, which may provide measures of multiple methylation CpG sites within each gene. We evaluated BDE-47’s effect on methylation at each CpG site as well as the average effect on all CpGs within each gene. To this end, we used mixed-effects regression models by adapting the methods described by Boeke et al. (Boeke et al., 2012). We used linear regression models to assess the effect of BDE-47 treatment (control, 0.002 mg /kg, and 0.2 mg/kg) on 5hmC methylation. We checked regression assumptions by performing diagnostic tests for each model, including the Shapiro-Wilk and Kolmogorov-Smirnov tests for normality of residuals and predicted values graph for assessing heteroscedasticity and presence of outlier values.
All tests were two-sided and a p-value less than 0.05 was considered statistically significant. False discovery rate (FDR) was computed with the Benjamini-Hochberg’s method. Average positions were not included in the FDR calculation. Results were considered significant at FDR < 0.20. All statistical analyses were performed in SAS (version 9.2; SAS Institute Inc., Cary, NC, USA).
1.3 RESULTS
1.3.1 Effect of Perinatal BDE-47 Exposure on Mitochondrial DNA Methylation
Mitochondria were isolated from the frontal lobe of BDE-47 exposed rats and Mt-co1 (1 CpG), Mt-co2 (2 CpGs), and Mt-co3 (3 CpGs) mitochondrial DNA methylation was measured. Mean methylation levels ranged from 0.74% (Mt-co3 in position 1 of 0.002 mg/kg BDE-47 dose) to 4.28% (Mt-co2 in position 1 of Control) (Table 1). One of the CpGs in the Mt-co2 gene (position 1 in Table 1) showed a significant decrease in methylation at the 0.2 mg/kg BDE-47 dose compared to controls [difference vs. control in %5mC= −0.68, 95% confidence interval (CI) −1.17;−0.19, p=0.01, and FDR=0.08]. Other positions showed changes that were not statistically significant. The Mt-co1 gene did not show any change in DNA methylation at the 0.002 mg/kg BDE-47 dose, nor at the 0.2 mg/kg BDE-47 dose (Table 1).
TABLE 1.
Effect of BDE-47 treatment on DNA methylation in the brain frontal lobe of mitochondrial Cox genes
| BDE-47 Dose |
||||||||
|---|---|---|---|---|---|---|---|---|
| Control | 0.002 mg kg−1 | 0.2 mg kg−1 | ||||||
|
| ||||||||
| Gene | CpG position |
Mean Estimate (95% CI) |
Mean Estimate (95% CI) |
Relative to the Control group |
Mean Estimate (95% CI) |
Relative to the Control group |
||
| Difference (95% CI) |
p | Difference (95% CI) |
p | |||||
| Mt-co1 | Position 1 | 1.78 (1.47; 2.09) |
1.59 (1.28; 1.89) |
−0.19 (−0.63; 0.24) |
0.37 | 1.45 (1.17; 1.72) |
−0.33 (−0.74; 0.08) |
0.11 |
| Position 1 | 4.28 (3.91; 4.65) |
3.96 (3.59; 4.33) |
−0.32 (−0.84; 0.20) |
0.21 | 3.60 (3.27;3.93) |
−0.68 (−1.17;−0.19) |
0.01 | |
| Mt-co2 | Position 2 | 1.11 (0.89; 1.32) |
1.13 (0.92; 1.34) |
