Abstract
Systems biology is a quantitative approach for understanding a biological system at its global level through systematic perturbation and integrated analysis of all its components. Simultaneous acquisition of information datasets pertaining to the system components (e.g., genome, proteome) is essential to implement this approach. There are limitations to such an approach in measuring gene expression levels and accounting for all proteins in the system. The success of genomic studies is critically dependent on PCR for its amplification, but PCR is very uneven in amplifying the samples, ineffective in scarce samples and unreliable in low copy number transcripts. On the other hand, lack of amplifying techniques for proteins critically limits their identification to only a small fraction of high concentration proteins. Atomic force microscopy (AFM), AFM cantilever sensors and AFM force spectroscopy in particular, could address these issues directly. In this article, we reviewed and assessed their potential role in systems biology.
Keywords: Atomic force microscopy (AFM), AFM force spectroscopy, Cantilever sensor array, Systems biology, Genomics, Proteomics
Introduction
The ultimate goal of systems biology is to build analytical models of biological systems, for a quantitative and comprehensive understanding of their functioning in normal and abnormal conditions. Systems biology takes a holistic (systematic, integrated and quantitative) approach to understand the causal relationship of a biological system’s dynamic behavior and organization at its global scale, in response to a stimulus.
Dynamic behavior of a biological system is determined by its individual components, their level of organization (physical topology), their interaction and differential flow of signals through their physical organization (logical topology). It is essential to have quantitative understanding of the causal relationship of a biological system’s (emergent) behavior so that analytical models could be built for predicting outcomes for a stimulus. Such an approach would provide better perspective of human physiology and pathology that would ultimately help in the delivery of better health care. A basic framework towards systems approach includes1:
Defining a system by breaking it down into its fundamental components- to make an initial analytical model
Systematic perturbation and simultaneous monitoring of the system, accounting for all the components of the system
Reconstructing the initial model based on the agreements between experimental observations (step 2) and model prediction (step 1)and
Verifying the reconstructed model (step 3) by carrying out new perturbations (step 2).
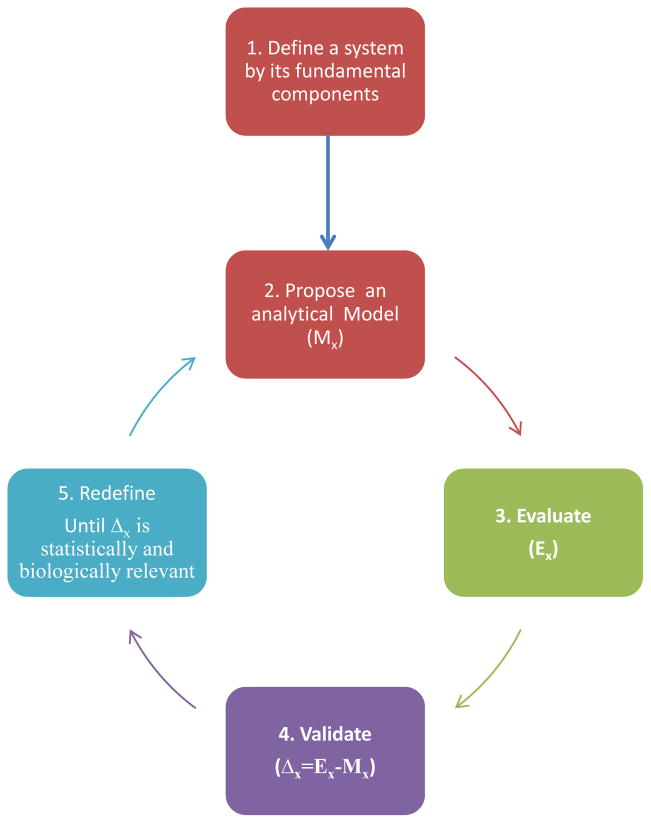
These steps are iterated until the goodness of fit between the predicted model and the experimental observations are statistically significant and biologically relevant (see Figure 1).
Fig. 1.
A simplified framework of systems approach. The first step involves defining a system that represents a structure or function of a biological entity, by breaking it down into its components to propose an analytical model (Mx). This is followed by experimental evaluation (Ex) of the model by simultaneous interrogation of the information pathways. Validate the analytical model in the light of experimental results (Δx=Ex-Mx) and redefine the model to maximize the goodness of fit between the experimental results and the model. These steps are repeated until the agreement between them is statistically significant and biologically relevant.
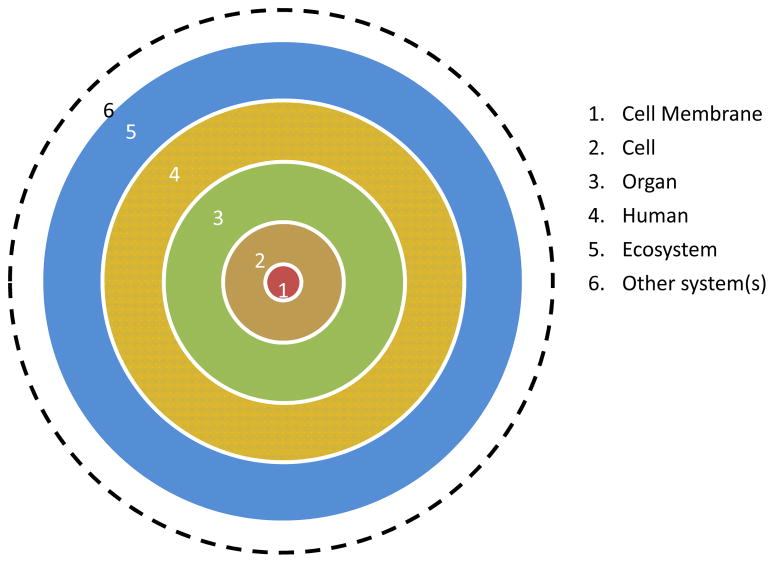
Structurally, biological systems are hierarchically arranged in complex layers of systems or subsystems. Thus, one system could be part of another system, which in turn could be part of another complex system and so forth. At meso-macro scale, the cell membrane is part of a cell as a whole, which in turn is a part of an organ system, which in turn part of a human being, who is in turn part of an ecosystem and so forth (see Figure 2). A simple biological system could be components that are necessary to represent a structure or a function of a biological entity. For example, a reconstituted lipid bilayer with a mixture of lipids in an aqueous environment could represent a model biological membrane system.
Fig. 2.
A simplified representation of the hierarchical nature of biological systems.
In a systems approach, system wide changes to a given stimulus are tracked using high-throughput techniques by monitoring all the components of the system simultaneously. The resulting massive data are registered, integrated, analyzed and/or modeled using computational tools to understand the system’s emergent behavior. This is very much in contrast to the conventional approaches where parts of the system are studied in isolation at the molecular level. The latter approach has the advantage of simplicity and manageability but lacks the understanding of the system as a whole. On the other hand, although the systems approach has the unique advantage of studying the system as a whole, it is highly complex in terms of experimental design, collecting, analyzing and managing the data. Such integrated approach of systems biology helps to understand the cause-effect relationship of a biological system in right perspective which is not possible in the traditional reductive approaches.
Why Atomic Force Microscopy for System Biology?
Advances in high throughput technologies and information sciences made possible to collect and analyze the massive and often parallel data sets to interrogate simultaneously the network of information highways of life (DNA→ mRNA→ Protein(s) →Structure & Function). Especially, high-throughput techniques like polymerase chain reaction (PCR), automated gene sequencing, microarrays (DNA/mRNA/protein-antibody), 2D gel electrophoresis, mass spectrometry (MS), supercomputing platforms, parallel processing algorithms to parse the massive data sets, etc., gave hope for systems approach. These developments are expected to evolve in time and continue our ability to understand biological systems in better perspective.
However, there are still significant bottlenecks in realizing these goals. The fundamental datasets essential for realizing systems approach are our genome and proteome maps. Human genome project (HGP) was successful in mapping our entire genome but the goals of human proteome project (HPP) are yet to be realized for several reasons (discussed later). Even though the atlas of our genome is available, researchers implementing the systems approach often encounter low abundant samples (typical of clinical samples, DNA extracted from fossils, etc.). In such scenarios, PCR amplification is ineffective and unreliable thus impeding genomic studies and systems approach. In addition, even in abundant samples, PCR fails or is ineffective in amplifying low copy number species like gene regulatory proteins. Under these circumstances, the scope and requirement for direct sequencing techniques become relevant and essential. Lack of amplifying techniques like PCR for proteomics resulted in proteomics to be at the mercy of the concentration sensitivity limits (CSL) of detection techniques. The current proteomics technologies allow detection of only 20% of protein species in the blood plasma 2, a large majority (about 80%) of the proteome is still at large. In this context, atomic force microscopy (AFM) in all its different modalities of imaging (contact, non-contact, tapping, lateral force, force volume, magnetic force, chemical force, surface potential, electrochemical, conductive-AFM, recognition force, etc.), as well as force spectroscopy, cantilever based sensing, and other Scanning Probe Microscopy (SPM) techniques, e.g. scanning near-field optical microscopy (SNOM), with their exquisite sensitivity to detect or discriminate single molecules, could play an indispensable role in achieving direct sequencing of genome on scarce materials as well as herald a new way to identify a large plethora of proteins below CSL, down to orders of magnitude comparable to the inverse of the Avogadro number. Above all, AFM per se as an imaging or as a spectroscopy tool, and AFM based techniques like cantilever sensors or multimodal AFM platform (see below) could be used to study simple model systems as well as verifying some of the refined hypothesis (step 4, of the systems framework) through single molecule studies whenever necessary.
