Abstract
Extracellular superoxide dismutase (SOD3), which catalyzes the dismutation of superoxide anions to hydrogen peroxide at the cell membranes, regulates the cellular growth in a dose-dependent manner. This enzyme induces primary cell proliferation and immortalization at low expression levels whereas it activates cancer barrier signaling through the p53-p21 pathway at high expression levels, causing growth arrest, senescence, and apoptosis. Because previous reports suggested that the SOD3–induced reduction in the rates of cellular growth and migration also occurred in the absence of functional p53 signaling, in the current study we investigated the SOD3-induced growth-suppressive mechanisms in anaplastic thyroid cancer cells. Based on our data, the robust over-expression of SOD3 increased the level of phosphorylation of the EGFR, ERBB2, RYK, ALK, FLT3, and EPHA10 receptor tyrosine kinases with the consequent downstream activation of the SRC, FYN, YES, HCK, and LYN kinases. However, pull-down experiments focusing on the small GTPase RAS, RAC, CDC42, and RHO revealed a reduced level of growth and migration signal transduction, such as the lack of stimulation of the mitogen pathway, in the SOD3 over-expressing cells, which was confirmed by MEK1/2 and ERK1/2 Western blotting analysis. Interestingly, the mRNA expression analyses indicated that SOD3 regulated the expression of guanine nucleotide-exchange factors (RHO GEF16, RAL GEF RGL1), GTPase-activating proteins (ARFGAP ADAP2, RAS GAP RASAL1, RGS4), and a Rho guanine nucleotide-disassociation inhibitor (RHO GDI 2) in a dose dependent manner, thus controlling signaling through the small G protein GTPases. Therefore, our current data may suggest the occurrence of dose-dependent SOD3–driven control of the GTP loading of small G proteins indicating a novel growth regulatory mechanism of this enzyme.
Introduction
Cell membrane-bound extracellular superoxide dismutase (SOD3) is one of the three SOD isoenzymes that catalyze the dismutation of superoxide radical (O2 .-) to hydrogen peroxide (H2O2) [1]. Notably, both O2 .- and H2O2 are second messengers in cell signaling [2,3] suggesting that the cellular effects of SOD3 are mediated by the local reduction of the O2 .- concentration and the simultaneous increase of the H2O2 concentration affecting the activation of the cell membrane-bound receptor tyrosine kinases (RTK), with a consequent impact on downstream growth and migration signaling [4,5,6]. Consistent with this hypothesis, several studies have demonstrated that SOD3 regulated growth and survival related signal transduction and gene expression [5,6,7,8,9,10,11], hence emphasizing the importance of this enzyme in growth regulation. Interestingly, SOD3 expression is mildly upregulated in benign tumor model systems whereas it is downregulated in several cancers and in transformed cell lines [12,13] suggesting that the enzyme might be involved in the initiation of benign hyperplasia.
Based on our recent data SOD3 has a dose-dependent effect on cellular proliferation; low expression levels of SOD3 induce GTP loading on small GTPase proteins, mitogen signaling and cell proliferation, whereas high levels of SOD3 activate the DNA-damage response and the p53-p21 growth arrest pathway [5,6]. Although activation of the p53-p21 signal transduction pathway is likely to play a major role in growth limitation, high-dose SOD3–inhibited proliferation was also observed in anaplastic thyroid cancer cells lacking functional p53 [5], indicating the existence of additional growth regulatory mechanisms.
Therefore, in the current study we focused on examining the effect of high-dose SOD3 on cell membrane-associated growth-regulatory molecules to identify the target molecules that translate the enzyme-based signaling to the cellular signal transduction network. According to our current data high-level SOD3 expression activated growth signaling through the cell membrane-bound receptor tyrosine kinases (RTKs) and cytoplasmic non-receptor tyrosine kinases (TKs). However, simultaneously with the increased phosphorylation of RTKs and TKs, high-dose SOD3 decreased the level of expression of a number of growth-promoting genes, downregulated the activation of the RAS, RHO, RAC, and CDC42 small GTPases, and controlled the expression of the small GTPase regulatory genes encoding guanine nucleotide-exchange factors (GEFs), GTPase-activating protein (GAPs) and Rho guanine nucleotide dissociation inhibitor (GDI). These results could therefore clarify how SOD3 controls cellular proliferation and may additionally suggest potential drug targets for reducing carcinogenic growth.