0.03 (−0.27; 0.32) |
0.86 | 1.20 (1.01; 1.39) |
0.09 (−0.19; 0.38) |
0.50 |
| Average | 2.96 (2.63; 3.28) |
2.83 (2.51; 3.15) |
−0.13 (−0.58;0.33) |
0.57 | 2.61 (2.32; 2.89) |
−0.35 (−0.78; 0.08) |
0.11 | |
| Position 1 | 0.85 (0.76; 0.93) |
0.74 (0.65; 0.83) |
−0.11 (−0.23; 0.02) |
0.09 | 0.76 (0.68; 0.84) |
−0.09 (−0.20; 0.03) |
0.14 | |
| Mt-co3 | Position 2 | 0.92 (0.80; 1.04) |
0.76 (0.64; 0.88) |
−0.16 (−0.33; 0.01) |
0.07 | 0.84 (0.72; 0.95) |
−0.08 (−0.25; 0.08) |
0.32 |
| Position 3 | 3.70 (3.39; 4.00) |
3.45 (3.15; 3.76) |
−0.24 (−0.67; 0.19) |
0.26 | 3.61 (3.34; 3.88) |
−0.09 (−0.49; 0.32) |
0.66 | |
| Average | 1.90 (1.74; 2.07) |
1.70 (1.54; 1.87) |
−0.20 (−0.43; 0.03) |
0.09 | 1.80 (1.65; 1.95) |
−0.10 (−0.32; 0.12) |
0.35 | |
1.3.2 Effect of Perinatal BDE-47 Exposure on Global Nuclear 5hmC Methylation
The mean level of global nuclear 5hmC methylation was 0.48% (95% CI 0.33; 0.63) in controls, 0.38% (95% CI 0.24; 0.53) at the 0.002 mg/kg BDE-47 dose, and 0.69% (95% CI 0.56;0.82) at the 0.2 mg/kg BDE-47 dose (Table 2). Global 5hmC levels in the 0.2 mg/kg group were non-significantly increased relative to the control group (difference in %5hmC=0.21, 95% CI −0.02; 0.41, p=0.08). The 5hmC level following the 0.002 mg/kg BDE-47 dose was not different from controls (difference in %5hmC=−0.10; 95% CI −0.31; 0.11, p=0.58) (Table 2).
TABLE 2.
Effect of BDE-47 treatment on DNA methylation in the brain frontal lobe of global 5-hydroxymethylcytosine
| BDE-47 Dose |
|||||||
|---|---|---|---|---|---|---|---|
| Control | 0.002 mg kg−1 | 0.2 mg kg−1 | |||||
|
| |||||||
| Mean Estimate (95% CI) |
Mean Estimate (95% CI) |
Relative to the Control group |
Mean Estimate (95% CI) |
Relative to the Control group |
|||
| Difference (95% CI) |
p | Difference (95% CI) |
p | ||||
|
Global
5hmc |
0.48 (0.33; 0.63) |
0.38 (0.24; 0.53) |
−0.10 (−0.31; 0.11) |
0.58 | 0.69 (0.56; 0.82) |
0.21 (0.02; 0.41) |
0.08 |
1.3.3 Effect of Perinatal BDE-47 Exposure on 5mC Methylation on Repetitive Elements L1Rn
We measured 5mC DNA methylation at two regions of repetitive elements L1Rn: the 5’ untranslated region (5’UTR) and open reading frame (ORF) 1. Mean methylation in the L1Rn 5’UTR region was significantly decreased at the 0.002 mg/kg BDE-47 dose compared to controls (difference %5mC=−1.23; 95% CI −2.27; −0.18, p=0.02). The L1Rn UTR region did not show significant changes in mean methylation at the 0.2 mg/kg BDE-47 dose relative to controls (difference in %5mC= −0.40; 95% CI −1.40; 0.59, p=0.41). L1 ORF1 did not show significant changes in methylation at either dose (difference in %5mC= −0.22; 95% CI −1.03; 0.59, p=0.58 at 0.002 mg/kg, FDR=0.58; and difference in %5mC=0.41; 95% CI −0.36; 1.18, p=0.28, FDR=0.42 at 0.2 mg/kg) (Table 3).
TABLE 3.