Atomic Force Microscope
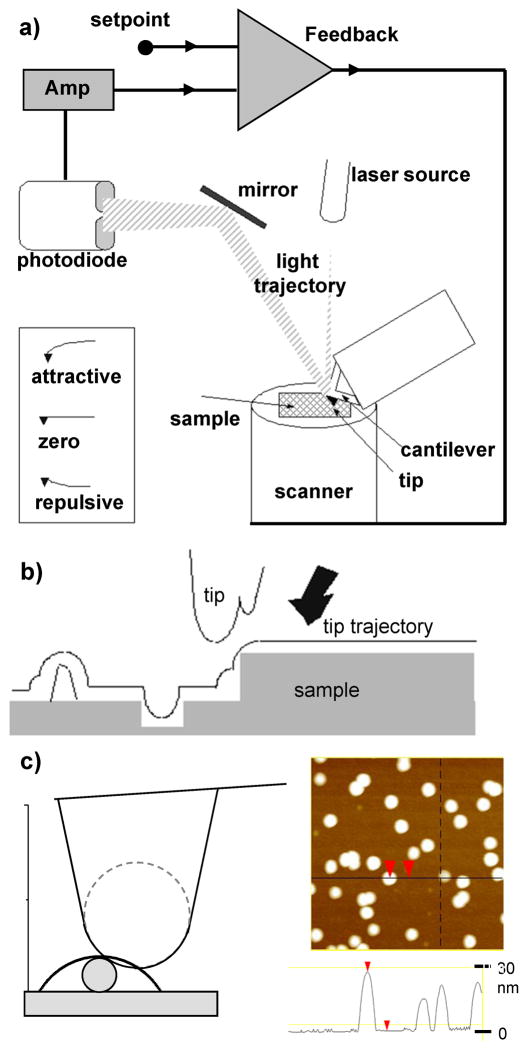
The atomic force microscope (AFM) (Fig. 3a) is a key member of a series of scanning probe microscopes (SPMs) that make use of a local measurable interaction between a very small probe and the sample as signal for image formation(Fig. 3b). In principle, any interaction that is spatially resolved in (x, y) and is a function of distance (z) from the sample can be used as SPM signal. Examples of interactions used as signals are: electric currents, forces, magnetic field, etc. As the probe scans over an area of the sample, data of the interaction strength is collected at a rectangular array of points, in which each data point is associated with the strength of the interaction between tip and sample at that particular point. A topographic relief of a surface is obtained (Fig. 3c) by introducing a feedback system that keeps the signal (interaction strength between tip and sample) constant by forcing the tip (or sample) to displace vertically and compensate the change in signal if the interaction strength changes (Fig. 3a). These displacements provide the heights in a three dimensional relief of the sample area scanned (Fig. 3c). Significantly, the resolution of these microscopes is not limited by the diffraction limit as in light or electron microscopes, since the interactions used are independent of wavelength.
Fig. 3.
a) Schematic diagram of an AFM with optical detection system of cantilever deflection. Only 2 quadrants of the photodetector are shown. A feedback system compares the amplified output of the photodetector with a setpoint value chosen by the user. When the difference between both values is different to zero, a voltage is sent to the piezoelectric scanner, which moves the sample to bring the difference value back to zero. The inset shows cantilever deflections as a result of attractive and repulsive forces. b) The probe plays a fundamental role in the image formation mechanism of scanning probe microscopies. The resulting image is a convolution of the topography of the sample and the geometrical features of the probe being scanned. If the probe has multiple protrusions close to one another, the resulting image will be distorted, displaying multiple features of the same object. The measured width of an object is also broadened due to the size of probe radius c). However, the measured height is independent of it. The figure shows images of gold nanoparticles with 30 nm diameter. The measured height is close to 30 nm, but the width is larger.
Shortly after the inception of the scanning tunnelling microscope (STM) by G. Binnig and H. Rohrer in 1982,3, 4 the first SPM, a potential for biological applications was noticed.5 An important motivation was the possibility to operate in liquids, close to the native environments of biological samples. A major limitation of the STM for biological specimens was the requirement to use conductive samples since the tunnelling current is used as STM signal. The development of the atomic force microscope (AFM) by Binnig, Quate and Gerber in 1986 was able to overcome this limitation by sensitively measuring the interaction forces between the sample and a sharp tip integrated in a flexible cantilever and using them as signal for image acquisition.6 Precise positioning of the probe with respect to the sample was, and still is, achieved by making use of piezoelectric materials due to their property of displacing objects with very high accuracy.
True atomic resolution is possible with the AFM,7, 8 although it has typically been achieved for hard samples in ultra-high vacuum conditions.9 Resolution also depends on the material properties of the sample under investigation. Biological samples are soft and thus easily deformable under the forces exerted by the tip.10 This creates a large contact area between the tip and the specimen under investigation and significantly limits the resolution that can achieved.11 Nevertheless, the resolution can still be remarkable with soft specimens. For example, AFM studies on reconstituted ion channels can resolve substructure of individual channels and, at times, the subunits within each channel.12–16 Spatial resolution also depends on the sensitivity of the piezoelectric scanner, which may be as high as 0.1 nm laterally and 0.01 nm vertically. The resolution achievable for a given sample also depends on the geometrical shape of the tip. Higher theoretical resolutions are expected with sharper tips because force interactions are confined to a smaller area. However, one should also bear in mind that the pressure on a sample increases for sharper tips due to the smaller contact area, and this may result in sample damage, especially for delicate samples. Although the same principles apply to the resolution limits of biological samples in wet or dry conditions, other factors, e.g. sample degradation, limit imaging of biological samples in air. Also, additional capillary forces exist in air due to the presence of a thin water layer on the sample surface originating from condensed humid air, which increases the contact area, thus further limiting resolution in air. For faithful imaging of biological samples, imaging in liquids is preferred.
Force detection is critical in AFM design since very small forces are measured. Even though other force detection schemes were used in early designs,6, 17 and some of these detection schemes are still investigated for particular applications,18 the optical deflection scheme has been adopted in most AFMs nowadays.19 In this setup, the light beam emitted by a laser is reflected from the cantilever and collected by a photodiode (Fig. 3a). The deflection of the cantilever is proportional to the interaction force between the sample and the AFM probe (Hooke’s law). The proportionality constant is the spring constant of the cantilever. Vertical (or lateral) deflections are detected by measuring the difference of light intensities that reach the upper and lower (or left and right) sectors of a four-quadrant photodiode. The vertical deflection is used as feedback control signal, from which the heights are measured. The lateral deflection of the AFM cantilever is used to measure friction forces and these data can be used to conduct tribological studies at the molecular scale20, 21 or provide information about the local surface chemistry.22
In addition to the high resolution that can be achieved, one of the most important advantages of the AFM compared to other high resolution microscopes, such as electron microscopes, is the possibility to operate in liquids,19 close to the native environments of biological samples. Important biomolecules, such as transmembrane proteins reconstituted in lipid bilayers or DNA molecules,23–26 can be investigated with nm resolution in quasi native environments. Another important feature is the possibility to use it as a force measuring device,17, 18 thus go beyond morphology and investigate the mechanical (such as elastic and adhesive),10, 27–29 electrical and magnetic properties of a sample, in addition to its structure. Since deflections of ~ 1 nm can be detected with the optical deflection method, forces in the range of 10−2–105 nN can be measured with the AFM by utilizing a cantilever with the appropriate spring constant. Further, AFM is able to provide kinetic information of dynamic processes with time scales slower than its image acquisition rate.30, 31 This is typically several minutes to tens of minutes in conventional AFMs for high resolution images of biomolecules or cells. However, new developments in high-speed AFM allow acquisition rates of a few seconds to tens of milliseconds per image.32–35
Force measurements are carried out by means of force-displacement curves during approach and retraction between the cantilever and the sample.17, 18 During data acquisition, the vertical deflection of the AFM cantilever is recorded as a function of the position of the piezo moving it. When the cantilever tip is far away from the sample and the interaction forces between them are negligible, the cantilever deflection is zero. As the tip comes closer to the surface and makes contact with it, repulsive forces produce an upwards deflection of the cantilever. These contact forces can produce reversible (elastic) or irreversible (plastic) deformations on the sample, which are detectable in the curves and can be used to determine mechanical properties of the sample, such as its elastic modulus or critical shear stress. Even in the case of elastic material deformations, hysteresis often occurs in the curves during retraction due to the presence of adhesion forces between tip and sample. These adhesion forces, observed as downward deflections of the cantilever, have different origins. In air, a thin water layer present on samples due to condensation of air humidity produces capillary forces with values of a few nN. In liquid, capillary forces are mostly absent, thus adhesion forces between the tip and solid samples are often negligible. Significantly, this allows the examination of mechanical properties of single biopolymer molecules via force measurements as a function of their elongation.36 These measurements opened the possibility of specifically recognizing, “fingerprinting”,37 single molecules by means of their force elongation curves. Furthermore, by attaching a ligand to the tip, specific interaction forces between these and receptors adsorbed on a surface can be determined.38
Several modes of imaging have been developed; however three are the most commonly employed imaging modes for biological applications: 1) contact mode, 2) tapping or intermittent contact mode and 3) force mapping. In contact mode, which was developed first,6 an electronic feedback circuit maintains a constant deflection, ensuring a constant force of interaction between cantilever tip and sample. The amount of z variation required to maintain the interaction force constant is plotted versus the x and y coordinate, thereby producing a topographic image. Images may not be strictly topographical at edges between two materials with different mechanical properties due to different deformations or adhesion.
