SUMMARY
Early detection of colonic polyps can prevent up to 90% of colorectal cancer deaths. Conventional colonoscopy readily detects the majority of pre-malignant lesions, which exhibit raised morphology. However, lesions that are flat and depressed are often undetected using this method. Therefore, there is a need for molecular-based contrast agents to improve detection rates over conventional colonoscopy. We evaluated a quenched fluorescent activity-based probe (qABP; BMV109) that targets multiple cysteine cathepsins that are overexpressed in intestinal dysplasia in a genetic model of spontaneous intestinal polyp formation and in a chemically induced model of colorectal carcinoma. We found that the qABP selectively targets cysteine cathepsins resulting in high sensitivity and specificity for intestinal tumors in mice and humans. Additionally, the qABP can be administered by either intravenous injection or by local delivery to the colon making it a highly valuable tool for improved detection of colorectal lesions using fluorescence-guided colonoscopy.
INTRODUCTION
Colorectal cancer is the second leading cause of cancer-related mortality in the United States (Jemal et al., 2008). Colonoscopy and sigmoidoscopy have been shown to reduce the incidence of colorectal cancer-related deaths (Blumenstein et al., 2013). During the last decade, the awareness of screening options has increased dramatically, helping to improve early detection (Amersi et al., 2005). Nevertheless, more than 20% of grossly visible colorectal polyps remain undetectable by standard white light colonoscopy (Cheng et al., 2002). These undetectable lesions include small and flat or depressed polyps with malignant potential. Thus, there is an unmet medical need for new tools to improve detection of colorectal polyps. Improvements in image contrast for visualizing pre-malignant lesions will potentially improve detection rates and consequently improve clinical outcomes. Additionally, such tools may be useful for evaluation of responses to therapeutic interventions.
Proteases support tumorigenesis by facilitating cell division and motility, mediating local invasion and promoting angiogenesis and metastasis (Lopez-Otin and Matrisian, 2007; Lopez-Otin and Overall, 2002). Cysteine cathepsins are a family of lysosomal proteases that play important roles in various aspects of tumorigenesis (Gounaris et al., 2008) (Mohamed and Sloane, 2006). In human cancers, particularly in colorectal tumors, cathepsins B, C, D, H, L, S, and X have been shown to be upregulated both in tumor cells and in associated stromal cells, including in immune cells (i.e. macrophages, lymphocytes and neutrophils), endothelial cells and fibroblasts (Herszenyi et al., 1999; Mohamed and Sloane, 2006; Troy et al., 2004). These observations have prompted efforts to target cathepsins with molecular imaging agents as a strategy for detecting colorectal malignancies. Over the past decade, numerous biosensors have been developed for the purpose of imaging protease activity in native environments (Bogdanov and Mazzanti, 2013; Sanman and Bogyo, 2014). These include fluorescent (Boonacker and Van Noorden, 2001) and bioluminescence protein substrates (O’Brien et al., 2005; Shinde et al., 2006), macromolecular and peptide substrate-based probes (Hu et al., 2014; Mahmood et al., 1999; Olson et al., 2012; Saravanakumar et al., 2012) and FRET protein reporters (Cummings et al., 2002). Although many of these methods have shown promise for contrast imaging, most have exhibited relatively poor selectivity and specificity. Furthermore, several of these methods require either the use of bulky cell-impermeable molecules with slow turn-on rates or genetic manipulation of cells or organisms to introduce reporters (Baruch et al., 2004). Substrate-based probes also do not allow direct identification of the protease target that activated the signal, thus making it difficult to assign function to specific targets. An alternative method for visualizing protease activity uses small-molecule probes that covalently attach to an enzyme-target through a chemical reaction that is specific for the target protease (Terai and Nagano, 2008). These probes, called activity-based probes (ABPs) (Fonovic and Bogyo, 2007; Sanman and Bogyo, 2014), are designed to be highly selective for the catalytically active form of a protease or protease family (Cravatt et al., 2008). Typical ABPs consist of three main elements: (i) a reactive functional group that covalently reacts with the active site of the enzyme, (ii) a linker region that confers enzyme specificity, directs binding to the target, and prevents steric congestion, and (iii) a tag used for direct visualization of the probe-labeled proteins (Deu et al., 2012). An additional element that improves the sensitivity and specificity of ABPs is a quencher that prevents fluorescent emission of unbound probe. Once the probe covalently binds to the target enzyme, the quencher is removed and the probe emits the desired fluorescent signal (Blum et al., 2005) (Fig. 1B). The benefit of the ABPs is that they are relatively small molecules that can directly diffuse into tissues resulting in rapid turn on rates in vivo (Edgington et al., 2011). This enables applications in which probes are topically applied and imaging can be performed within a time frame of minutes rather than hours (Cutter et al., 2012).
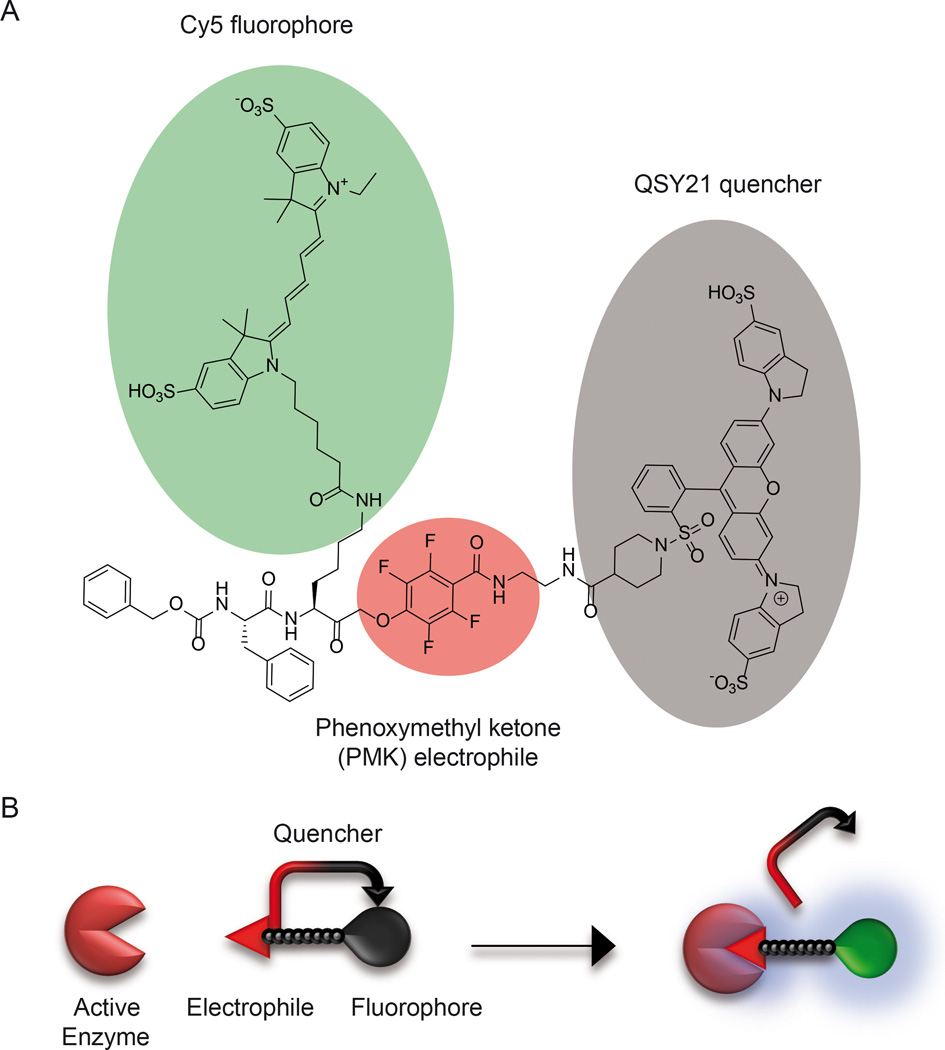
Figure 1. Structure and mechanism of action of the qABP.