Effect of BDE-47 treatment on DNA methylation in the brain frontal lobe of repetitive elements L1Rn (long interspersed repeated)
| BDE-47 Dose |
||||||||
|---|---|---|---|---|---|---|---|---|
| Control | 0.002 mg kg−1 | 0.2 mg kg−1 | ||||||
|
| ||||||||
| Gene | CpG position |
Mean Estimate (95% CI) |
Mean Estimate (95% CI) |
Relative to the Control group |
Mean Estimate (95% CI) |
Relative to the Control group |
||
| Difference (95% CI) |
p | Difference (95% CI) |
p | |||||
| Position 1 | 69.05 (68.43; 69.67) |
68.37 (67.75; 68.99) |
−0.68 (−1.55; 0.19) |
0.12 | 68.54 (67.99; 69.09) |
−0.51 (−1.33; 0.32) |
0.22 | |
|
L1Rn
(5'UTR) |
Position 2 | 40.51 (39.44; 41.58) |
38.84 (37.77; 39.91) |
−1.67 (−3.18;−0.16) |
0.03 | 40.30 (39.34; 41.25) |
−0.21 (−1.64; 1.22) |
0.76 |
| Average | 54.81 (54.07; 55.55) |
53.59 (52.84; 54.33) |
−1.23 (−2.27;−0.18) |
0.02 | 54.41 (53.75; 55.07) |
−0.40 (−1.40; 0.59) |
0.41 | |
|
L1Rn
(ORF1) |
Position 1 | 82.27 (81.69; 82.84) |
82.05 (81.48; 82.62) |
−0.22 (−1.03;0.59) |
0.58 | 82.68 (82.17; 83.20) |
0.41 (−0.36; 1.18) |
0.28 |
1.3.4 Effect of Perinatal BDE-47 Exposure on Methylation of Neurodevelopment and Stress Nuclear Genes
We examined DNA methylation in genes associated with neuron development and neurotoxicity (three CpGs in Bdnf and two CpGs in Snca) and stress-related genes (five CpGs in Crhr1, two CpGs in Mc2r, and six CpGs in Nr3c1) in the frontal lobes of rats exposed to BDE-47. Of the three CpGs measured in Bdnf, position 2 showed significantly decreased methylation at both BDE-47 doses relative to controls (difference in %5mC= −1.34; 95% CI −2.61; −0.06, p=0.04, FDR=0.45 at 0.002 mg/kg and difference in %5mC= −2.13; 95% CI −3.34; −0.91, p=0.001, FDR=0.02 at 0.2 mg/kg) (Table 4). The average of the three Bdnf positions showed a marginal decrease in DNA methylation at the 0.2 mg/kg BDE-47 dose (difference in %5mC= −1.63; 95% CI −3.44; 0.17, p=0.07) (Table 4). Among all the CpG positions in Nr3c1, one CpG (position 4 in Table 4) showed a significant increase in DNA methylation at the 0.2 mg/kg BDE-47 dose relative to control (difference in %5mC= 0.41; 95% CI 0.13; 0.68, p=0.01, FDR=0.09); however, Nr3c1 average methylation was not significantly changed at the same dose (difference in %5mC= 0.26; 95% CI −0.55; 1.07, p=0.50). None of the CpGs in Nr3c1 showed significant changes in DNA methylation at the 0.002 mg/kg BDE-47 dose (difference in %5mC= 0.35; 95% CI −0.39; 1.08 p=0.32 for the average methylation of the six CpG sites). Crhr1, Mc2r, and Snca did not show any changes in DNA methylation in response to BDE-47 treatment (Table 4).
TABLE 4.
Effect of BDE-47 treatment on DNA methylation in the brain frontal lobe of candidate nuclear genes
| BDE-47 Dose |
||||||||
|---|---|---|---|---|---|---|---|---|
| Control | 0.002 mg kg−1 | 0.2 mg kg−1 | ||||||
|
| ||||||||
| Gene | CpG position |
Mean Estimate (95% CI) |
Mean Estimate (95% CI) |
Relative to the Control group |
Mean Estimate (95% CI) |
Relative to the Control group |
||
| Difference (95% CI) |
p | Difference (95% CI) |
p | |||||
| Position 1 | 2.91 (1.78; 4.04) |
1.28 (0.15; 2.41) |
−1.62 (−3.22; −0.03) |