In tapping or intermittent contact mode, 39, 40 the cantilever oscillates vertically near its resonance frequency by a piezoelectric element on the tip holder. The amplitude of oscillation is kept constant during scanning by a feedback mechanism. The image is produced by plotting the displacement of the sample relative to the base of the cantilever necessary to keep the amplitude of the vibration constant. The phase lag between the driving circuit and the actual tip vibration is also measured and used for imaging viscoelastic properties. An advantage of tapping mode is that it reduces the lateral forces present in contact mode and this translates into more consistent and reproducible images for relatively poorly adsorbed biomolecules, such as DNA 41–43.
The force displacement curves described above can be utilized for mapping additional properties of the sample in addition to its topography. Experimentally, this is performed by acquiring N×N curves in an array fashion inside the area of interest and fitting each one of the curves to a theoretical model, from which important local magnitudes of the sample can be extracted. This type of measurement provides information about inhomogeneities of the property of interest, which can then be correlated to the structural information obtained via contact or tapping modes.
As discussed in early review papers,44 sample preparation is paramount in obtaining high quality images of biological samples. It is essential to have high purity samples to define the structure and function of the molecules. In addition, it’s desirable to possess as much a priori information as possible about the molecule’s dimensions as well as its morphological and mechanical properties. In the case of force spectroscopy, it’s necessary to pay special attention to limitations related to low sample concentrations (~0.01–1 μM),37 speed of data acquisition and data analysis. Use of low sample concentrations is necessary to prevent aggregation in single molecule experiments and it poses a challenge for subsequent data analysis. At first, it is therefore important to analyze samples with high purity, possibly complemented by analytical and computational modeling of the results, to create a “library” of force-extension curves, which can be subsequently used to interpret data from unknown samples.37, 45–47 According to reports for polysaccharides, only 10% or less of the data produces results which can be interpreted unambiguously and in the remaining 90% either no molecules or too many molecules are lifted by the AFM tip, thus producing un-interpretable results. 37 Data acquisition is also slow, requiring typically ~ 10–30 minutes to collect 1000 curves. For further thorough information about force spectroscopy, see for instance the review paper by Gianotti and Vancso.36
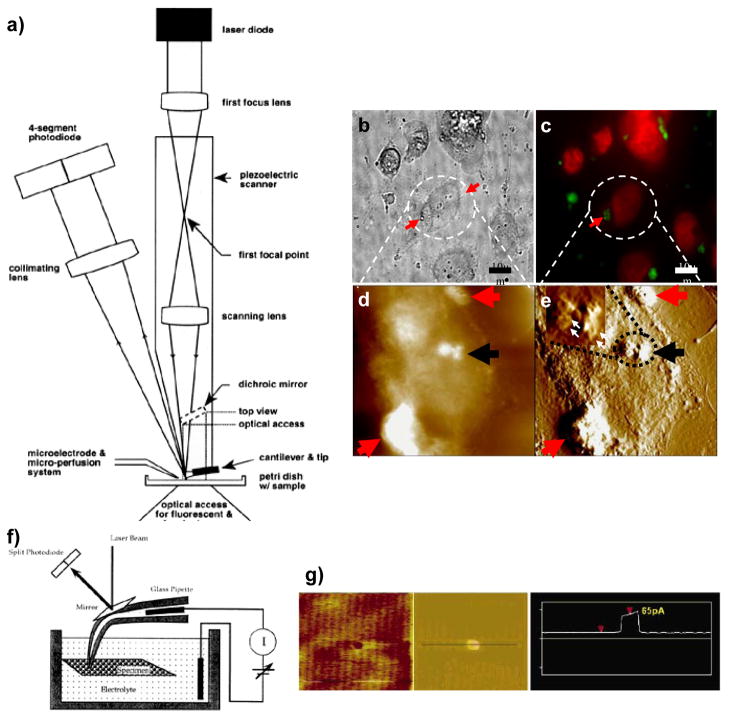
In addition, the open architecture of AFM allows integration with other complementary techniques, including conventional light and fluorescence microscopy, total internal reflection fluorescence (TIRF), Förster resonance energy transfer (FRET) microscopies, electrophysiology, opto-electronic manipulators, nano micro fluidics, etc., to obtain thorough structure–function information of biomolecules and their assemblies at molecular resolution in their native physiological environment. This flexibility makes it possible to convert the AFM into a powerful multidimensional AFM platform (Fig. 4)48.
Fig. 4.
a) Optical components of an AFM combined with a light fluorescence microscope. The optical focusing of the laser light is integrated within the piezotube which provides the lateral movement of the cantilever. The scanning lens is located inside the lower segment, which provides the vertical movement of the cantilever. After the positions of the lenses are adjusted, the scanning focused spot accurately tracks the cantilever, and the zero deflection signal from the four-segment photodiode is independent of position within the scan area. The AFM is placed on the stage of an inverted microscope (adapted from Lal and Proksch106. For details see Hansma et al. 107). Bottom right Internalization of small liposomes: light microscopy (b), fluorescence (c), and tapping mode AFM (d–e) imaging of fixed cells at different time intervals after incubating with cisplatin-encapsulated liposomes. Height mode image (d) and error mode image (e) of the same cells. Error mode image shows better contrast and ultrastructural details. The liposomal position with respect to the cell membrane was determined from the height mode images. For cells incubated with cisplatin-encapsulated liposomes for only 1-h incubation sample, many large liposomes are seen on the cell surface (red arrows in b–e). On the other hand, small cisplatin-encapsulated liposomes appear to be internalized in the cell cytoplasm (black arrows in d, e). The inset in e) shows a zoomed-in portion of the encircled area: the internalized liposomes are clusters of small liposomes (white arrows) of ~250 nm diameter. After 16 h of incubation, no defined liposomes were observed Scan size of AFM images is 25 μm, adapted from Ramachandran et al. (17). f) Schematics of a combined Scanning Ion Conductance Microscope (SICM) and AFM setup able to obtain simultaneous current and topographical information. A laser beam reflecting off a mirror glued to the back of a pipette with a nanometer provides the deflection signal for the topographic image (adapted from Lal and Proksch 106). Intrapipette and bath electrodes measure electrical currents. g) Simultaneous AFM and conductivity images of a nanopore fabricated in a silicon chip (adapted from Quist et al. 2007108).
AFM in Genomics
The journey of AFM applications in genomics started with a humble beginning of imaging DNA in ambient conditions. But the initial images were marred from poor resolution due to movement of DNA samples from (a) poor adsorption to the substrate and (b) high imaging forces exerted on the sample by the AFM probe. Developments in the sample preparation methods 23 improved the resolution of DNA in air, but adhesive forces in ambient air were still a limitation. Further improved images were obtained when imaging under liquid which minimized the adhesive forces 49, 50. Still poor adhesion of DNA to the mica substrate prevented obtaining higher resolution images in liquid, since both mica and DNA are negatively charged. This was overcome by modification of the substrate with divalent cations (Mg++, Ni++ etc.,) 26, 51, 52, by a thin layer of organic molecules, such as aminopropyltriethoxysilane (APTES), deposited on mica 53, 54, etc. Further, development of newer imaging techniques allowed routine imaging of DNA in physiological buffer solutions to obtain 2–3 nm resolutions in a reproducible manner 55.
Some of the important milestones in AFM imaging of DNA towards genomic applications are: Protein-DNA interactions investigated by Bustamante in 1993;56 wherein he showed changes in DNA conformation following protein interaction. Until then it was believed that DNA undergoes structural changes following protein interaction, this was confirmed by Pietrasanta et al., 57 by employing conformation specific DNA-antibodies to determine the DNA conformation changes with AFM. Wyman et al., 58 showed the specific interaction between heat-shock transcription factor 2 and DNA, its stoichiometry with AFM. Real time digestion of DNA by enzymes 43 and DNA repair by photolyase 59. These studies enabled to understand the molecular mechanisms of transcription which are fundamental in understanding how a gene responds to stimuli.
After initial encouraging results in sequencing DNA with STM 60, the idea of using AFM for DNA sequencing was pushed forward by several groups 61, 62. For the sake of simplicity we could classify the sequencing techniques into amplified and non-amplified techniques. Currently, amplification based techniques represent the mainstream sequencing methodology. It works best for abundant samples, high enough to moderate copy number transcripts. However, in scarce and low copy number transcripts serious drawbacks remain. There is a poor correlation between pre and post amplification in the above cases 63, 64. It was also found out that random effects prevent even amplification of low copy number transcripts in PCR. Finally, the rising interest in personalized medicine which warrants individual genome sequencing with scarce clinical specimens within reasonable costs and time necessitates development of direct sequencing techniques.