(A) Structure of BMV109 highlighting the reactive phenoxymethyl ketone electrophile that forms a covalent bond with the active site cysteine (red) as well as the fluorophore (green) and quencher (grey). (B) Diagram of the mechanism of un-quenching of the probe upon binding to an active cysteine protease target.
In this study we evaluated a recently reported, biocompatible, optically quenched activity-based probe (qABP), BMV109, that becomes fluorescent upon covalently binding to active cysteine cathepsins (Verdoes et al., 2013) for the detection of intestinal cancer. This qABP selectively labels a broad spectrum of cathepsins in the intestines, primarily in the tumor microenvironment. In multiple mouse models, the probe exhibited strong labeling of polyps following not only intravenous injection (i.v.) but also by local application using intra-rectal administration. Furthermore, because the probe covalently labels target proteases, we were able to biochemically confirm selective labeling of cysteine cathepsins. Direct histological analysis of the whole intestine enabled us to measure the selectivity and specificity of the probe for polyps. Using this method, we confirmed rapid detection of lesions with high selectivity and specificity ratios following either i.v. or intra-rectal administration. We used the probe to topically label human tissue sections, demonstrating the ability of the probe to detect human polyps. These unique features make our qABP a strong candidate for clinical translation as a tool for use in fluorescence-guided colonoscopy to enhance detection and surgical treatment of colon cancer.
RESULTS
The qABP preferentially labels polyps of APCmin/+ mice
We previously reported the synthesis and evaluation of a qABP that was designed to selectively target a broad spectrum of cysteine cathepsins (Verdoes et al., 2013). The probe is composed of a phenoxymethyl ketone (PMK) electrophile, a linker, a Cy5 fluorophore and a Sulfo-QSY21 quencher (Fig 1A, 1B). As an initial system to evaluate the ability of the probe to label intestinal polyps, we used the well-established APCmin/+ (multiple intestinal neoplasia) mouse model (Moser et al., 1990). These mice carry a mutation resulting in a premature truncation of one allele of the APC tumor suppressor, analogous to the human disease familial adenomatous polyposis. In both the mouse model and human disease, spontaneous loss of the other allele results in the development of multiple adenomas in the intestine. In addition, this model has been used to evaluate other classes of protease beacons (Clapper et al., 2011; Gounaris et al., 2008). The qABP was injected i.v. into both APCmin/+ and wild type (WT) mice. A group of the APCmin/+ mice was treated with vehicle as a control. Mice were euthanized 1 hour later and intestines were removed, flushed and prepared for ex vivo fluorescence imaging. Multiple variable-sized intestinal polyps were observed in the tissues from APCmin/+ mice with strong fluorescent labeling of tumors in mice treated with the qABP (Fig. 2A). We also assessed whether cathepsin activity levels correlated with polyp size by comparing fluorescent signal intensity with polyp diameter. This analysis confirmed a strong correlation between fluorescence signal and polyp size (Fig 2B).
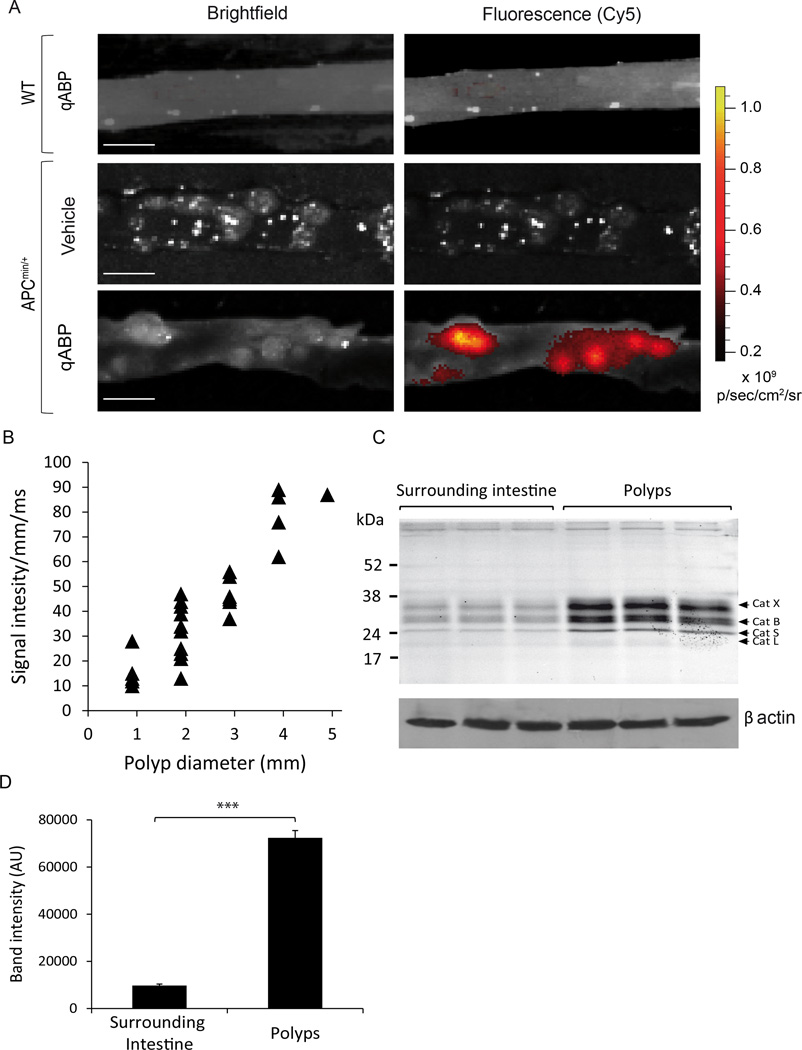
Figure 2. The qABP preferentially labels polyps in APCmin/+ mice.
(A) Ex vivo bright field (left) and fluorescence optical images (right) of the intestine tissue of wild-type (WT; top) and APCmin/+ (center and bottom) mice 1 hour post i.v. administration of the probe (scale bar represents 5mm). (B) Correlation of signal intensity/mm/exposure time (ms) with polyp diameters (mm). Calculated correlation coefficient – R2 = 0.823 (C) Analysis of cathepsin labeling in samples from surrounding normal tissues (left) compared to polyp tissue (right) from three individual mice. Samples were analyzed by SDS-PAGE followed flatbed laser scanning of the gel to visualize probe labeled proteins. (D) Quantification of the intensity of labeled proteins in (C). P values are two-sided (analysis of variance). Error bars represent the mean ± s.d. * P<0.05, ** P<0.03, *** P<0.01.
To identify the specific targets of the qABP, we performed SDS-PAGE analysis of homogenized samples of polyps and matched surrounding intestine dissected from APCmin/+ mice (Fig. 2C). The intensity of total cysteine cathepsin labeling was 7-fold higher in polyps compared to uninvolved surrounding tissue (Fig. 2D; p=0.00065), similar to the ratio of polyp signal to background observed in the fluorescence optical images. The labeling profile also confirmed that the probe is pan-reactive towards cysteine cathepsins and that cathepsins X, B, S and L are all upregulated in the polyp tissues.