0.05 | 1.74 (0.72; 2.77) |
−1.16 (−2.69; 0.36) |
0.13 | |
| Bdnf | Position 2 | 3.63 (2.73; 4.53) |
2.29 (1.39; 3.19) |
−1.34 (−2.61; −0.06) |
0.04 | 1.50 (0.68; 2.31) |
−2.13 (−3.34; −0.91) |
0.001 |
| Position 3 | 10.28 (6.77; 13.80) |
9.73 (6.21; 13.24) |
−0.56 (−5.52; 4.41) |
0.82 | 7.92 (4.8; 11.04) |
−2.36 (−7.06; 2.34) |
0.31 | |
| Average | 5.76 (4.42; 7.10) |
4.49 (3.15; 5.83) |
−1.27 (−3.17; 0.63) |
0.18 | 4.13 (2.92; 5.33) |
−1.63 (−3.44; 0.17) |
0.07 | |
| Position 1 | 1.93 (79.62; 91.72) |
3.59 (1.35; 5.82) |
1.66 (−1.50; 4.82) |
0.29 | 1.50 (−0.61; 3.60) |
−0.43 (−3.50; 2.64) |
0.78 | |
| Position 2 | 1.91 (76.80; 95.52) |
1.58 (−0.12; 3.27) |
−0.33 (−2.73; 2.07) |
0.78 | 0.31 (−1.29; 1.91) |
−1.60 (−3.93; 0.73) |
0.17 | |
| Crhr1 | Position 3 | 1.65 (93.05; 98.92) |
1.11 (−1.12; 3.34) |
−0.54 (−3.70; 2.61) |
0.72 | 1.12 (−0.98; 3.23) |
−0.53 (−3.59; 2.53) |
0.72 |
| Position 4 | 2.68 (1.29; 3.13) |
1.92 (0.43; 3.41) |
−0.77 (−2.87; 1.34) |
0.46 | 2.57 (1.17; 3.98) |
−0.11 (−2.16; 1.94) |
0.91 | |
| Position 5 | 1.73 (0.49; 2.96) |
1.09 (−0.14; 2.33) |
−0.63 (−2.37; 1.11) |
0.46 | 1.84 (0.68; 3.00) |
0.11 (−1.58; 1.81) |
0.89 | |
| Average | 2.21 (1.20; 4.17) |
1.62 (0.69; 2.54) |
−0.59 (−1.89; 0.71) |
0.36 | 1.59 (0.72; 2.46) |
−0.62 (−1.88; 0.65) |
0.33 | |
| Position 1 | 95.99 (−0.58; 3.88) |
95.82 (92.89; 98.75) |
−0.17 (−4.31; 3.98) |
0.93 | 94.53 (91.77; 97.3) |
−1.45 (−5.48; 2.58) |
0.46 | |
| Mc2r | Position 2 | 86.16 (0.21; 3.61) |
91.72 (82.36; 101.07) |
5.56 (−7.67; 18.79) |
0.39 | 89.50 (80.68; 98.32) |
3.34 (−9.52; 16.20) |
0.60 |
| Average | 85.67 (−0.31; 4.16) |
90.56 (84.50; 96.61) |
4.88 (−3.67; 13.44) |
0.25 | 89.74 (84.03; 95.44) |
4.06 (−4.25; 12.38) |
0.32 | |
| Position 1 | 0.56 (0.35; 0.77) |
0.53 (0.33; 0.72) |
−0.03 (−0.32; 0.26) |
0.82 | 0.71 (0.47; 0.95) |
0.15 (−0.17; 0.47) |
0.34 | |
| Position 2 | 1.42 (1.19; 1.65) |
1.46 (1.25; 1.67) |
0.05 (−0.27; 0.36) |
0.75 | 1.64 (1.39; 1.90) |
0.23 (−0.12; 0.57) |
0.18 | |
| Position 3 | 1.51 (1.19; 1.83) |
1.51 (1.22; 1.81) |
0.00 (−0.43; 0.44) |
0.99 | 1.54 (1.18; 1.91) |
0.03 (−0.45; 0.52) |
0.89 | |
| Nr3cl | Position 4 | 0.18 (0.00; 0.37) |
0.17 (0.00; 0.34) |
−0.01 (−0.26; 0.24) |
0.93 | 0.59 (0.38; 0.80) |
0.41 (0.13; 0.68) |
0.01 |
| Position 5 | 1.90 (−1.64; 5.44) |
3.84 (0.61; 7.08) |
1.94 (−2.85; 6.74) |
0.39 | 0.92 (−3.04; 4.88) |
−0.98 (−6.29; 4.33) |
0.69 | |
| Position 6 | 1.22 (0.91; 1.53) |
1.21 (0.94; 1.49) |
0.00 (−0.42; 0.41) |
0.98 | 1.12 (0.78; 1.46) |
−0.10 (−0.55; 0.36) |
0.66 | |
| Average | 0.94 (0.39; 1.48) |
1.28 (0.79; 1.78) |
0.35 (−0.39; 1.08) |
0.32 | 1.20 (0.59; 1.80) |
0.26 (−0.55; 1.07) |
0.50 | |
| Position 1 | 3.73 (−0.19; 7.66) |
4.73 (0.54; 8.93) |
1.00 (−4.75; 6.74) |
0.72 | 1.84 (−2.38; 6.06) |
−1.89 (−7.66; 3.87) |
0.50 | |
| Snca | Position 2 | 6.69 (5.69; 7.69) |
5.71 (4.64; 6.79) |
−0.97 (−2.44; 0.50) |
0.18 | 5.73 (4.72; 6.74) |
−0.96 (−2.38; 0.47) |
0.17 |
| Average | 4.77 (2.95; 6.58) |
5.48 (3.54; 7.42) |
0.71 (−1.95; 3.37) |
0.58 | 4.31 (2.36; 6.25) |
−0.46 (−3.12; 2.20) |
0.72 | |
1.4 DISCUSSION