In this context, non-amplified single molecule sequencing techniques like nanopores, fluorescent detection of single molecules 65, 66 using optical traps, using STM 60 are being considered. Each one of these methods has their merits and demerits (for review see 67). For example, nanopores have low signal to noise ratio issues, optical traps are far from being practical and STM methods are inconsistent 68. Earlier Allison et al.,69, 70 applied AFM to image the binding sites of mutated EcoRI endonuclease to plasmid DNA molecules. They achieved 50–100 nucleotides resolution that made AFM a promising technique for direct genomic applications. Later in 1997 70 they mapped 35kb cosmid DNA using mutant EcoRI and demonstrated the accuracy of mapping to better than 1%. Similarly, in 1999 Nakamura et al., 71 used the rutile TiO2(110) single crystal surface, as an optimal substrate for DNA mapping using Pvu II and Hinc II restriction enzymes.
Reed et al., 72 have used AFM for profiling single transcripts using ordered restriction mapping. They utilized type II restriction enzymes to elegantly profile DNA molecules of about 2kb or longer (roughly about 40% of all cDNA 73, 74. It is expected that improvements in size resolution and digestion efficiency would improve profiling up to 1kb fragments (80% of all transcripts). Improvements in probe design and advanced metrology methods are expected to improve continuously the sizing accuracy to account for the remaining DNA molecules less than 1 kb. These studies exemplify the scope and application of AFM as a highly promising direct sequencing technique where amplified sequencing techniques like PCR have no scope.
Recently, Bailo et al. 75 have applied AFM tip enhanced Raman spectroscopy (TERS) for sequencing single strand RNA at high lateral resolution with excellent contrast and sensitivity. They successfully sequenced a single stranded RNA cytosine homopolymer and demonstrated that single base resolution is not required as long as the tip is moved at single base-to-base distances and collecting the Raman spectra. The variations in spectra could then be ascribed to the variations in the sequence. This technique holds potential for direct sequencing of biopolymers (RNA/DNA/protein) at single base/amino acid resolution without any labeling procedures.
Another very important area of application in genomics is AFM force spectroscopy for the detection of single nucleotide polymorphisms (SNP). SNPs account for about 90% of human genetic variations which determine an individual’s unique response to various physical/chemical/biological insults. In other words, they determine someone’s likelihood of developing a disease following exposure to a pathogen or their response to a drug treatment. Therefore mapping SNP database of the human genome has enormous implications in health care delivery (diagnosis, drug treatment and biomedical research) towards realizing the goals of personalized medicine.
There are several scanning techniques available for detecting DNA polymorphisms, but the need for direct measurements independent of PCR technologies is essential, as PCR based techniques result in uneven amplification leading to underscoring of SNPs. Direct measurements based on DNA’s physical characteristics like mass is a preferred choice 76. Intermolecular forces resulting from adsorption of few biomolecules on the surface of AFM cantilever induce mechanical stress resulting in its deflection which is detected by an efficient optical lever system. They are capable of sensing ultra low intermolecular forces in the regime of piconewtons and they already have been used to understand the energy landscape of receptor-ligand or interaction between complementary molecules at single molecular level (see Reviews by 77, 78).
Using similar principles, AFM force spectroscopy was used to detect SNP’s in high throughput format 79, 80. Fritz et al successfully demonstrated single base pair mismatches by designing a simple dual cantilever system functionalized with oligonucleotides and detected their complements by analyzing the differential signals from the cantilevers. The advantages of cantilever based detection are that they are direct, no labeling required and they can be implemented in high throughput schemes (massive parallel sequencing) under both at static and dynamic conditions. Force resolution of cantilever detection is limited by its thermal vibrations (noise); by a careful design of experimental conditions like ultralow temperatures and noise free environs, one could improve the sensitivity of detection. Stowe et al.,81 achieved a force resolution of ~6 attonewton using ultrathin cantilevers at 4.8 K in vacuum. This level of sensitivity would certainly lower the detection limits to just a few molecules in a sample.
AFM in Proteomics
After the successful completion of HGP, in the post genomic era proteomics has become the new genomics 82, but an organism’s proteome is much more complex than its genome, even though it is the product of the genome (DNA→RNA→Protein(s)). To apprise this better, let us consider these facts: there are about 30,000 genes and about 2 million proteins in the human body. Therefore there is more than one protein per gene and this is due to several reasons including: (a) differential splicing of the gene (b) post translational modifications, (c) interaction with the message itself (Protein-DNA/Protein-RNA) and altering its states, (d) ability to interact with itself or with others and evolve into new functional units, all these events lead to production of different functional entities (protein products) from a single gene.
It is evident that mapping human proteome (HP) is a natural extension of the HGP, but mapping HP is more complex and challenging due to the aforementioned reasons. HGP was possible due to (a) structural simplicity and static nature of our genome. In contrast, proteome is highly variable both spatially (varies from one cell type to the other, varies from one organ to the other) and temporally (depending on the state of the organism). (b) Genetic material is easily soluble in physiological buffers and relatively stable thus amenable for biochemical processing and automated sequencing. On the other hand, proteins are highly variable in solubility and stability. (c) Most important of all is the availability of amplification techniques like PCR made HGP possible and lack of such amplification methods for proteins is the single most important hurdle in mapping HP. As a direct consequence of this, mapping HP is limited by the concentration sensitivity limits (CSL) of the current technologies (see 2 for an excellent treatment on this subject). For example, the widely used mainstream protein sequencing tool, including all variants of mass spectrometry (MS), have CSL of >10−8 M. Figure 5 2 shows the concentration dependence of the number of human plasma proteins. MS could account only few hundreds of protein species in the plasma in this CSL range. In addition, these techniques had to deal with detecting majority (>100,000) of the low concentration (<10−12) proteins against a background of small number (<100) of high concentration proteins (>10−6). Microcantilever sensors have a CSL of <10−15 M which covers about 10,000 proteins. In summary, all the existing proteomic techniques could account for only about 20% proteins in the plasma.
Fig. 5.
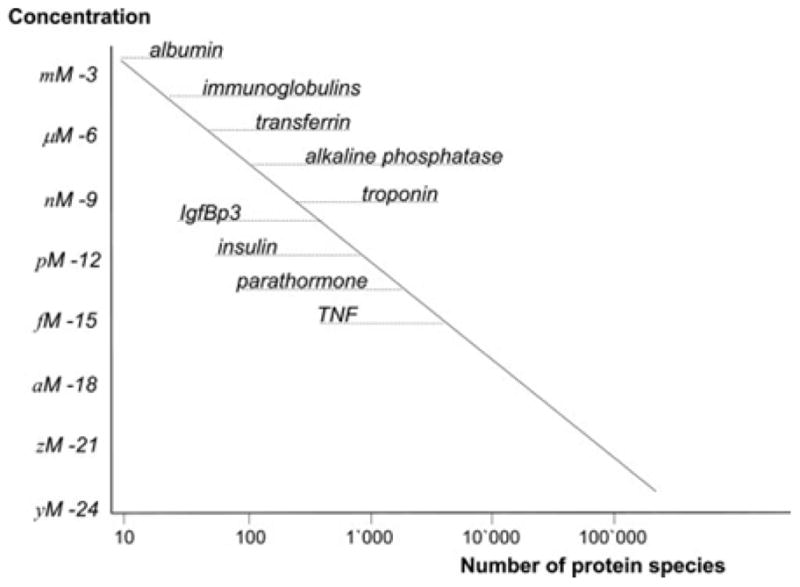
Distribution of human plasma proteins and their concentration. The dynamic range of proteins in the plasma ranges from millimolar (10−3) to yactomolar (10−24) concentrations. It also shows the concentration dependence of the number of protein species. Current mainstream proteomic methods account for few hundreds of proteins only leaving the large majority (~80%) unaccounted [adapted from 2].
Genomics provides the structural basis of disease/state of an organism but it is proteomics that determines the functional basis or defines the ultimate state. A defective gene may not necessarily result in a defective phenotype, but a defective protein without any associated genetic lesion could produce a defective phenotype. Therefore proteomics is of more clinical significance over genomics, in identifying a biomarker of a disease, drug target and more importantly in understanding the mechanistic details of disease. Much of the future of proteomics depends on breaking the CSL of detecting and identifying 1 molecule per Liter of solution (10−24) for mapping the human proteome. Such CSL free proteomic technique will empower realizing the goals of human proteome project.
One such promising technique is AFM based molecular sensors (single molecule force spectroscopy) that are capable of detecting single molecule force interactions down to 10−12 N 51, 52, 77. Advances in the development of ultrasensitive cantilevers 81, 83, 84 promise a force resolution (10−18N) enough to detect an average single protein molecule (~50,000 Da) in a not so remote future. Although screening of unknown molecules with AFM is still in its infancy and for reliable identification of biomolecules additional techniques (e.g. immunochemical staining) should be used for comparison, a few studies have been able to identify the presence of biomolecules in live and fossil samples. Dugdale et al., 85, 86 applied AFM as a mechanical fingerprinting tool in identifying modular proteins that help diatoms to secure their attachment to a substrate at single molecular level. In a similar approach87, 88, AFM was used to identify collagen molecule in fossilized samples. Also employing single molecule force spectroscopy, the contour lengths of short lipopolysaccharides (LPS) molecules present on Haemophilus influenzae could be correlated with the lengths of LPS observed in TEM images.89 This opens up very exciting possibilities of fingerprinting single protein molecule in a sample thus breaking the CSL down to 10−24 M.