To confirm specificity of probe labeling, APCmin/+ mice were treated with a broad-spectrum inhibitor of the cysteine cathepsins for 5 days to reduce activity before treatment with the qABP. This inhibitor, K11777, is a dipeptide vinyl sulfone that has been previously reported to block cathepsin activity in vivo (Blum et al., 2007). Fluorescence images from mice treated with K11777 showed a substantial drop in probe fluorescence in polyps when compared to the vehicle treated samples (Fig. 3A). This drop in signal within the polyps correlated with a drop in the total amount of labeled cathepsins, as assessed by SDS-PAGE analysis and quantification of labeling of resected colon tissue (Fig 3B, C; p=0.0126), thus confirming that the qABP labels polyps by selectively modification of active cathepsins.
Figure 3. Probe signal intensity correlates with the activity of the cysteine cathepsinsin vivo.
(A) Ex vivo fluorescence optical images overlaid on bright field images of intestines from APCmin/+ mice pretreated with either vehicle (top) no inhibitor (middle) or the K11777 inhibitor (bottom) 1 hour post i.v. administration of the probe (scale bar represents 5mm). (B) Analysis of cathepsin labeling by the probe in polyps from the APCmin/+ mice administered with either vehicle (left), the probe and no inhibitor (middle) or the probe and K11777 (right). Samples were analyzed by SDS-PAGE followed flatbed laser scanning of the gel to visualize probe labeled proteins. (C) Quantification of the intensity of labeled polyps from (A; left) and labeled cathepsins in (B; right). P values are two-sided (analysis of variance). Error bars represent the mean ± s.d. * P<0.05, ** P<0.03, *** P<0.01.
The qABP labels colitis-associated colonic neoplasms following either systemic or local administration
Chronic inflammatory bowel disease (IBD), particularly ulcerative colitis, strongly predisposes individuals to develop colorectal cancer. Therefore, we tested the ability of the probe to image neoplasms in a mouse model of colitis-associated colorectal cancer induced by azoxymethane and dextran sodium sulfate (AOM/DSS). We chose this model because it allows assessment of the probe in a different model with clinical relevance to colorectal cancer and because others have recently use a similar colitis model to demonstrate the value of a substrate based cathepsin probe for detecting dysplasia that is induced by colitis (Gounaris et al., 2013). Mice with colitis induced by AOM/DSS were treated with the probe by either i.v. injection as performed for the APCmin/+ mice, or by direct intra-rectal administration into the colon. We reasoned that, since the probe is relatively low molecular weight and can diffuse into tissues, local administration may be effective and a clinically valuable strategy. Control groups included AOM/DSS mice administered i.v. or intra-rectally with vehicle and non-induced mice treated with the probe i.v. or intra-rectally. In colons dissected from AOM/DSS mice, we observed mainly macroscopic polyps in the distal colon that were highly labeled with the qABP upon either intravenous or intra-rectal administration (Fig. 4A). Auto-fluorescence was not detected in AOM/DSS mice treated with the vehicle and non-specific labeling in non-induced mice treated with the probe was minimal. Furthermore, similar to our results in the APCmin/+ mice, we observed a strong correlation between fluorescence signal and polyp size for both the i.v. and intra-rectal administration, demonstrating their equivalent detection capabilities (Fig. 4B).
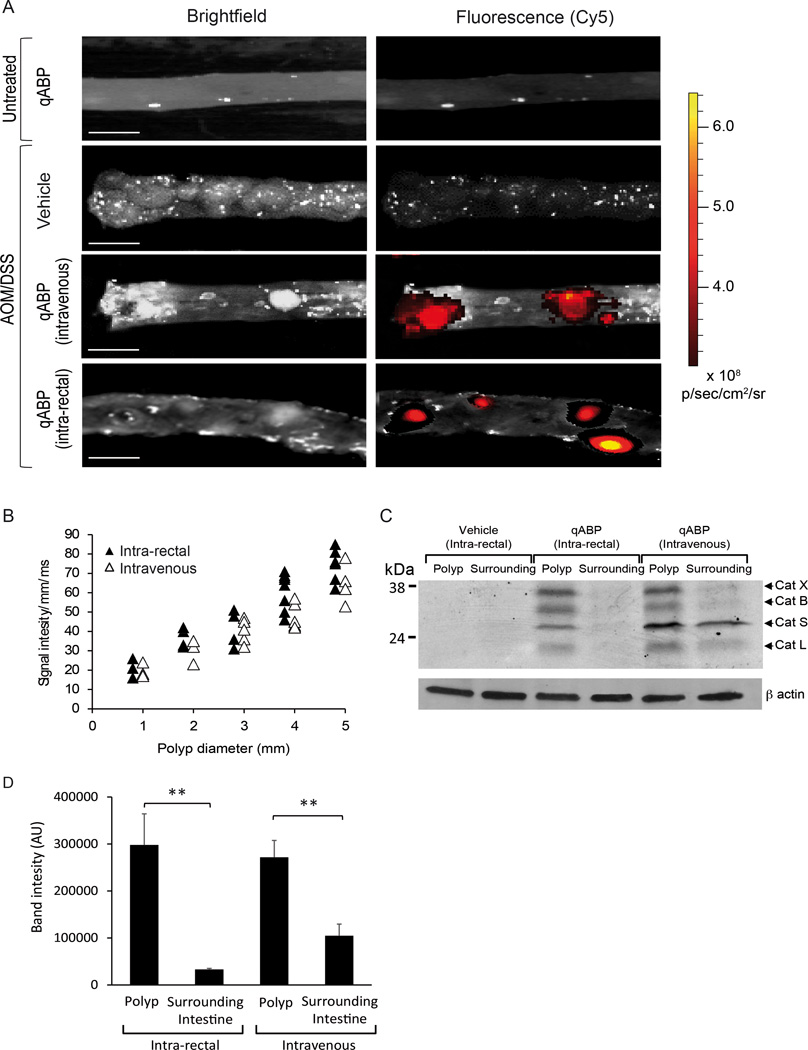
Figure 4. The probe selectively labels cysteine cathepsins up-regulated in intestinal adenomas and adenocarcinomas.
(A) Ex vivo bright field (left) and fluorescence optical images (right) of the colon of untreated controls (top) and AOM/DSS (bottom) mice 1 hour post i.v. or intra-rectal administration of the probe (scale bar represents 5mm). (B) Correlation of signal intensity/mm/exposure time (ms) with polyp diameters (mm) for multiple polyp samples from mice treated i.v. (open triangles) or intra-rectally (filled triangles). Calculated correlation coefficients – intra-rectal R2 = 0.810, intravenous R2 = 0.843. (C) Analysis of cathepsin labeling by the probe in surrounding normal tissue and polyps from the intra-rectal vehicle (left), intra-rectal probe (middle) and intravenous probe (right). Samples were analyzed by SDS-PAGE followed flatbed laser scanning of the gel to visualize probe labeled proteins. (D) Quantification of intensity of labeled cathepsins from multiple samples (see figure S1) from each treatment group (n=3 ). P values are two-sided (analysis of variance). Error bars represent the mean ± s.d. * P<0.05, ** P<0.03, *** P<0.01. (see also figure S1)
To verify the targets of the probe and exclude the possibility that signals observed in the fluorescence images resulted from non-specific interactions with other protein targets, we performed SDS-PAGE analysis on labeled tissues (Fig. 4C). Samples of surrounding tissue and colonic polyps from AOM/DSS mice treated with the probe (i.v. or intra-rectally) and samples from AOM/DSS mice treated intra-rectally with vehicle were collected, homogenized and analyzed. In tissues from mice that were administered the probe intra-rectally, polyps showed strong labeling of cathepsin X, B, S and L, while no labeling of cathepsins was observed in healthy mice. No labeling of cathepsins was observed in mice administered vehicle control intrarectally. In mice administered the probe intravenously, polyps showed strong labeling of cathepsin X and B, and attenuated labeling of cathepsin S and L, while some labeling of only cathepsin X, B, L and S was observed in the surrounding tissue (Fig. 4C). Signal in surrounding tissue of mice treated intravenously may be the result of labeling of populations of immune cells in the circulation or endothelial cells that are less directly targeted when the probe is applied locally. When multiple samples from polyp and normal surrounding tissues were analyzed (Fig. S1) and quantified (Fig. 3D), the total levels of cysteine cathepsin labeling were 9-fold and 2.6 fold higher in polyps compared to the surrounding tissue in mice treated with the probe intrarectally (p=0.0245) or intraveneously (p=0.0362) respectively. These findings demonstrate that probe administered either locally or systemically results in high selectivity for cysteine cathepsins in colonic polyps.