We investigated changes in DNA methylation in the frontal lobe of rats that were perinatally exposed to BDE-47. The prefrontal cortex of rats treated with BDE-47 showed decreased 5mC levels of the mitochondrially-encoded gene Mt-co2; decreased L1Rn 5mC levels, and alterations in DNA methylation in 5mC of nuclear encoded genes related to behavioral and brain functions (Bdnf and Nr3c1) at some of the CpGs. However, PBDE exposure did not affect the majority of DNA methylation patterns in the brain. These results indicate that perinatal exposure to BDE-47 at low doses comparable to those found in human populations (Suvorov and Takser 2011) does not have significant effects upon brain nuclear and mitochondrial DNA methylation patterns.
PBDE exposure has been shown to increase reactive oxygen species production (Blanco et al., 2014), to which mitochondria are particularly sensitive. Cytochrome c oxidase (COX) has three subunits encoded by mitochondrial DNA, and defects in COX structure can result in severe, and often fatal, metabolic disorders that predominantly affect tissues with high energy demands, such as the brain. We observed decreased methylation of mitochondrial COX-encoding genes with perinatal BDE-47 exposure. Although some regions showed significant and more robust decreases in methylation levels, the methylation changes were subtle.
DNA cytosine methylation is a key marker controlling gene transcription and has been increasingly studied in relation to environmental exposures such as air particles (Byun et al., 2013), metals (Doi et al., 2011), endocrine disruptors (Patel et al., 2013), and persistent organic pollutants (Rusiecki et al., 2009). DNA methylation, particularly 5mC, has been of primary interest for such studies. 5hmC has recently emerged as an alternative and functionally different form of methylation. Unlike 5mC, 5hmC has been associated with actively expressed genes (Chia et al., 2011). Relevant to this study, 5hmC is highly abundant in the brain, which may suggest a role in the epigenetic control of neuronal functions. In our study, perinatal exposures to low dose BDE-47 did not affect global 5hmC levels in the frontal lobe. However, further studies need to be done to understand the gene-specific changes in 5hmC levels.
Of the active repetitive elements in the brain, L1 elements are the most widely spread retrotransposons and constitute almost ~20% of the mammalian genome (Lander et al., 2001). L1 elements have retrotransposable ability in the central nervous system and are active during adult neurogenesis (Permiakov and Tskhovrebova 1988). A substantial number of newly inserted L1 elements in differentiated neurons are observed in the mouse brain in vivo (Muotri et al., 2009), and insertions can dramatically impact on neuronal differentiation affecting neural plasticity (Thomas and Muotri 2012). We observed that BDE-47 exposure did not decrease majority of L1 methylation in the frontal lobe of the brain. In a previous study, however, we showed decreased L1 RNA expression in the rat frontal lobe after BDE-47 exposure (Suvorov and Takser 2011). The source of this discrepancy should be further investigated in studies measuring both L1 methylation and expression in the same samples.