Apart from fishing out single molecules from complex solutions, AFM has been applied as an imaging tool to study the size, shape, volume, molecular interactions, energy landscape of protein folding-unfolding, structure-function correlations of proteins at single molecular level at physiological environments (for reviews see 44, 77, 78, 90, 91). All these would help us to understand the structure and function of individual proteins of the proteome. This will enrich the atlas of human proteome which in turn will help us to fingerprint unknown proteins from a complex sample.
Other potential applications of AFM in systems biology is in the validation and cross-correlation of hypothesis that associates a protein with a structure and function that is measurable by AFM. The following are representative studies to illustrate their application.
Single molecule architecture
The structure of connexin hemichannels were unknown until AFM studies12, 13, 16, 92 delineated their molecular structure for the first time. It revealed their molecular organization, size, shape and response to external stimuli such as extracellular calcium levels (Fig. 6A-B). These studies heralded a new paradigm for studying native membrane proteins, 93 (for review see 94). It helped to understand gap junctional coupling and their role in electrical conduction in the heart. Similarly, the ion channel hypothesis of amyloid disorders were originally derived from electrophysiological studies but there were no structural correlates available to support this hypothesis. High resolution AFM in model lipid bilayers provided the structural evidence for variety of amyloid peptides including amyloid-β(1–42)(Fig. 6C), α-synuclein, ABri, ADan, serum amyloid A, amylin, K3, truncated amyloid peptides (p3, Fig. 6D) for the first time 15, 95, 96. This provided a common structural link to protein mis-folding diseases and identified a potential target in amyloid disorders.
Fig. 6.
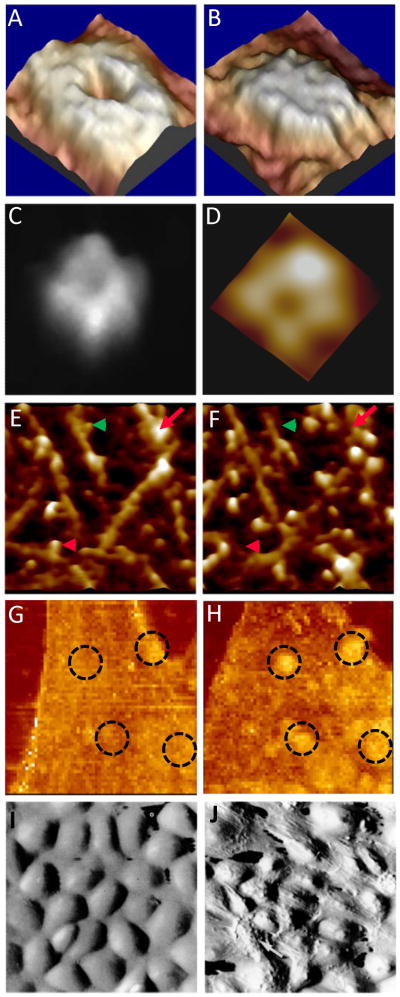
High resolution AFM images of pores formed by amyloid beta 1–42 (A)48 and amyloid beta 17–42 (B)95 oligomers in lipid bilayers. Open (C) and close (D) conformations of connexin-43 hemichannels in the absence and presence of 1.8 mM Ca++ respectively.16 Real time aggregation of VEGF receptors on the surface of a endothelial cell before (E) and after (F) the addition of VEGF-antibody.29 Real time proteolysis of collagen molecules before (G) and after (H) the addition of collagenase.98 Shear stress induced reorganization of cytoskeleton in living endothelial cells imaged by AFM.105 Under static conditions (I), the cells topography is smooth and under unidirectional shear stress the topography is reorganized (J) to minimize the shear stress gradient to reduce resistance to flow.
Single molecule interactions
Lin et al. 97, 98, have demonstrated molecular interactions in real time by imaging proteolysis of collagen I by high resolution AFM (Fig. 6E-F). It showed the association of collagenase molecules to single triple-helical collagen, its site of action and its subsequent proteolysis in real time. The proteolysis was inhibited by reduced calcium and acidification validating collagenase action. It was the first report of real-time proteolysis and its inhibition at single molecular level. Such real-time studies of single molecule interaction would serve as a valuable tool in drug research and development.
Protein interactions with other biomolecules (protein-protein/protein-DNA etc.,) are studied by either direct imaging or by applying force spectroscopy. In one such study, force volume imaging was used to understand the distribution and clustering of surface receptors following its specific ligand interaction and the resultant change in the membrane biophysical properties 99. The effect of vascular endothelial growth factor (VEGF) receptor clustering on local cell mechanics was obtained from maps of interaction forces between VEGF antibody conjugated tips and its receptor on cell membrane. The technique allowed simultaneous measurement of the real-time lateral diffusion of receptors and their effect on local rheological properties like elasticity. VEGF receptors are found to concentrate toward the cell boundaries and cluster rapidly upon addition of its ligand (Fig. 6G-H). The local stiffness reductions are proportional to receptor density and, being concentrated near the cell edges, this study provided the insights into the mechanism of cell growth and angiogenesis occurring at the edges.
Protein-DNA interaction was studied between Photolyase 8, a DNA repair protein and DNA. DNA-photolyase interactions and the annealing of restriction fragment ends and movement of photolyase over the DNA were directly visualized. For the first time, AFM revealed that a sliding mechanism by which photolyase can scan DNA for damaged sites.
Structure-Function relationships
Another classical study that correlated a macromolecule’s structure to its function was polysaccharide elasticity by the chair-boat transitions of glucopyranose ring structure47. It was found that the elasticity of the three polysaccharides results from a force-induced elongation of the ring structure and a final transition from a chair-like to a boat-like conformation. Till then, it was assumed that pyranose ring is inelastic and locked structure. In another example, Liu et al., 45 used AFM force spectroscopy to unravel the connexin channels gating mechanism. They attached an antibody specific to the C-terminus of a connexin and approached hemichannels immobilized on a substrate. Specific binding and unbinding events were observed upon approaching and retracting the tip. The force spectroscopy revealed the mechanism of hemichannels gating supporting the particle and receptor model. Until then there was no evidence to support this hypothesis for connexin hemichannels.
Correlative studies
A classic representative study that linked the effect of altered proteome on the biomechanical properties of a cell was carried out by Vergara et al. 100, using AFM and proteomics to analyze and correlate the modifications induced in the mechanical properties of astrocytes to the changes induced in protein expression after interferon- β treatment (IFN-β). The results indicated that IFN-β treatment reduced the Young’s modulus, a measure of cell elasticity. The molecular mechanisms that underlie these changes were found to be involved in cytoskeleton organization through cofilin-1 and profilin-1.
Multidimensional AFM
The open architecture of AFM allows easy integration of other complementary modalities like light microscopy, electrical measurement system, microfluidics, optical/magnetic tweezers, etc. This flexibility transforms it into a powerful multidimensional platform (m-AFM) for studying molecules structure-function relationships(for reviews see 101, 102).
Microcantilever arrays are utilized to identify specific biomarkers using defined ligands/antibodies, resolving intermolecular affinity, as high throughput sensors. Also, AFM tips were employed as fluid force sensors to study microrheology of body fluids. In one such study, Barbee et al. 103–105, used AFM to image topography of vascular endothelial cells under different flow conditions. For the first time, flow induced alterations in surface topography in real time were visualized and correlated with cytoskeletal rearrangements (Fig. 6I-J). Such studies provided powerful insights into understanding the mechanism of atherosclerosis.
Conclusions
For successful implementation of systems approach, genome and proteome are the two fundamental datasets essential. Even though entire human genome atlas is available, large majority of human proteome is still unaccounted due to inherent complexities in their biosynthesis and regulation as well as due to technical limitations. Lack of amplification methods like PCR for proteins is a major handicap for proteomics. As a result, the number of proteins that could be detected is severely restricted by the CSL of protein measurement techniques. At present, only 20 % of human plasma proteomeis accounted and it is even less in cellular proteomes. PCR, on the other handis unreliable and ineffective in faithfully amplifying scarce samples (typical of clinical specimens, forensic and fossil DNA) and low copy number transcripts.
Therefore, there is a potential for direct sequencing (non-amplifying) techniques in implementing genomics and huge scope for techniques that could achieve yactomolar (10−24) sensitivity thereby accounting for almost every protein in the proteome. Under these circumstances, AFM and AFM based techniques have huge potential to meet the above demands. Recent breakthroughs in AFM based single molecule transcript profiling, TERS based sequencing, SNP discrimination had shown promise to meet the above demands in genomics. Similarly, micro/nano mechanical cantilever sensors have successfully demonstrated breaking the current CSL limits from nanomolar to attomolar concentrations. AFM force spectroscopy based protein fingerprinting in particular has the potential to break away proteomics from CSL limits all the way to the inverse Avagadro’s number.