Probe labeling is associated with regions of cytological and architectural abnormalities of the intestinal epithelium
Increased proliferation is believed to be an early event of the adenoma-carcinoma sequence (Jiang et al., 2013). Additionally, the rate and distribution of proliferating epithelial and mucosal cells has been used as a biomarker of neoplasia progress (Lipkin, 1988). We therefore examined if probe labeling correlates with abnormal patterns of the intestinal epithelium such as cell dysplasia (i.e. atypia, elongation, hyperchromatic nuclei and stratified nuclei). Because the probe permanently labels target proteases, it was possible to perform direct histology and fluorescence imaging of tissues sections to determine the extent of probe accumulation in cancer lesions. We also evaluated the distribution of CD68, a cellular marker for macrophages, in samples from APCmin/+ and WT mice intestines by histology. To collect and analyze data from the entire small intestine, we used a wide-field tile scan technique to generate a mosaic image of H&E and immunofluorescence staining (Fig. S2). The intestinal mucosa from APCmin/+ mice showed areas of increased proliferation at intestinal crypts. In addition, intestinal microadenomas and adenomatous polyps in APCmin/+ samples showed consistently increased CD68 staining compared to normal mucosa. Importantly, the probe did not accumulate in the intestine of WT control mice, verifying the specificity for polyps. Most regions of increased epithelial proliferation, including adenomas, exhibited increased accumulation of the probe in APCmin/+ mice. Using the ex vivo bright field and fluorescence images (Fig. 2 and Fig. S3) we were able to calculate the sensitivity and specificity of the probe for detecting adenomas as 87.5% and 76.9% respectively (Table S1). In addition, the positive predictive and the negative predictive values were determined to be 90.3% and 71.4% respectively (Table S1). We were also able to calculate the sensitivity and specificity of the probe in the AOM/DSS model using the ex vivo bright field and fluorescence images (Fig. 4 and S4). The sensitivity and specificity for detecting colonic adenomas in mice intravenously treated with the probe were 88.9% and 71.5%, respectively. The positive predictive and the negative predictive values were 89.5% and 81.2%, respectively (Table S2). The sensitivity and specificity in detecting colonic adenomas in mice treated with the probe locally by intra-rectal administration were 93.6% and 80.0%, respectively (Table S2). The positive predictive and the negative predictive values were 94.9% and 81.2%, respectively (Table S2). These results demonstrate that the probe can be used by i.v. or intra-rectal administration to selectively label colitis-associated colonic polyps.
Cathepsins labeled by the probe co-localize with tumor-associated immune cells in both models of colon cancer
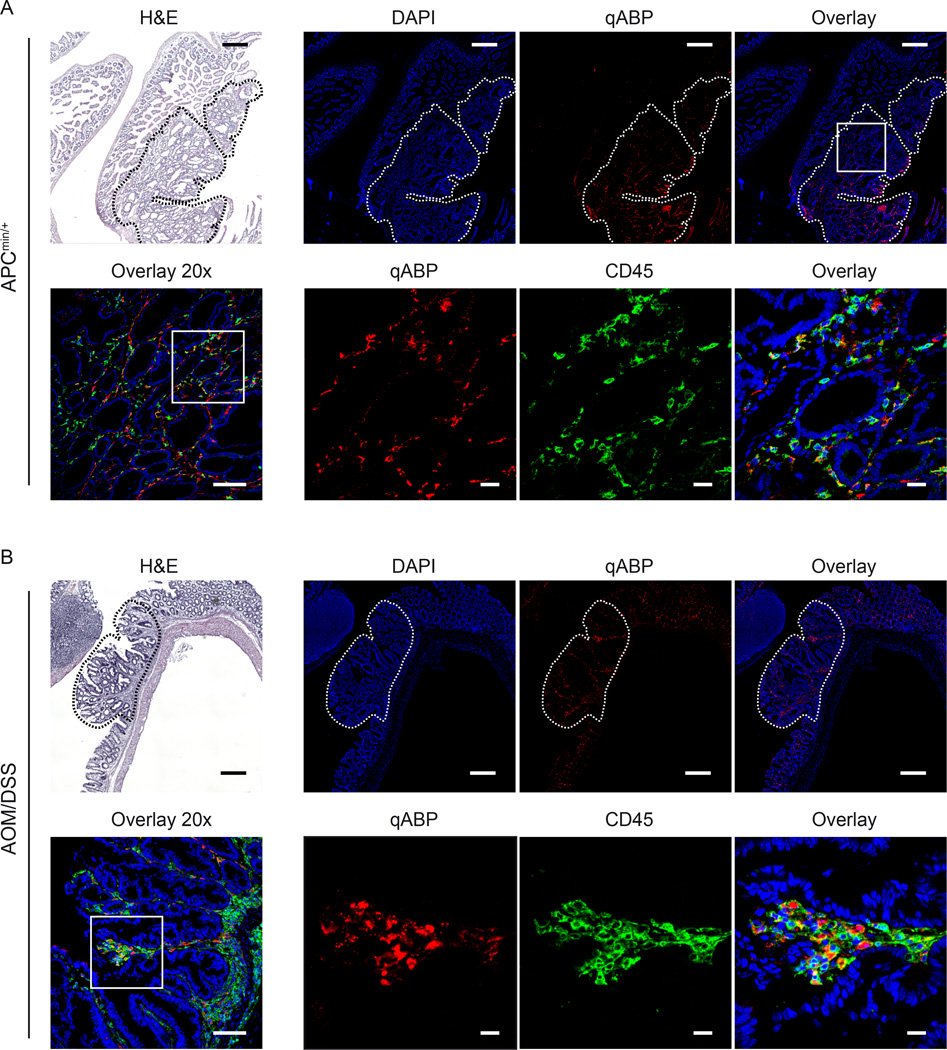
Numerous cell types in the tumor microenvironment express cathepsins. To determine whether specific immune cells are the major targets of the qABP in mouse tissues, we performed immunofluorescence staining with the cell marker CD45 on intestine sections from both the APCmin/+ and AOM/DSS mice treated with the qABP (Fig. 5). These images show that although signal is present in normal tissue, the probe accumulates primarily in polyps. We observed that the probe localized mainly to the lysosomal compartments of mononuclear cells in the lamina propria. As cysteine cathepsins are mainly lysosomal, the staining pattern matched expected observations. Furthermore, the probe was found to accumulate mainly in tumor-associated immune cells, as indicated by the labeling with CD45 membrane protein staining. Importantly, these results were similar in both models, confirming that the probe is able to highlight general regions of inflammation in and around developing lesions in the intestine.