Methods
Cell lines
8505c cells (DSMZ, German collection of microorganism and cell cultures, Braunschweig, Germany) modeling anaplastic thyroid cancer, were cultured in RPMI medium (Sigma, St. Louis, MO, USA) supplemented with 10% FBS. Cell lines stably expressing human SOD3 (kindly provided by Professor Stefan L. Marklund of the University of Umeå, Sweden), human RHO GEF16 (Applied Biological Materials, Richmond, Canada), H-RasV12 or the pcDNA3 GFP control plasmid were utilized. Cell lines were generated by nucleofection of 5 μg of the SOD3 expression plasmid or the control plasmid into 5x105 cells, 5 μg of SOD3/1.5 μg of H-RasV12, and 5 μg of SOD3/1.5 μg of RHO GEF16 transfection into 5x105 cells. The appropriate antibiotic selection was applied 48 hours after transfection and was continued for six weeks to create stable mixed cell populations. N-acetyl-cysteine (NAC) (Sigma) was applied to the 8505c cells at 2.5 mM daily.
Growth analysis
For the growth curve analyses 5x103 cells were seeded in the wells of 6-well plates in triplicate, and were counted daily until the cells reached a maximum of 70% confluence to avoid artifacts caused by cellular overgrowth. For the BrdU cell proliferation cell proliferation analyses, 10 mM bromodeoxyuridine (BrdU) (Roche, Basel, Switzerland) was added to the growth medium for 15 min. Subsequently, the cells were fixed using an ethanol fix solution. The BrdU-positive cells were detected using FITC–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories Inc., West Grove, PA). The nuclei were counter stained using Hoechst (Sigma) staining. Each cell count was performed in triplicate: each point represents the mean value for 3 samples.
Invasive growth in Matrigel
For outgrowth in Matrigel, 1x103 stably transfected SOD3 and control plasmid were mixed with 160 μl of Matrigel (BD Biosciences, San Jose, CA, USA) and were plated in 35 mm dishes containing glass coverslips. After the gel hardened, the cells were overlaid with 2 ml of growth medium and were incubated at 37°C for up to 8 days.
Soft agar assay
GFP control and SOD3 transfected 8505c cells (5x103) were plated in 6 cm gridded dishes with a bottom layer of 1% agarose and a top layer of 0.4% agarose constructed using complete growth medium. The plates were refreshed with complete medium twice a week. The number of colonies in each grid was counted at the 6-week time point. Each count was performed in triplicate.
Protein arrays
Proteins were isolated from 8505c cells stably expressing SOD3 or GFP control. Human phospho-RTK and phospho-kinase arrays (R&D Systems, Minneapolis, MN USA) were performed using 300 μg of protein according to the manufacturer’s instructions. Signal density analysis was performed using ImageJ software GEL blot software.
Pull-down analysis
RAS, RAC, CDC42, and RHO pull-down analysis was performed using cells at 40% confluence in 10 cm dishes. After serum starvation for 24 h, the cells were lysed using ice-cold Rho-lysis buffer containing 20 mM HEPES (pH 7.4), 0.1 M NaCl, 1% Triton X-100, 10 mM EGTA, 40 mM glycerophosphate, 20 mM MgCl2, 1 mM Na3VO4, 1 mM dithiothreitol, a mixture of protease inhibitors, and 1 mM phenylmethylsulfonyl fluoride. The lysates were incubated for 15 min with a purified, bacterially expressed GST-fusion protein containing the CRIB domain of PAK1 (p21 activated kinase) that had been previously bound to glutathione-Sepharose beads, followed by three washes using Rho-lysis buffer. The GTP-bound forms of RAC1 or CDC42 associated with GST-CRIB were quantified through Western blotting analysis. GST-Rhotekin-RBD and GST-RAF1-RBD beads were used for RHO and RAS pull-down assay, respectively, and analyzed by Western blotting.
Western blotting analysis
The cells were homogenized in lysis buffer (50 mM/L HEPES pH 7.5, 150 mM/L NaCl, 10% glycerol, 1% Triton X-100, 1 mM/L EGTA, 1.5 mM/L MgCl2, 10 mM/L NaF, 10 mM/L sodium pyrophosphate, 1mM/L Na3VO4, 10 μg approtinin/ml, 10 μg leupeptin/ml) (Sigma). The following antibodies were used: α-RAS, α-RAC, α-CDC42, α-RHO, α-pEGFR, α-EGFR, α-pFAK, α-FAK, α-pSCR, α-SCR, α-pMEK, α-MEK, α-pERK1/2, α-ERK1/2, α-pAKT, α-AKT, α-pGSK3 α /β, α-GSK3 α /β, α-β catenin, α-pCHK2, α-CHK2, α-tubulin, α-SP1, α-pRET, α-RET (Cell Signaling, Danvers, MA, USA), and α-SOD3 (Santa Cruz, Santa Cruz, CA, USA). Signal density analysis was performed using ImageJ software GEL blot software.