AFM is the only imaging technique that is currently available that can provide unprecedented molecular resolution at physiological environments. AFM per se or as anm-AFM platform, contributed a wealth of information in characterizing single molecules in terms of their size, shape, volume, molecular organization, energy landscape, their interactions, etc., enriching the proteome atlas. Such an atlas would further accelerate fingerprinting of single molecules by just comparing an unknown molecules’ fingerprint from the atlas. Also, information about single molecules would accelerate development of newer bioassays which in turn would aid in pushing the CSL barrier down. Continued developments in AFM research would make both genomics and proteomics a total reality in near future which in turn would accelerate realization of systems approach to biological systems.
Acknowledgments
This work is supported by grants from NIH (DA024871 and DA025296) to R.L.
Abbreviations
- AFM
Atomic force microscopy
- HGP
Human Genome Project
- HPP
Human Proteome Project
References
- 1.Ideker T, Galitski T, Hood L. A new approach to decoding life: Systems biology. Annual Review of Genomics and Human Genetics. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 2.Archakov AI, Ivanov YD, Lisitsa AV, Zgoda VG. AFM fishing nanotechnology is the way to reverse the Avogadro number in proteomics. Proteomics. 2007;7:4–9. doi: 10.1002/pmic.200600467. [DOI] [PubMed] [Google Scholar]
- 3.Binnig G, Rohrer H. Scanning Tunneling Microscopy. Helvetica Physica Acta. 1982;55:726–735. [Google Scholar]
- 4.Binnig G, Rohrer H. Scanning Tunneling Microscopy -from birth to adolescence (Nobel Lecture) Available at: http://nobelprize.org/nobel_prizes/physics/laureates/1986/
- 5.Binnig G, Rohrer H. The Scanning Tunneling Microscope. Vol. 253. Scientific American; 1985. p. 50. [Google Scholar]
- 6.Binnig G, Quate CF, Gerber C. Atomic Force Microscope. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 7.Giessibl FJ. Atomic-Resolution of the Silicon (111)-(7×7) Surface by Atomic-Force Microscopy. Science. 1995;267:68–71. doi: 10.1126/science.267.5194.68. [DOI] [PubMed] [Google Scholar]
- 8.Giessibl FJ, Hembacher S, Bielefeldt H, Mannhart J. Subatomic features on the silicon (111)-(7×7) surface observed by atomic force microscopy. Science. 2000;289:422–425. doi: 10.1126/science.289.5478.422. [DOI] [PubMed] [Google Scholar]
- 9.Giessibl FJ. Advances in atomic force microscopy. Rev Mod Phys. 2003;75:949–983. [Google Scholar]
- 10.Kasas S, Dietler G. Probing nanomechanical properties from biomolecules to living cells. Pflugers Arch -Eur J Physiol. 2008;456:13–27. doi: 10.1007/s00424-008-0448-y. [DOI] [PubMed] [Google Scholar]
- 11.Weihs TP, Nawaz Z, Jarvis SP, Pethica JB. Limits of Imaging Resolution for Atomic Force Microscopy of Molecules. Appl Phys Lett. 1991;59:3536–3538. [Google Scholar]
- 12.Hoh JH, Lal R, John SA, Revel JP, Arnsdorf MF. Atomic Force Microscopy and Dissection of Gap-Junctions. Science. 1991;253:1405–1408. doi: 10.1126/science.1910206. [DOI] [PubMed] [Google Scholar]
- 13.Hoh JH, Sosinsky GE, Revel JP, Hansma PK. Structure of the Extracellular Surface of the Gap Junction by Atomic-Force Microscopy. Biophysical Journal. 1993;65:149–163. doi: 10.1016/S0006-3495(93)81074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller DJ, Fotiadis D, Scheuring S, Muller SA, Engel A. Electrostatically balanced subnanometer imaging of biological specimens by atomic force microscope. Biophysical Journal. 1999;76:1101–1111. doi: 10.1016/S0006-3495(99)77275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quist A, Doudevski L, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Amyloid ion channels: A common structural link for protein-misfolding disease. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thimm J, Mechler A, Lin H, Rhee S, Lal R. Calcium-dependent open/closed conformations and interfacial energy maps of reconstituted hemichannels. Journal of Biological Chemistry. 2005;280:10646–10654. doi: 10.1074/jbc.M412749200. [DOI] [PubMed] [Google Scholar]
- 17.Cappella B, Dietler G. Force-distance curves by atomic force microscopy. Surf Sci Rep. 1999;34:1. [Google Scholar]
- 18.Butt HJ, Cappella B, Kappl M. Force measurements with the atomic force microscope: Technique, interpretation and applications. Surf Sci Rep. 2005;59:1–152. [Google Scholar]
- 19.Drake B, Prater CB, Weisenhorn AL, Gould SAC, Albrecht TR, Quate CF, Cannell DS, Hansma HG, Hansma PK. Imaging Crystals, Polymers, and Processes in Water with the Atomic Force Microscope. Science. 1989;243:1586–1589. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- 20.Barrena E, Kopta S, Ogletree DF, Charych DH, Salmeron M. Relationship between friction and molecular structure: Alkylsilane lubricant films under pressure. Phys Rev Lett. 1999;82:2880–2883. [Google Scholar]
- 21.Carpick RW, Salmeron M. Scratching the surface: Fundamental investigations of tribology with atomic force microscopy. Chem Rev. 1997;97:1163–1194. doi: 10.1021/cr960068q. [DOI] [PubMed] [Google Scholar]
- 22.Hansma PK, Cleveland JP, Radmacher M, Walters DA, Hillner PE, Bezanilla M, Fritz M, Vie D, Hansma HG, Prater CB, et al. Tapping Mode Atomic-Force Microscopy in Liquids. Appl Phys Lett. 1994;64:1738–1740. [Google Scholar]
- 23.Bustamante C, Vesenka J, Tang CL, Rees W, Guthold M, Keller R. Circular DNA-Molecules Imaged in Air by Scanning Force Microscopy. Biochem. 1992;31:22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay SM, Nagahara LA, Thundat T, Knipping U, Rill RL, Drake B, Prater CB, Weisenhorn AL, Gould SAC, Hansma PK. Stm and Afm Images of Nucleosome DNA under Water. J Biomol Struct Dyn. 1989;7:279–287. doi: 10.1080/07391102.1989.10507771. [DOI] [PubMed] [Google Scholar]
- 25.Lyubchenko YL, Jacobs BL, Lindsay SM. Atomic Force Microscopy of Reovirus Dsrna - a Routine Technique for Length Measurements. Nucleic Acids Res. 1992;20:3983–3986. doi: 10.1093/nar/20.15.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vesenka J, Guthold M, Tang CL, Keller D, Delaine E, Bustamante C. Substrate Preparation for Reliable Imaging of DNA-Molecules with the Scanning Force Microscope. Ultramicroscopy. 1992;42:1243–1249. doi: 10.1016/0304-3991(92)90430-r. [DOI] [PubMed] [Google Scholar]
- 27.Arce FT, Carlson R, Monds J, Veeh R, Hu FZ, Stewart PS, Lal R, Ehrlich GD, Avci R. Nanoscale Structural and Mechanical Properties of Nontypeable Haemophilus influenzae Biofilms. J Bacteriol. 2009;191:2512–2520. doi: 10.1128/JB.01596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arce FT, Whitlock JL, Birukova AA, Birukov KG, Arnsdorf MF, Lal R, Garcia JGN, Dudek SM. Regulation of the micromechanical properties of pulmonary endothelium by S1P and thrombin: Role of cortactin. Biophys J. 2008;95:886–894. doi: 10.1529/biophysj.107.127167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almqvist N, Bhatia R, Primbs G, Desai N, Banerjee S, Lal R. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys J. 2004;86:1753–1762. doi: 10.1016/S0006-3495(04)74243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang H, Arce FT, Mustata M, Ramachandran S, Capone R, Nussinov R, Lal R. Antimicrobial Protegrin-1 Forms Amyloid-Like Fibrils with Rapid Kinetics Suggesting a Functional Link. Biophys J. 2011;100:1775–1783. doi: 10.1016/j.bpj.2011.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellermayer MSZ, Karsai A, Benke M, Soos K, Penke B. Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:141–144. doi: 10.1073/pnas.0704305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fantner GE, JBR, SGD, Belcher AM. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nature Nanotechnology. 2010;5:280–285. doi: 10.1038/nnano.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansma PK, Schitter G, Fantner GE, Prater C. High-speed atomic force microscopy. Science. 2006;314:601–602. doi: 10.1126/science.1133497. [DOI] [PubMed] [Google Scholar]
- 34.Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphris ADL, Miles MJ, Hobbs JK. A mechanical microscope: High-speed atomic force microscopy. Applied Physics Letters. 2005:86. [Google Scholar]
- 36.Giannotti MI, Vancso GJ. Interrogation of single synthetic polymer chains and polysaccharides by AFM-based force spectroscopy. Chemphyschem. 2007;8:2290–2307. doi: 10.1002/cphc.200700175. [DOI] [PubMed] [Google Scholar]