Figure 5. Cathepsins labeled by the probe colocalize with macrophages in intestinal dysplasia and colitis-related colonic neoplasms.
Sections of confocal macroscopic mosaic images of intestine dissected from APCmin/+ (A) and AOM/DSS (B) mice. H&E (top left panels, scale bar represents 250 µm) are shown along with DAPI (blue), and probe (red) and anti-CD45 (green). Images are shown are mosaic images from tile scanning (top), 20x (bottom left, scale bar represents 100 µm) and 63x magnification of the region indicated with a white box (lower right images, scale bar represents 20 µm). Polyps are indicated with white dashed lines. (see also figure S2)
The qABP labels tumor-associated immune cells in human polyps
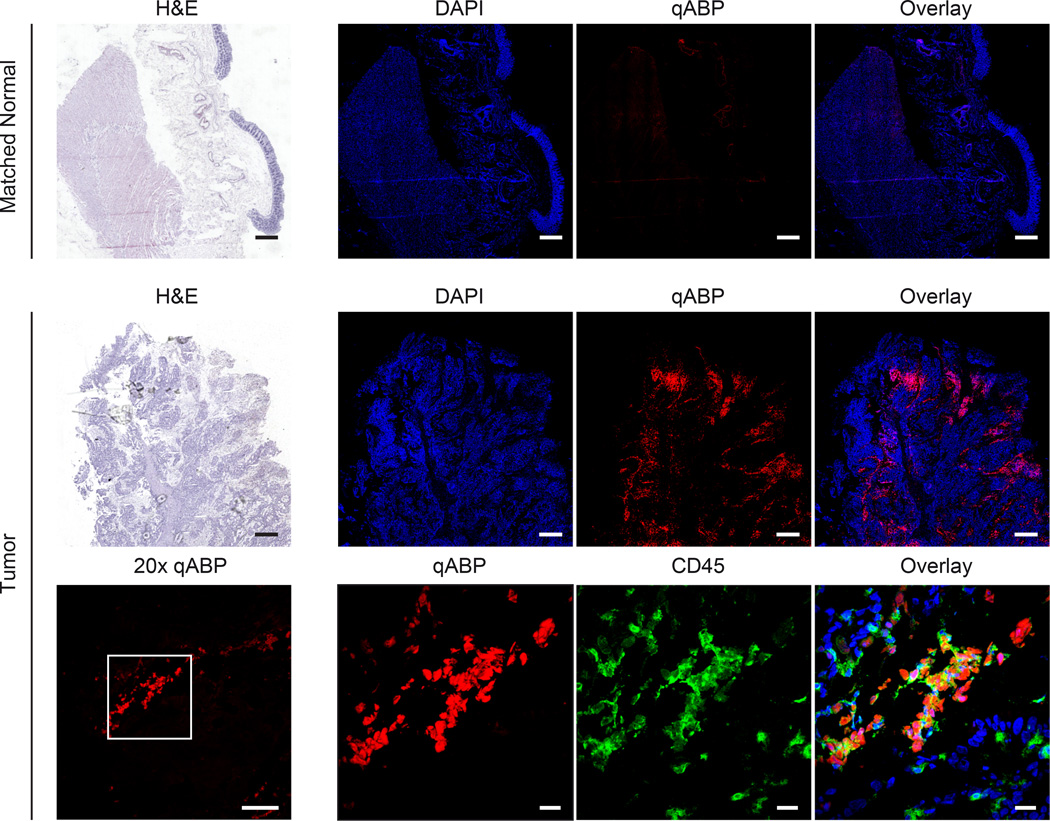
To assess whether the probe labeling of early cancer lesions in mice was relevant to human disease, we tested the probe on human clinical samples. In order to do this, we obtained fresh frozen tissue sections of matched human polyps and healthy surrounding tissue. Because the probe could be applied topically, we directly labeled tissues sections following brief fixation in acetone. We found that the probe very rapidly and selectively labeled cells in tumors. We analyzed a total of five matched polyp and normal tissues by staining with the probe and generated tile scan images, as we had performed for the mouse tissues (Fig. 6). Although the signal intensity and frequency of cells with probe staining varied between samples, there were consistently increased levels of probe fluorescence in tumor tissue compared to the matched healthy surrounding tissues. At a cellular level, the probe was found to show rather diffuse labeling of mononuclear cells, with many of them exhibiting CD45 membrane staining. Similar to results in mouse tissues, these data confirm that the probe is specific for inflammatory cells that are found in cancerous lesions and suggest a great potential for translation to clinical studies.
Figure 6. The qABP probe labels cathepsins in human polyps with similar labeling specificity observed in mouse models.
Bright field and confocal macroscopic mosaic images of matched normal colon tissue and tumor tissue. The top row shows matched normal colon stained with H&E (left panel, scale bar represents 1 mm) and DAPI (blue) or qABP (red) or overlay on right. The middle row shows matched tumor with H&E (left panel, scale bar represents 1 mm) and DAPI (blue) or qABP (red) or overlay on right. In the bottom row, the left panel shows a 20x image (left, scale bar represents 100 µm) of the qABP and 63x images (right, scale bar represents 20 µm) of qABP (red), CD45 (green) or overlay with DAPI (blue) staining of the region designated by the white box on the left.
DISCUSSION
Removal of adenomatous colonic polyps during colonoscopy significantly minimizes the risk of future malignancy and cancer-related death (Zauber et al., 2012). Conventional colonoscopy is limited to the detection of pedunculated and visible mucosal lesions, whereas flat or depressed polyps often escape detection and have the potential to transform to malignant carcinoma in asymptomatic patients (Buie and MacLean, 2008). In addition, patients with chronic inflammatory bowel disease are at increased risk of developing malignancy due to undetected dysplastic lesions (Mayer et al., 1999). Unfortunately, at present there is no clinically approved molecular-based tool to improve detection of colorectal adenomas and adenocarcinomas during optical colonoscopy procedures. In this study, we have validated a novel method for detection of intestinal and colorectal adenomas using a Cy5-labeled qABP with broad-spectrum reactivity for the cysteine cathepsins. These results demonstrate that cysteine cathepsin activity is a reliable biomarker of intestinal neoplasia and demonstrate that our probe can be used to detect intestinal polyps with high sensitivity and specificity when administered either intravenously or intrarectally. The fluorescent signal generated by the bound probe correlates with polyp size and allows detection of adenomas from matched surrounding non-neoplastic mucosa. These results demonstrate that the qABP is a promising tool for improving detection of colorectal adenomas using colonoscopy.
To enhance detection of colorectal adenomas, several optical techniques have been developed including chromoendoscopy, auto-fluorescence imaging and narrow-band imaging (Hurlstone and Sanders, 2006). While they are valuable additions to the imaging toolbox, the readouts of these methods are subjected to image interpretation by operator and cannot be applied to determine polyp histology. Moreover, there have been technical issues with false positives in patients with inflammatory bowel disease and misinterpretation of mucosal folds forming shadows in healthy subjects (Taylor et al., 2007). Targeted molecular labeling has the potential to overcome these challenges by probing for specific biomarkers that distinguish dysplastic and neoplastic lesions from healthy tissue.