DNA damage assays
For γH2AX staining (Millipore, Billerica, MA, USA) the cells were grown on coverslips, fixed using 4% PFA/PBS for 20 min at room temperature, permeabilized using 0.5% TritonX-100 and incubated with a primary antibody directed against γH2AX for 1h at RT. After several washes and labeling using the secondary antibody (30 min at RT), the cells were counterstained using Hoechst dye and were analyzed using fluorescence microscopy. Cells containing more than ten nuclear foci were considered positive.
Microarray
For the microarray expression analysis, the RNA was extracted from the 8505c cells in triplicate for use in cDNA synthesis. The analysis was performed at the Finnish Microarray and Sequencing Centre (FMSC), Turku Centre for Biotechnology, University of Turku, Finland. In brief, the array analysis was performed using an Illumina Human HT-12 v.3 Expression BeadChip that contained more than 48 000 types of probes according to the instructions of the manufacturer. Washing and scanning were performed according to the instructions in the Illumina BeadStation 500x manual (revision C). The Illumina expression data were extracted using BeadStudio version 1.5.0.34 software applying the default settings. The following quality assessments were applied to the samples: RNA quality control, quintile normalization of the sample data, inspection of the signal-intensity distribution, and sample correlation analysis. The threshold values for positive hits were selected based on the MA plot, and volcano blot comparisons were used to identify list the most differentially expressed array features. Three functional pathway analytical tools, the KEGG pathway signaling (http://www.genome.jp/kegg/pathway.html), GOrilla GO signaling pathway (http://www.ebi.ac.uk/QuickGO/), and GSEA gene enrichment programs were used to connect the gene expression patterns to cellular functions. The statistical analysis was performed using the R/Bioconductor6 open software package. The resulting gene list was filtered according to P G 0.001 (without adjustment for multiple testing). The genes were annotated, and output files were created using the biomaRt package. The selected positive hits were double-checked using quantitative RT-PCR.
Gene expression analysis using real-time RT-PCR
The total RNA was isolated from the cells using an RNeasy minikit (Qiagen, Hilden, Germany). The first strand synthesis was performed using a QuantiTect Reverse Transcription kit (Qiagen) and quantitative PCR was performed using the SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA). The primers are listed in S1 Table.
Analysis of reactive oxygen species
Human 8505c control, 8505c SOD3, 8505c SOD3 Ras, and 8505c SOD3 GEF16 cells were grown on coverslips. Dihydroethidium (Sigma) and MitoTracker RED CMX (Invitrogen, Waltham, MA, USA) were added to the cells in 100 nM final concentrations. Cells were incubated 45 minutes at 37°C and washed with PBS. Images were analyzed with fluorescence microscope.
Mouse transplantation
Human 8505c cells (10x106 cells) that were stably transfected with luciferase or SOD3 were transplanted subcutaneously into two locations of the dorsal portion of female BALB/c nude (nu/nu) mice. The tumor size was assessed using a caliper at regular intervals and the tumor volume was calculated according to the following formula: (LxW 2 )/2 (where L = length and W = width of tumor). The mice were sacrificed four weeks after transplantation and the tumors were isolated for RNA preparation (5). All of the animal procedures were approved by the Southern Finland Regional Experimental Animal Committee (License STH349A), and were performed according to the European Commission guidelines.
Statistical analyses
The experiments were repeated at least three times. All of the results were expressed as the mean values ±SD. The p-values (* = p<0.05, ** = p<0.01, *** = p<0.001) were determined using a one-way ANOVA with a Tukey-Kramer multiple comparison post-analysis test or, when appropriate, using two-tailed independent sample t-tests.