- 37.Marszalek PE, Li HB, Fernandez JM. Fingerprinting polysaccharides with single-molecule atomic force microscopy. Nature Biotechnology. 2001;19:258–262. doi: 10.1038/85712. [DOI] [PubMed] [Google Scholar]
- 38.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong Q, Inniss D, Kjoller K, Elings VB. Fractured Polymer Silica Fiber Surface Studied by Tapping Mode Atomic-Force Microscopy. Surf Sci. 1993;290:L688–L692. [Google Scholar]
- 40.Stark M, Bercu BN, Marchi F, Chevrier J, Huant S. AIP Conf Proc. Eindhoven, the Netherlands: American Institute of Physics; 2003. Marking the Difference: Interferometric Detection vs Optical Beam Deflection in AFM. [Google Scholar]
- 41.Hansma PK, Cleveland JP, Radmacher M, Walters DA, Hillner PE, Bezanilla M, Fritz M, Vie D, Hansma HG, Prater CB, et al. Tapping Mode Atomic-Force Microscopy in Liquids. Applied Physics Letters. 1994;64:1738–1740. [Google Scholar]
- 42.Putman CAJ, Vanderwerf KO, Degrooth BG, Vanhulst NF, Greve J. Tapping Mode Atomic-Force Microscopy in Liquid. Applied Physics Letters. 1994;64:2454–2456. [Google Scholar]
- 43.Bezanilla M, Drake B, Nudler E, Kashlev M, Hansma PK, Hansma HG. Motion and Enzymatic Degradation of DNA in the Atomic-Force Microscope. Biophysical Journal. 1994;67:2454–2459. doi: 10.1016/S0006-3495(94)80733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal R, John SA. Biological Applications of Atomic-Force Microscopy. American Journal of Physiology. 1994;266:C1. doi: 10.1152/ajpcell.1994.266.1.C1. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, Arce FT, Ramachandran S, Lal R. Nanomechanics of hemichannel conformations -Connexin flexibility underlying channel opening and closing. Journal of Biological Chemistry. 2006;281:23207–23217. doi: 10.1074/jbc.M605048200. [DOI] [PubMed] [Google Scholar]
- 46.Marszalek PE, Lu H, Li HB, Carrion-Vazquez M, Oberhauser AF, Schulten K, Fernandez JM. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 47.Marszalek PE, Oberhauser AF, Pang YP, Fernandez JM. Polysaccharide elasticity governed by chair-boat transitions of the glucopyranose ring. Nature. 1998;396:661–664. doi: 10.1038/25322. [DOI] [PubMed] [Google Scholar]
- 48.LIN H, BHATIA R, LAL R. Amyloid β protein forms ion channels: implications for Alzheimer’s disease pathophysiology. The FASEB Journal. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 49.Hansma HG, Vesenka J, Siegerist C, Kelderman G, Morrett H, Sinsheimer RL, Elings V, Bustamante C, Hansma PK. Reproducible Imaging and Dissection of Plasmid DNA under Liquid with the Atomic Force Microscope. Science. 1992;256:1180–1184. doi: 10.1126/science.256.5060.1180. [DOI] [PubMed] [Google Scholar]
- 50.Weisenhorn AL, Maivald P, Butt HJ, Hansma PK. Measuring Adhesion, Attraction, and Repulsion between Surfaces in Liquids with an Atomic-Force Microscope. Physical Review B. 1992;45:11226–11232. doi: 10.1103/physrevb.45.11226. [DOI] [PubMed] [Google Scholar]
- 51.Allison DP, Bottomley LA, Thundat T, Brown GM, Woychik RP, Schrick JJ, Jacobson KB, Warmack RJ. Immobilization of DNA for Scanning Probe Microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10129–10133. doi: 10.1073/pnas.89.21.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansma HG, Bezanilla M, Zenhausern F, Adrian M, Sinsheimer RL. Atomic Force Microscopy of DNA in Aqueous-Solutions. Nucleic Acids Res. 1993;21:505–512. doi: 10.1093/nar/21.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyubchenko YL, Gall AA, Shlyakhtenko LS, Harrington RE, Jacobs BL, Oden PI, Lindsay SM. Atomic Force Microscopy Imaging of Double-Stranded DNA and Rna. Journal of Biomolecular Structure & Dynamics. 1992;10:589–606. doi: 10.1080/07391102.1992.10508670. [DOI] [PubMed] [Google Scholar]
- 54.Hegner M, Wagner P, Semenza G. Immobilizing DNA on Gold Via Thiol Modification for Atomic-Force Microscopy Imaging in Buffer Solutions. Febs Letters. 1993;336:452–456. doi: 10.1016/0014-5793(93)80854-n. [DOI] [PubMed] [Google Scholar]
- 55.Hansma HG, Laney DE, Bezanilla M, Sinsheimer RL, Hansma PK. Applications for Atomic-Force Microscopy of DNA. Biophysical Journal. 1995;68:1672–1677. doi: 10.1016/S0006-3495(95)80343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rees WA, Keller RW, Vesenka JP, Yang GL, Bustamante C. Evidence of DNA Bending in Transcription Complexes Imaged by Scanning Force Microscopy. Science. 1993;260:1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- 57.Pietrasanta LI, Schaper A, Jovin TM. Probing Specific Molecular-Conformations with the Scanning Force Microscope - Complexes of Plasmid DNA and Anti-Z-DNA Antibodies. Nucleic Acids Res. 1994;22:3288–3292. doi: 10.1093/nar/22.16.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyman C, Grotkopp E, Bustamante C, Nelson HCM. Determination of Heat-Shock Transcription Factor-2 Stoichiometry at Looped DNA Complexes Using Scanning Force Microscopy. Embo Journal. 1995;14:117–123. doi: 10.1002/j.1460-2075.1995.tb06981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Noort SJT, van der Werf KO, Eker APM, Wyman C, de Grooth BG, van Hulst NF, Greve J. Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophysical Journal. 1998;74:2840–2849. doi: 10.1016/S0006-3495(98)77991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindsay SM, Philipp M. Can the Scanning Tunneling Microscope Sequence DNA. Genetic Analysis-Biomolecular Engineering. 1991;8:8–13. doi: 10.1016/1050-3862(91)90003-a. [DOI] [PubMed] [Google Scholar]
- 61.Hansma HG, Weisenhorn AL, Gould SAC, Sinsheimer RL, Gaub HE, Stucky GD, Zaremba CM, Hansma PK. Progress in Sequencing Deoxyribonucleic-Acid with an Atomic Force Microscope. Journal of Vacuum Science & Technology B. 1991;9:1282–1284. [Google Scholar]
- 62.Lindsay SM, Nagahara LA, Thundat T, Knipping U, Rill RL, Drake B, Prater CB, Weisenhorn AL, Gould SAC, Hansma PK. Stm and Afm Images of Nucleosome DNA under Water. Journal of Biomolecular Structure & Dynamics. 1989;7:279–287. doi: 10.1080/07391102.1989.10507771. [DOI] [PubMed] [Google Scholar]
- 63.Nygaard V, Holden M, Loland A, Langaas M, Myklebost O, Hovig E. Limitations of mRNA amplification from small-size cell samples. Bmc Genomics. 2005:6. doi: 10.1186/1471-2164-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nygaard V, Loland A, Holden M, Langaas M, Rue H, Liu F, Myklebost O, Fodstad O, Hovig E, Smith-Sorensen B. Effects of mRNA amplification on gene expression ratios in cDNA experiments estimated by analysis of variance. Bmc Genomics. 2003:4. doi: 10.1186/1471-2164-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen DC, Keller RA, Jett JH, Martin JC. Detection of Single Molecules of Phycoerythrin in Hydrodynamically Focused Flows by Laser-Induced Fluorescence. Analytical Chemistry. 1987;59:2158–2161. doi: 10.1021/ac00144a032. [DOI] [PubMed] [Google Scholar]
- 66.Jett JH, Keller RA, Martin JC, Marrone BL, Moyzis RK, Ratliff RL, Seitzinger NK, Shera EB, Stewart CC. High-Speed DNA Sequencing - an Approach Based Upon Fluorescence Detection of Single Molecules. Journal of Biomolecular Structure & Dynamics. 1989;7:301–309. doi: 10.1080/07391102.1989.10507773. [DOI] [PubMed] [Google Scholar]
- 67.Treffer R, Deckert V. Recent advances in single-molecule sequencing. Current Opinion in Biotechnology. 2010;21:4–11. doi: 10.1016/j.copbio.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 68.Rosenthal A, Brenner S. DNA Sequencing Method. 2000;6,087,095:4. [Google Scholar]
- 69.Allison DP, Kerper PS, Doktycz MJ, Spain JA, Modrich P, Larimer FW, Thundat T, Warmack RJ. Direct atomic force microscope imaging of EcoRI endonuclease site specifically bound to plasmid DNA molecules. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8826–8829. doi: 10.1073/pnas.93.17.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Allison DP, Kerper PS, Doktycz MJ, Thundat T, Modrich P, Larimer FW, Johnson DK, Hoyt PR, Mucenski ML, Warmack RJ. Mapping individual cosmid DNAs by direct AFM imaging. Genomics. 1997;41:379–384. doi: 10.1006/geno.1997.4686. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura T, Maeda Y, Oka T, Tabata H, Futai M, Kawai T. Atomic force microscope observation of plasmid deoxyribose nucleic acid with restriction enzyme. Journal of Vacuum Science & Technology B. 1999;17:288–293. [Google Scholar]