BDNF is a member of the nerve growth factor family. BDNF has important functions in the development of the nervous system and in brain plasticity-related processes such as memory, learning (Aid et al., 2007), and drug addiction, and it may play a role in the regulation of stress response (Roth et al., 2009) and in the biology of mood disorder (Aid et al., 2007). Altered brain Bdnf DNA methylation has been suggested as a cellular mechanism underlying the persistent cognitive deficits that are associated with the pathophysiology of post-traumatic stress disorder (Roth et al., 2011). Altered BDNF expression has been observed in perinatal hypothyroidism (Abedelhaffez and Hassan 2013). We observed decreased Bdnf promoter methylation at some CpGs following BDE-47 exposure, which may underlie the development of cognitive deficits and perinatal hypothyroidism. However, there was no effect of BDE-47 exposure on overall Bdnf promoter methylation.
Disruption of the Nr3c1 gene expression is associated with neurodegeneration in rats (Murphy et al., 2002). A significant correlation between maternal prenatal stress, newborn birth weight, and newborn methylation in the promoter region of the glucocorticoid receptor Nr3c1 has been observed in humans (Mulligan et al., 2012). Increased DNA methylation of the Nr3c1 promoter region is also associated with increased stress in rat pups raised by dams exhibiting poor maternal care behaviors (McGowan et al., 2009). Thus, epigenetic alterations of NR3C1 genes are associated with early neurobehavioral outcomes. In this study, perinatal exposure to BDE-47 was not linked to DNA methylation changes in Nr3c1 in rat frontal lobe.
One of the limitations of our study, as in most bisulfite conversion-based DNA methylation studies, is that we were not able to discriminate 5hmC from 5mC levels. When we report 5mC levels, it includes a trace amount of 5hmC that technically cannot be distinguished by conventional bisulfite conversion. Another limitation is that we did not investigate gene expression or mitochondrial function with BDE-47 exposure in this study.
In summary, we demonstrated altered mitochondrial and nuclear DNA methylation in the frontal brain of rats treated with low doses of BDE-47. However, significant findings were observed and did not show consistent dose-response relationships. Further studies are needed to substantiate the role of DNA methylation—including both 5mC and 5hmC—as potential mechanism for PBDE toxicity on neuronal activity and development, and to link gene-specific changes in methylation levels to effects upon gene transcription, as well as to mitochondrial function.
Supplementary Material
Acknowledgements
This work was supported by ANSES (Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travail, France), [grant number EST-08-46]. Dr. L. Takser is a member of the Centre de Recherche Clinique Étienne-Le Bel, which is supported by the FRQ-S. Dr. L. Takser is supported by a CIHR (the Canadian Institutes of Health Research) New Investigator Award. N. Benachour is supported by FRQ-S (Fonds de recherche du Québec - Santé) postdoctoral fellowship. Other support comes from grants from the United States National Institute of Environmental Health Sciences [grants number R21ES022694, ES021357 and ES021733].
Footnotes
Competing/Financial Interest Declaration: The authors have no financial or competing interests.
REFERENCES
- Abedelhaffez AS, Hassan A. Brain derived neurotrophic factor and oxidative stress index in pups with developmental hypothyroidism: neuroprotective effects of selenium. Acta. Physiol. Hung. 2013;100:197–210. doi: 10.1556/APhysiol.100.2013.2.7. [DOI] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Mulero M, Domingo JL, Sanchez DJ. Perinatal exposure to BDE-99 causes decreased protein levels of cyclin D1 via GSK3beta activation and increased ROS production in rat pup livers. Toxicol. Sci. 2014;137:491–498. doi: 10.1093/toxsci/kft257. [DOI] [PubMed] [Google Scholar]