Over the last decade numerous biomarkers have been reported to play an important role in colorectal cancer. Examples of promising biomarkers for colorectal cancer with suitable characteristics such as high expression rates, specificity and high signal-to-background ratio include CXCR4, MMPs, EGFR, VEGF-A, Muc-1, EpCAM (van Oosten et al., 2011). Recent studies described an essential role of cysteine cathepsins in colorectal cancer and a strong correlation between cathepsin activity and clinical and pathological features (Kuester et al., 2008). More specifically, cathepsins B, D, L, S and X are generally upregulated in colorectal carcinomas both in epithelial cells and in tumor supporting cells such as macrophages, fibroblasts and endothelial cells (Herszenyi et al., 2008; Kirana et al., 2012; Sebzda et al., 2005). In agreement with these studies, we show strong labeling of cathepsins X, B, S and L in samples of adenomas from APCmin/+ and AOM/DSS mice treated with the qABP. Cathepsin S has been suggested as a relevant biomarker in colorectal cancer and is expressed in 95% of cases of primary human colorectal tumors (Gormley et al., 2011). Furthermore, cathepsin S has significantly higher expression rates in colorectal tumors compared with matched healthy surrounding colonic mucosa. Here, we report that in samples from AOM/DSS mice intravenously treated with the probe, we observed strong labeling of cathepsin S both in adenomas and in matched normal intestinal mucosa, whereas in AOM/DSS mice locally treated with the probe, we observed strong labeling of cathepsin S in adenomas but not in surrounding intestinal mucosa. Since cathepsin S is mostly expressed by macrophages, it is possible that when administered intravenously, the probe is more accessible to macrophages than when administered locally due to only partial penetration through the colonic mucosal layer. This might also explain the improved sensitivity and specificity that we achieved in the AOM/DSS model when the probe was administered locally to the colon by intra-rectal injection.
Several optical molecular probes with different functional characteristics for detection of colorectal adenomas have been described previously (Sheth and Mahmood, 2010). These include non-specific, targeted, and activatable probes, such as MMPSense 680, which is a near-infrared substrate-based probe that labels a broad array of MMP enzymes to detect early colorectal adenomas (Clapper et al., 2011). Optimal signal-to-noise ratio was achieved 66 hours after administration of the probe to Apcmin/+ mice and sensitivity and specificity, were 67% and 97%. Other examples include a targeted heptapeptide for colonic dysplasia that was isolated using a phage library screening approach (Hsiung et al., 2008), a near-infrared octapeptide for dysplastic lesions (Miller et al., 2011) and ProSense 680 that was used to target cysteine cathepsins upregulated in colon polyps (Gounaris et al., 2008). Even though these probes accumulate at colorectal lesions, the long pretreatment times needed to achieve optimal signal-to-background, combined with the relatively low sensitivity and specificity, hamper their use in the clinical setting.
SIGNIFICANCE
Colon cancer remains one of the most treatable forms of cancer if detected early. Currently, screening by endoscopic methods can virtually eliminate the mortality associated with this disease yet it is currently not possible to achieve comprehensive screening of all adults over the age of 50. Therefore, simple methods to detect early stage lesions will eventually enable rapid and simple identification of those at risk for having this disease so that more advanced interventional methods can be initiated. Here, we described a highly selective Cy5-labeled qABP that binds covalently to the catalytic active site of cysteine cathepsin proteases. These proteases are highly expressed in immune inflammatory cells that infiltrate into a nascent tumor tissue thus making them ideal imaging biomarkers. In contrast to the substrate-based probes that have been used to image protease activities in various forms of cancer, the qABP presented here generates fluorescence rapidly (<1 hour) and labels intestinal polyps in both mouse models of colon cancer and in human polyp tissues with high sensitivity and selectivity. Furthermore, the probe is effective when administered directly to the colon, thereby reducing accumulation in other organs. These unique functional characteristics make this qABP a clinically promising tool that could be used for detection of colonic polyps either directly in combination with conventional colonoscopy or for prescreening that could allow identification of at-risk patients in need of interventional procedures. The studies presented here serve as proof of concept studies that should enable advancement of the current optical probe into clinical trials for these applications.
EXPERIMENTAL PROCEDURES
General
Unless otherwise noted, all reagents were purchased from commercial suppliers and used without further purifications. All solvents used were HPLC grade. Fluorescent gels were scanned using a Typhoon 9400 flatbed laser scanner (GE Healthcare).
Animal models
For Genetic mouse model of intestinal cancer we used C57BL/6J–ApcMin/J (APCmin/+) strain that susceptible to spontaneous intestinal adenoma formation (Jackson Laboratory). Eight to ten weeks male APCmin/+ mice were injected intravenously with 10 nmol of the probe. One hour later mice were euthanized and intestines were dissected, flushed with PBS and processed for further analysis. Inflammation-related mouse colon carcinogenesis model was established as previously described (Neufert et al., 2007). Briefly, 6- to 8-week-old mice were injected i.p. with AOM (Sigma-Aldrich) at a concentration of 10 mg/kg. On day 5 after AOM injection, mice were treated with 2% DSS (MW, 36,000–50,000 Da; MP Biomedicals) in drinking water for 5 consecutive days, which was followed by 16 days of regular drinking water administration. This DSS treatment was repeated for two additional cycles. During the course of the experiment, mice were monitored for body weight, diarrhea, and macroscopic bleeding. On day 35 of the regime, mice were injected intravenously or intra-rectally with 10nmol of the probe. One hour later mice were euthanized and colons were dissected, flushed with PBS and processed for further analysis. For inhibitor studies, 100 mg/kg of K11777 or vehicle were injected intra-peritoneally twice daily in 40% DMSO/sterile PBS in a total volume of 100 µl each day for 5 days before probe administration.
Ex vivo imaging of colons and intestines
Mice were euthanized and intestines or colons were removed, flushed with PBS and opened longitudinally. Colons or intestines were imaged using IVIS 100 system with a Cy5.5 filter (Xenogen). Images were evaluated using Living Image software (PerkinElmer). Macroscopic polyp diameters was measured by caliper and signal intensity/mm/ms (exposure time) of labeled polyps was calculated using IVIS software.
SDS PAGE gel analysis
Samples were collected and sonicated (1 min on ice) in citrate buffer (50mM Citrate buffer pH 5.5, 5 mM DDT, 0.5% CHAPS, 0.1% Triton X). After centrifugation at 4 degree C for 30 min, the supernatants were collected, and protein concentration was determined using a BCA kit (pierce). Protein (40 µg total) was denatured in SDS-sample buffer for 2 min at 100 °C and analyzed. Samples were resolved by SDS-PAGE (15%) and labeled proteases were visualized by scanning the gel with a Typhoon fluorescence imager (GE Healthcare).
Gel staining with Coomassie® Blue
SDS PAGE gels were stained with 0.1% Coomassie® Blue Staining (Life Technologies™) in 10% acetic acid, 50% methanol, and 40% H2O for the minimum time (typically one hour) necessary to visualize the bands with shaking and at least three solvent changes to ensure adequate removal of SDS. Gels were then de-stained by soaking for at least 2 hours in 10% acetic acid, 50% methanol and 40% H2O with at least two changes of this solvent.