- 72.Reed J, Mishra B, Pittenger B, Magonov S, Troke J, Teitell MA, Gimzewski JK. Single molecule transcription profiling with AFM. Nanotechnology. 2007:18. doi: 10.1088/0957-4484/18/4/044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sommer SS, Cohen JE. The Size Distributions of Proteins, Messenger-Rna and Nuclear-Rna. Journal of Molecular Evolution. 1980;15:37–57. doi: 10.1007/BF01732582. [DOI] [PubMed] [Google Scholar]
- 74.Hames BD. In: Gene expression 2, eukaryotic chromosomes. 2. Lewin B, editor. John Wiley & Sons; Chichester: 1980. p. 1160. £25.65 (hardback)/£13.25 (paperback) [Google Scholar]; Biochemical Education. 1982;10:158–158. [Google Scholar]
- 75.Bailo E, Deckert V. Tip-enhanced Raman spectroscopy of single RNA strands: Towards a novel direct-sequencing method. Angewandte Chemie-International Edition. 2008;47:1658–1661. doi: 10.1002/anie.200704054. [DOI] [PubMed] [Google Scholar]
- 76.Schafer AJ, Hawkins JR. DNA variation and the future of human genetics. Nature Biotechnology. 1998;16:33–39. doi: 10.1038/nbt0198-33. [DOI] [PubMed] [Google Scholar]
- 77.Clausen-Schaumann H, Seitz M, Krautbauer R, Gaub HE. Force spectroscopy with single bio-molecules. Current Opinion in Chemical Biology. 2000;4:524–530. doi: 10.1016/s1367-5931(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 78.Hinterdorfer P, Dufrene YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nature Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 79.Fritz J, Baller MK, Lang HP, Rothuizen H, Vettiger P, Meyer E, Guntherodt HJ, Gerber C, Gimzewski JK. Translating biomolecular recognition into nanomechanics. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 80.Hansen KM, Ji HF, Wu GH, Datar R, Cote R, Majumdar A, Thundat T. Cantilever-based optical deflection assay for discrimination of DNA single-nucleotide mismatches. Analytical Chemistry. 2001;73:1567–1571. doi: 10.1021/ac0012748. [DOI] [PubMed] [Google Scholar]
- 81.Stowe TD, Yasumura K, Kenny TW, Botkin D, Wago K, Rugar D. Attonewton force detection using ultrathin silicon cantilevers. Applied Physics Letters. 1997;71:288–290. [Google Scholar]
- 82.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 83.Abadal G, Davis ZJ, Helbo B, Borrise X, Ruiz R, Boisen A, Campabadal F, Esteve J, Figueras E, Perez-Murano F, et al. Electromechanical model of a resonating nano-cantilever-based sensor for high-resolution and high-sensitivity mass detection. Nanotechnology. 2001;12:100–104. [Google Scholar]
- 84.Davis ZJ, Abadal G, Kuhn O, Hansen O, Grey F, Boisen A. Fabrication and characterization of nanoresonating devices for mass detection. Journal of Vacuum Science & Technology B. 2000;18:612–616. [Google Scholar]
- 85.Dugdale TM, Dagastine R, Chiovitti A, Mulvaney P, Wetherbee R. Single adhesive nanofibers from a live diatom have the signature fingerprint of modular proteins. Biophysical Journal. 2005;89:4252–4260. doi: 10.1529/biophysj.105.062489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fernandez JM. Fingerprinting single molecules in vivo. Biophysical Journal. 2005;89:3676–3677. doi: 10.1529/biophysj.105.072223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Avci R, Schweitzer MH, Boyd RD, Wittmeyer JL, Arce FT, Calvo JO. Preservation of bone collagen from the late cretaceous period studied by immunological techniques and atomic force microscopy. Langmuir. 2005;21:3584–3590. doi: 10.1021/la047682e. [DOI] [PubMed] [Google Scholar]
- 88.Schweitzer MH, Suo Z, Avci R, Asara JM, Allen MA, Arce FT, Horner JR. Analyses of soft tissue from Tyrannosaurus rex suggest the presence of protein. Science. 2007;316:277–280. doi: 10.1126/science.1138709. [DOI] [PubMed] [Google Scholar]
- 89.Arce FT, Carlson R, Monds J, Veeh R, Hu FZ, Stewart PS, Lal R, Ehrlich GD, Avci R. Nanoscale Structural and Mechanical Properties of Nontypeable Haemophilus influenzae Biofilms. Journal of Bacteriology. 2009;191:2512–2520. doi: 10.1128/JB.01596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kasas S, Thomson NH, Smith BL, Hansma PK, Miklossy J, Hansma HG. Biological applications of the AFM: From single molecules to organs. International Journal of Imaging Systems and Technology. 1997;8:151–161. [Google Scholar]
- 91.Parot P, Dufrene YF, Hinterdorfer P, Le Grimellee C, Navajas D, Pellequer JL, Scheuring S. Past, present and future of atomic force microscopy in life sciences and medicine. Journal of Molecular Recognition. 2007;20:418–431. doi: 10.1002/jmr.857. [DOI] [PubMed] [Google Scholar]
- 92.Muller DJ, Hand GM, Engel A, Sosinsky GE. Conformational changes in surface structures of isolated connexin 26 gap junctions. Embo Journal. 2002;21:3598–3607. doi: 10.1093/emboj/cdf365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bahatyrova S, Frese RN, Siebert CA, Olsen JD, van der Werf KO, van Grondelle R, Niederman RA, Bullough PA, Otto C, Hunter CN. The native architecture of a photosynthetic membrane. Nature. 2004;430:1058–1062. doi: 10.1038/nature02823. [DOI] [PubMed] [Google Scholar]
- 94.Frederix PLTM, Bosshart PD, Engel A. Atomic Force Microscopy of Biological Membranes. Biophysical Journal. 2009;96:329–338. doi: 10.1016/j.bpj.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s Disease and Down syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mustata M, Capone R, Jang H, Arce FT, Ramachandran S, Lal R, Nussinov R. K3 Fragment of Amyloidogenic beta(2)-Microglobulin Forms Ion Channels: Implication for Dialysis Related Amyloidosis. Journal of the American Chemical Society. 2009;131:14938–14945. doi: 10.1021/ja9049299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin H, Lal R, Clegg DO. Imaging and mapping heparin-binding sites on single fibronectin molecules with atomic force microscopy. Biochem. 2000;39:3192–3196. doi: 10.1021/bi991624o. [DOI] [PubMed] [Google Scholar]
- 98.Lin H, Clegg DO, Lal R. Imaging real-time proteolysis of single collagen I molecules with an atomic force microscope. Biochem. 1999;38:9956–9963. doi: 10.1021/bi990800q. [DOI] [PubMed] [Google Scholar]
- 99.Almqvist N, Bhatia R, Primbs G, Desai N, Banerjee S, Lal R. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophysical Journal. 2004;86:1753–1762. doi: 10.1016/S0006-3495(04)74243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vergara D, Martignago R, Leporatti S, Bonsegna S, Maruccio G, De Nuccio F, Santino A, Cingolani R, Nicolardi G, Maffia M, et al. Biomechanical and proteomic analysis of INF-beta-treated astrocytes. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/45/455106. [DOI] [PubMed] [Google Scholar]
- 101.Ramachandran S, Lal R. Scope of atomic force microscopy in the advancement of nanomedicine. Indian Journal of Experimental Biology. 2010;48:1020–1036. [PubMed] [Google Scholar]
- 102.Lal R, Ramachandran S, Arnsdorf M. Multidimensional Atomic Force Microscopy: A Versatile Novel Technology for Nanopharmacology Research. The AAPS Journal. 2010;12:716–728. doi: 10.1208/s12248-010-9232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davies PF, Barbee KA, Lal R, Robotewskyj A, Griem ML. Hemodynamics and Atherogenesis -Endothelial Surface Dynamics in Flow Signal-Transduction. Atherosclerosis Iii: Recent Advances in Atherosclerosis Research. 1995;748:86–103. [PubMed] [Google Scholar]
- 104.Barbee KA, Mundel T, Lal R, Davies PF. Altered Subcellular Shear-Stress Distribution by the Flow-Realignment Response of Endothelial-Cells. Faseb Journal. 1995;9:A588–A588. [Google Scholar]
- 105.Barbee KA, Davies PF, Lal R. Shear Stress-Induced Reorganization of the Surface-Topography of Living Endothelial-Cells Imaged by Atomic-Force Microscopy. Circulation Research. 1994;74:163–171. doi: 10.1161/01.res.74.1.163. [DOI] [PubMed] [Google Scholar]
- 106.Lal R, Proksch R. Multimodal atomic force microscopy: Biological imaging using atomic force microscopy combined with light fluorescence and confocal microscopies and electrophysiologic recording. Int J Imag Syst Tech. 1997;8:293–300. [Google Scholar]
- 107.Hansma PK, Drake B, Grigg D, Prater CB, Yashar F, Gurley G, Eling SV, Feinstein S, Lal R. A New, Optical-Lever Based Atomic-Force Microscope. J Appl Phys. 1994;76:796–799. [Google Scholar]
- 108.Quist AP, Chand A, Ramachandran S, Daraio C, Jin S, Lal R. Atomic force microscopy imaging and electrical recording of lipid bilayers supported over microfabricated silicon chip nanopores: Lab-on-a-chip system for lipid membranes and ion channels. Langmuir. 2007;23:1375–1380. doi: 10.1021/la062187z. [DOI] [PubMed] [Google Scholar]