- Boeke CE, Baccarelli A, Kleinman KP, Burris HH, Litonjua AA, Rifas-Shiman SL, Tarantini L, Gillman M. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: prospective results from a folate-replete population. Epigenetics. 2012;7:253–260. doi: 10.4161/epi.7.3.19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchi I, Alleva E, Costa LG. Effects of perinatal exposure to a polybrominated diphenyl ether (PBDE 99) on mouse neurobehavioural development. Neurotoxicology. 2002;23:375–384. doi: 10.1016/s0161-813x(02)00078-5. [DOI] [PubMed] [Google Scholar]
- Byun HM, Baccarelli AA. Environmental exposure and mitochondrial epigenetics: study design and analytical challenges. Hum. Genet. 2014;133:247–257. doi: 10.1007/s00439-013-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Panni T, Motta V, Hou L, Nordio F, Apostoli P, Bertazzi PA, Baccarelli AA. Effects of airborne pollutants on mitochondrial DNA methylation. Part. Fibre. Toxicol. 2013;10:18. doi: 10.1186/1743-8977-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N, Wang L, Lu X, Senut MC, Brenner C, Ruden DM. Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics. 2011;6:853–856. doi: 10.4161/epi.6.7.16461. [DOI] [PubMed] [Google Scholar]
- Cui C, Tian Y, Zhang L, Gao Y, Jin J, Wang P, Ding W, Wang X, Shi R, Wang Y. Polybrominated diphenyl ethers exposure in breast milk in Shanghai, China: levels, influencing factors and potential health risk for infants. Sci. Total Environ. 2012;433:331–335. doi: 10.1016/j.scitotenv.2012.06.075. [DOI] [PubMed] [Google Scholar]
- de Wit CA. An overview of brominated flame retardants in the environment. Chemosphere. 2002;46:583–624. doi: 10.1016/s0045-6535(01)00225-9. [DOI] [PubMed] [Google Scholar]
- Doi T, Puri P, McCann A, Bannigan J, Thompson J. Epigenetic effect of cadmium on global de novo DNA hypomethylation in the cadmium-induced ventral body wall defect (VBWD) in the chick model. Toxicol. Sci. 2011;120:475–480. doi: 10.1093/toxsci/kfr022. [DOI] [PubMed] [Google Scholar]
- Eriksson P, Viberg H, Jakobsson E, Orn U, Fredriksson A. A brominated flame retardant, 2,2',4,4',5-pentabromodiphenyl ether: uptake, retention, and induction of neurobehavioral alterations in mice during a critical phase of neonatal brain development. Toxicol. Sci. 2002;67:98–103. doi: 10.1093/toxsci/67.1.98. [DOI] [PubMed] [Google Scholar]
- Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol. Med. Rep. 2012;6:125–130. doi: 10.3892/mmr.2012.870. [DOI] [PubMed] [Google Scholar]
- Fischer D, Hooper K, Athanasiadou M, Athanassiadis I, Bergman A. Children show highest levels of polybrominated diphenyl ethers in a California family of four: a case study. Environ. Health. Perspect. 2006;114:1581–1584. doi: 10.1289/ehp.8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JM. Frontal lobe and cognitive development. J. Neurocytol. 2002;31:373–385. doi: 10.1023/a:1024190429920. [DOI] [PubMed] [Google Scholar]
- Gascon M, Fort M, Martinez D, Carsin AE, Forns J, Grimalt JO, Santa Marina L, Lertxundi N, Sunyer J, Vrijheid M. Polybrominated diphenyl ethers (PBDEs) in breast milk and neuropsychological development in infants. Environ. Health. Perspect. 2012;120:1760–1765. doi: 10.1289/ehp.1205266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JE. Neurodevelopmental effects of decabromodiphenyl ether (BDE-209) and implications for the reference dose. Regul. Toxicol. Pharmacol. 2009;54:91–104. doi: 10.1016/j.yrtph.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Griguer CE, Cantor AB, Fathallah-Shaykh HM, Gillespie GY, Gordon AS, Markert JM, Radovanovic I, Clement-Schatlo V, Shannon CN, Oliva CR. Prognostic relevance of cytochrome C oxidase in primary glioblastoma multiforme. PLoS One. 2013;8:e61035. doi: 10.1371/journal.pone.0061035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YL, Zhang LW, Ma N, Ye LL, Wang de X, Gao X. Cognitive improvement of mice induced by exercise prior to traumatic brain injury is associated with cytochrome c oxidase. Neurosci. Lett. 2014;570:86–91. doi: 10.1016/j.neulet.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Huang SC, Giordano G, Costa LG. Comparative cytotoxicity and intracellular accumulation of five polybrominated diphenyl ether congeners in mouse cerebellar granule neurons. Toxicol. Sci. 2010;114:124–132. doi: 10.1093/toxsci/kfp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat. Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lengyel A, Adam E, Nasz I, Erdei J, Fachet J. A sensitive method for detection of polyclonal and monoclonal antibodies against the adenovirus hexon. Acta. Virol. 1985;29:362–372. [PubMed] [Google Scholar]
- Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids. 20112011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MM, Woods R, Chi LH, Schmidt RJ, Pessah IN, Kostyniak PJ, LaSalle JM. Levels of select PCB and PBDE congeners in human postmortem brain reveal possible environmental involvement in 15q11-q13 duplication autism spectrum disorder. Environ. Mol. Mutagen. 2012;53:589–598. doi: 10.1002/em.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan CJ, D'Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7:853–857. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EK, Spencer RL, Sipe KJ, Herman JP. Decrements in nuclear glucocorticoid receptor (GR) protein levels and DNA binding in aged rat hippocampus. Endocrinology. 2002;143:1362–1370. doi: 10.1210/endo.143.4.8740. [DOI] [PubMed] [Google Scholar]
- Napoli E, Hung C, Wong S, Giulivi C. Toxicity of the flame-retardant BDE-49 on brain mitochondria and neuronal progenitor striatal cells enhanced by a PTEN-deficient background. Toxicol. Sci. 2013;132:196–210. doi: 10.1093/toxsci/kfs339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novgorodov SA, Riley CL, Yu J, Borg KT, Hannun YA, Proia RL, Kindy MS, Gudz TI. Essential roles of neutral ceramidase and sphingosine in mitochondrial dysfunction due to traumatic brain injury. J. Biol. Chem. 2014;289:13142–13154. doi: 10.1074/jbc.M113.530311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BB, Raad M, Sebag IA, Chalifour LE. Lifelong exposure to bisphenol a alters cardiac structure/function, protein expression, and DNA methylation in adult mice. Toxicol. Sci. 2013;133:174–185. doi: 10.1093/toxsci/kft026. [DOI] [PubMed] [Google Scholar]
- Pereira LC, de Souza AO, Dorta DJ. Polybrominated diphenyl ether congener (BDE-100) induces mitochondrial impairment. Basic Clin. Pharmacol. Toxicol. 2013;112:418–424. doi: 10.1111/bcpt.12046. [DOI] [PubMed] [Google Scholar]
- Permiakov EA, Tskhovrebova LA. [Effect of temperature and pH on the environment of tryptophan residues in alpha-actinin] Biofizika. 1988;33:754–757. [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol. Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J. Psychiatr. Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze E, Meijer L, Bakker A, Van Braeckel KN, Sauer PJ, Bos AF. Prenatal exposure to organohalogens, including brominated flame retardants, influences motor, cognitive, and behavioral performance at school age. Environ. Health. Perspect. 2009;117:1953–1958. doi: 10.1289/ehp.0901015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Patel R, Koutros S, Beane-Freeman L, Landgren O, Bonner MR, Coble J, Lubin J, Blair A, Hoppin JA, Alavanja MC. Cancer incidence among pesticide applicators exposed to permethrin in the Agricultural Health Study. Environ. Health. Perspect. 2009;117:581–586. doi: 10.1289/ehp.11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer. Biol. Ther. 2008;7:1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A, Battista MC, Takser L. Perinatal exposure to low-dose 2,2',4,4'-tetrabromodiphenyl ether affects growth in rat offspring: what is the role of IGF-1? Toxicology. 2009;260:126–131. doi: 10.1016/j.tox.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Suvorov A, Takser L. Delayed response in the rat frontal lobe transcriptome to perinatal exposure to the flame retardant BDE-47. J. Appl. Toxicol. 2011;31:477–483. doi: 10.1002/jat.1667. [DOI] [PubMed] [Google Scholar]
- Thomas CA, Muotri AR. LINE-1: creators of neuronal diversity. Front Biosci (Elite Ed) 2012;4:1663–1668. doi: 10.2741/e488. [DOI] [PubMed] [Google Scholar]
- Vizcaino E, Grimalt JO, Lopez-Espinosa MJ, Llop S, Rebagliato M, Ballester F. Polybromodiphenyl ethers in mothers and their newborns from a non-occupationally exposed population (Valencia, Spain) Environ. Int. 2011;37:152–157. doi: 10.1016/j.envint.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.