Immunohistochemistry
Immunohistochemistry of intestine samples and polyps was performed using 5 µm thick formalin-fixed, paraffin-embedded tissue sections. Paraffin sections were de-paraffinized, rehydrated, and stained by hematoxylin and eosin (H & E). For immunofluorescence, intestines were flushed with 2% paraformaldehyde and then fixed for 2 hours in 2% paraformaldehyde at 4°C. Tissues were then transferred to 30% sucrose solution and incubated overnight at 4°C for cryopreservation. Tissues were washed with 50% OCT medium in PBS prior to embedding in OCT and freezing. 6-µm sections cut and were fixed in acetone, blocked with 5% goat serum in 1% BSA/PBS followed by incubation with FITC rat anti-mouse CD45 antibody (1:200; Biolegend) for 1 hour at room temperature. AlexaFluor488 goat-anti FITC (1:1000; Invitrogen) was incubated for 30 minutes at room temperature in order to improve FITC signal. Sections were then stained with DAPI (2µg/mL; Invitrogen) for five minutes and then mounted in ProLong Gold Mounting Medium (Invitrogen).
Freshly isolated human tissues were frozen in OCT prior to sectioning. 6-µm sections were fixed for 10 minutes in acetone at −20°C, sections were blocked in 1% bovine serum albumin with 5% goat serum and then stained for 1 hour with 1µM BMV109 in citrate buffer (50mM citrate buffer pH 5.5, 5 mM DDT, 0.5% CHAPS, 0.1% Triton X) and 1:25 diluted FITC mouse anti-human CD45 antibody (BD) at room temperature. Slides were washed and FITC signal was amplified using 1:2000 diluted Alexa Fluor 488-conjugated goat anti-FITC antibody (Life Technologies) for 20 minutes at room temperature.
Confocal microscopy
Localization of the probe in mouse intestines was imaged using Zeiss LSM 700 confocal imaging systems equipped with 63× oil objective and motorized stage. All images were taken using a multi-track channel acquisition to prevent emission cross-talk between fluorescent dyes. Single XY, XZ plane-images were acquired in 1024×1024 resolution. Images were processed as separate channels using Huygens® deconvolution software or ImageJ and merged as a single image. Mosaic images of H&E and fluorescence labeling were taken using 20x objective and stitched using 15% overlay.
Sensitivity and specificity analysis
Brightfield and fluorescence ex vivo images of colons and intestines were acquired as described above. The number and size of polyps and matched fluorescence intensity were analyzed using ImageJ analysis software. First, the threshold was set for each grayscale binary image. True scale was set based on the scale bar of the images acquired by IVIS 100 system. The number of polyps was quantified using ‘Analyze particles’ command and “Show Masks” was selected to display a drawing of the detected objects. Fluorescence intensity for each individual polyp was calculated by the analysis of the gray levels across the entire colons or intestines. Information of signal intensity, polyp number and diameter was used for sensitivity and specificity analysis. Calculations were based on the following definitions: (a) true positives are polyps correctly labeled, (b) false positives are healthy tissue incorrectly labeled, (c) true negatives are healthy tissue not labeled and (d) false negatives are polyps incorrectly not labeled. We used common equations for calculation of sensitivity a/(a+c), specificity d/(b+d), positive likelihood ratio [a/(a+c)]/[b/(b+d)], negative likelihood ratio [c/(a+c)]/[d/(b+d)], positive predictive value a/(a+b), negative predictive value d/(c+d).
Statistical analysis
Statistical analysis was performed using Microsoft Excel, and s.e.m. was calculated by dividing the s.d. by the square root of n. Statistical significance was determined using an unpaired t-test. P<0.05 was considered statistically significant. All statistical tests were two-sided.
Animal protocol approval
All animal care and experimentation was conducted in accord with current National Institutes of Health and Stanford University Institutional Animal Care and Use Committee guidelines under protocol APLAC-18026.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Yaara Segal for her help with the graphics and illustrations and Tina Oresic Bender for her help with SDS PAGE gels analysis. This work was supported by NIH grants EB005011 (to M.B.), T32GM007365 with additional support from Stanford School of Medicine and its Medical Scientist Training Program (to TRP), 5 U01 CA141468 and 1 R01 CA163441 (to EGE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
ES, TRP, YC, NB, EE and MB Conceived and designed experiments. WAL and MV synthesized materials used in the studies. ES, TRP, YC and NW performed the in vitro experiments. ES, TRP, YC, NB and NW performed the in vivo experiments:. ES, TRP, NW, EE and MB analyzed the data. ES, TRP, WAL, NB, EE and MB contributed reagents/materials/analysis tools: ES, TRP, YC, NB, AH, EE and MB wrote the paper.
REFERENCES
- Amersi F, Agustin M, Ko CY. Colorectal cancer: epidemiology, risk factors, and health services. Clin Colon Rectal Surg. 2005;18:133–140. doi: 10.1055/s-2005-916274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch A, Jeffery DA, Bogyo M. Enzyme activity--it’s all about image. Trends Cell Biol. 2004;14:29–35. doi: 10.1016/j.tcb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nature Chemical Biology. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nature Chemical Biology. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- Blumenstein I, Tacke W, Bock H, Filmann N, Lieber E, Zeuzem S, Trojan J, Herrmann E, Schroder O. Prevalence of colorectal cancer and its precursor lesions in symptomatic and asymptomatic patients undergoing total colonoscopy: results of a large prospective, multicenter, controlled endoscopy study. Eur J Gastroenterol Hepatol. 2013;25:556–561. doi: 10.1097/MEG.0b013e32835d1ef4. [DOI] [PubMed] [Google Scholar]
- Bogdanov AA, Mazzanti ML. Fluorescent macromolecular sensors of enzymatic activity for in vivo imaging. Prog Mol Biol Transl Sci. 2013;113:349–387. doi: 10.1016/B978-0-12-386932-6.00009-0. [DOI] [PubMed] [Google Scholar]
- Boonacker E, Van Noorden CJ. Enzyme cytochemical techniques for metabolic mapping in living cells, with special reference to proteolysis. J Histochem Cytochem. 2001;49:1473–1486. doi: 10.1177/002215540104901201. [DOI] [PubMed] [Google Scholar]
- Buie WD, MacLean AR. Polyp surveillance. Clin Colon Rectal Surg. 2008;21:237–246. doi: 10.1055/s-0028-1089938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TI, Wong JM, Hong CF, Cheng SH, Cheng TJ, Shieh MJ, Lin YM, Tso CY, Huang AT. Colorectal cancer screening in asymptomaic adults: comparison of colonoscopy, sigmoidoscopy and fecal occult blood tests. J Formos Med Assoc. 2002;101:685–690. [PubMed] [Google Scholar]
- Clapper ML, Hensley HH, Chang WC, Devarajan K, Nguyen MT, Cooper HS. Detection of colorectal adenomas using a bioactivatable probe specific for matrix metalloproteinase activity. Neoplasia. 2011;13:685–691. doi: 10.1593/neo.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Cummings RT, Salowe SP, Cunningham BR, Wiltsie J, Park YW, Sonatore LM, Wisniewski D, Douglas CM, Hermes JD, Scolnick EM. A peptide-based fluorescence resonance energy transfer assay for Bacillus anthracis lethal factor protease. Proc Natl Acad Sci U S A. 2002;99:6603–6606. doi: 10.1073/pnas.062171599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter JL, Cohen NT, Wang J, Sloan AE, Cohen AR, Panneerselvam A, Schluchter M, Blum G, Bogyo M, Basilion JP. Topical application of activity-based probes for visualization of brain tumor tissue. PLoS One. 2012;7:e33060. doi: 10.1371/journal.pone.0033060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deu E, Verdoes M, Bogyo M. New approaches for dissecting protease functions to improve probe development and drug discovery. Nat Struct Mol Biol. 2012;19:9–16. doi: 10.1038/nsmb.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington LE, Verdoes M, Bogyo M. Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Curr Opin Chem Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonovic M, Bogyo M. Activity based probes for proteases: applications to biomarker discovery, molecular imaging and drug screening. Curr Pharm Des. 2007;13:253–261. doi: 10.2174/138161207779313623. [DOI] [PubMed] [Google Scholar]
- Gormley JA, Hegarty SM, O’Grady A, Stevenson MR, Burden RE, Barrett HL, Scott CJ, Johnston JA, Wilson RH, Kay EW, et al. The role of Cathepsin S as a marker of prognosis and predictor of chemotherapy benefit in adjuvant CRC: a pilot study. Br J Cancer. 2011;105:1487–1494. doi: 10.1038/bjc.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris E, Martin J, Ishihara Y, Khan MW, Lee G, Sinh P, Chen EZ, Angarone M, Weissleder R, Khazaie K, et al. Fluorescence endoscopy of cathepsin activity discriminates dysplasia from colitis. Inflammatory bowel diseases. 2013;19:1339–1345. doi: 10.1097/MIB.0b013e318281f3f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris E, Tung CH, Restaino C, Maehr R, Kohler R, Joyce JA, Ploegh HL, Barrett TA, Weissleder R, Khazaie K. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PLoS One. 2008;3:e2916. doi: 10.1371/journal.pone.0002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszenyi L, Farinati F, Cardin R, Istvan G, Molnar LD, Hritz I, De Paoli M, Plebani M, Tulassay Z. Tumor marker utility and prognostic relevance of cathepsin B, cathepsin L, urokinase-type plasminogen activator, plasminogen activator inhibitor type-1, CEA and CA 19-9 in colorectal cancer. BMC Cancer. 2008;8:194. doi: 10.1186/1471-2407-8-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszenyi L, Plebani M, Carraro P, De Paoli M, Roveroni G, Cardin R, Tulassay Z, Naccarato R, Farinati F. The role of cysteine and serine proteases in colorectal carcinoma. Cancer. 1999;86:1135–1142. doi: 10.1002/(sici)1097-0142(19991001)86:7<1135::aid-cncr6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, et al. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–458. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, Vats D, Vizovisek M, Kramer L, Germanier C, Wendt KU, Rudin M, Turk B, Plettenburg O, Schultz C. In vivo imaging of mouse tumors by a lipidated cathepsin S substrate. Angewandte Chemie. 2014;53:7669–7673. doi: 10.1002/anie.201310979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlstone DP, Sanders DS. Recent advances in chromoscopic colonoscopy and endomicroscopy. Curr Gastroenterol Rep. 2006;8:409–415. doi: 10.1007/s11894-006-0027-3. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirana C, Shi H, Laing E, Hood K, Miller R, Bethwaite P, Keating J, Jordan TW, Hayes M, Stubbs R. Cathepsin D Expression in Colorectal Cancer: From Proteomic Discovery through Validation Using Western Blotting, Immunohistochemistry, and Tissue Microarrays. Int J Proteomics. 2012;2012:245819. doi: 10.1155/2012/245819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuester D, Lippert H, Roessner A, Krueger S. The cathepsin family and their role in colorectal cancer. Pathol Res Pract. 2008;204:491–500. doi: 10.1016/j.prp.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Lipkin M. Biomarkers of increased susceptibility to gastrointestinal cancer: new application to studies of cancer prevention in human subjects. Cancer Res. 1988;48:235–245. [PubMed] [Google Scholar]
- Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- Mahmood U, Tung CH, Bogdanov A, Jr, Weissleder R. Near-infrared optical imaging of protease activity for tumor detection. Radiology. 1999;213:866–870. doi: 10.1148/radiology.213.3.r99dc14866. [DOI] [PubMed] [Google Scholar]
- Mayer R, Wong WD, Rothenberger DA, Goldberg SM, Madoff RD. Colorectal cancer in inflammatory bowel disease: a continuing problem. Dis Colon Rectum. 1999;42:343–347. doi: 10.1007/BF02236351. [DOI] [PubMed] [Google Scholar]
- Miller SJ, Joshi BP, Feng Y, Gaustad A, Fearon ER, Wang TD. In vivo fluorescence-based endoscopic detection of colon dysplasia in the mouse using a novel peptide probe. PLoS One. 2011;6:e17384. doi: 10.1371/journal.pone.0017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- O’Brien MA, Daily WJ, Hesselberth PE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, Wood KV. Homogeneous, bioluminescent protease assays: caspase-3 as a model. J Biomol Screen. 2005;10:137–148. doi: 10.1177/1087057104271865. [DOI] [PubMed] [Google Scholar]
- Olson ES, Whitney MA, Friedman B, Aguilera TA, Crisp JL, Baik FM, Jiang T, Baird SM, Tsimikas S, Tsien RY, et al. In vivo fluorescence imaging of atherosclerotic plaques with activatable cell-penetrating peptides targeting thrombin activity. Integr Biol (Camb) 2012;4:595–605. doi: 10.1039/c2ib00161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman LE, Bogyo M. Activity-based profiling of proteases. Annu Rev Biochem. 2014;83:249–273. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- Saravanakumar G, Jo DG, Park JH. Polysaccharide-based nanoparticles: a versatile platform for drug delivery and biomedical imaging. Curr Med Chem. 2012;19:3212–3229. doi: 10.2174/092986712800784658. [DOI] [PubMed] [Google Scholar]
- Sebzda T, Saleh Y, Gburek J, Andrzejak R, Gnus J, Siewinski M, Grzebieniak Z. Cathepsin D expression in human colorectal cancer: relationship with tumour type and tissue differentiation grade. J Exp Ther Oncol. 2005;5:145–150. [PubMed] [Google Scholar]
- Sheth RA, Mahmood U. Optical molecular imaging and its emerging role in colorectal cancer. Am J Physiol Gastrointest Liver Physiol. 2010;299:G807–G820. doi: 10.1152/ajpgi.00195.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde R, Perkins J, Contag CH. Luciferin derivatives for enhanced in vitro and in vivo bioluminescence assays. Biochemistry. 2006;45:11103–11112. doi: 10.1021/bi060475o. [DOI] [PubMed] [Google Scholar]
- Taylor JC, Kendall CA, Stone N, Cook TA. Optical adjuncts for enhanced colonoscopic diagnosis. Br J Surg. 2007;94:6–16. doi: 10.1002/bjs.5628. [DOI] [PubMed] [Google Scholar]
- Terai T, Nagano T. Fluorescent probes for bioimaging applications. Curr Opin Chem Biol. 2008;12:515–521. doi: 10.1016/j.cbpa.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Troy AM, Sheahan K, Mulcahy HE, Duffy MJ, Hyland JM, O’Donoghue DP. Expression of Cathepsin B and L antigen and activity is associated with early colorectal cancer progression. Eur J Cancer. 2004;40:1610–1616. doi: 10.1016/j.ejca.2004.03.011. [DOI] [PubMed] [Google Scholar]
- van Oosten M, Crane LM, Bart J, van Leeuwen FW, van Dam GM. Selecting Potential Targetable Biomarkers for Imaging Purposes in Colorectal Cancer Using TArget Selection Criteria (TASC): A Novel Target Identification Tool. Transl Oncol. 2011;4:71–82. doi: 10.1593/tlo.10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoes M, Oresic Bender K, Segal E, van der Linden WA, Syed S, Withana NP, Sanman LE, Bogyo M. Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J Am Chem Soc. 2013;135:14726–14730. doi: 10.1021/ja4056068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber AG, Winawer SJ, O’Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.