Results
Robust SOD3 over-expression reduced the rates of anaplastic thyroid cancer cell proliferation, invasion, and anchorage independent growth in vitro
We previously showed that robust SOD3 over-expression reduced the rate of cell proliferation by causing DNA damage, inducing the DNA-damage response, and activating the downstream p53-p21 signal transduction pathway of TPC1 papillary thyroid cancer cells, which have an intact growth-arrest signaling pathway. Notably, robust SOD3 over-expression also reduced the rate of cell proliferation of anaplastic thyroid cancer 8505c cells, which lack functional growth-arrest signaling [5]. Therefore, in the current study, we sought novel SOD3–related signal transduction routes and the molecules that could transmit the SOD3-based extracellular signals to the cytoplasmic signaling network.
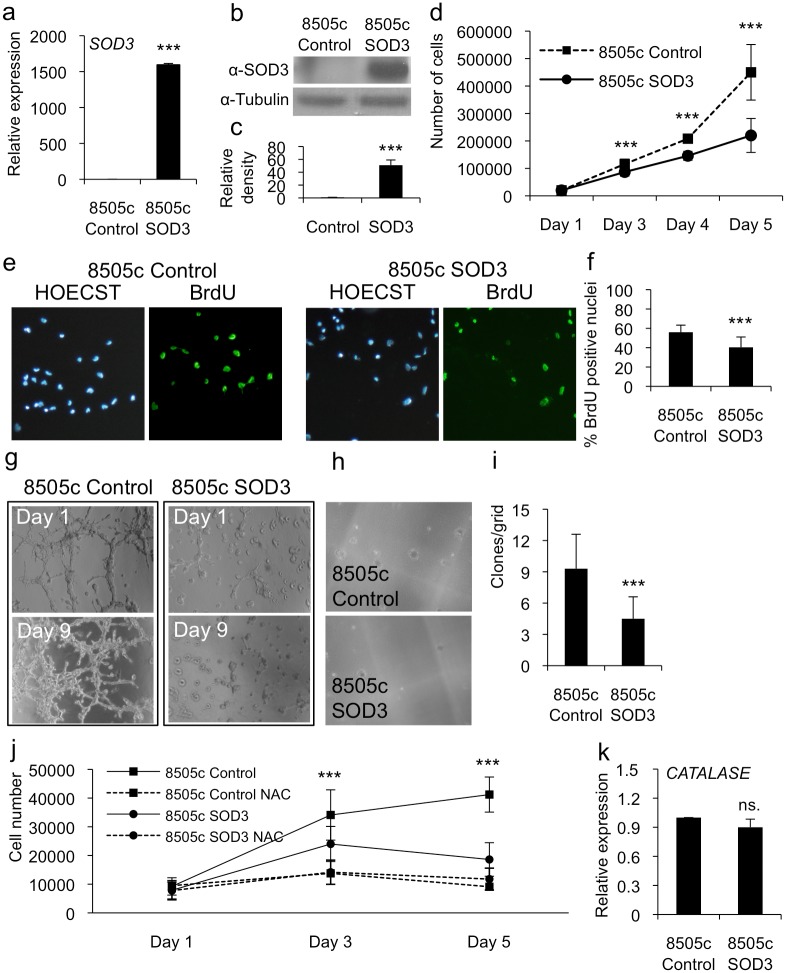
To evaluate the level of SOD3 synthesis, we determined the expression level of transfected SOD3 using real-time RT-PCR and Western blotting which demonstrated high-level mRNA and protein production in anaplastic 8505c cells (Fig. 1a-1c). To evaluate the effect of SOD3 over-expression on biological processes we studied the effect of SOD3 on the aggressiveness of these cancer cells. The growth curve analysis of anaplastic 8505c cells robustly over-expressing SOD3 demonstrated significantly (p<0.001) reduced cell numbers (Fig. 1d) and BrdU positive proliferating nuclei (Fig. 1e and 1f) compared with those of the control cells, consistent with our previous data [5]. Furthermore, the SOD3 over-expressing cells showed a reduced level of invasive growth in Matrigel (Fig. 1g) and colony formation in soft agar (p<0.001) as compared with those of the control cells (Fig. 1h and 1i). The data therefore suggested a reduced aggressiveness of the anaplastic thyroid cancer cells caused by high SOD3 expression.
Fig 1. Robust SOD3 expression reduced anaplastic thyroid cancer cell growth.
(a) Real time RT-PCR analysis showed a high-level mRNA production of SOD3 transgene in anaplastic thyroid cancer 8505c cells. (b,c) Western blotting and related band intensity analysis showed high-level SOD3 production caused by SOD3 transgene. (d) The growth curve analysis suggested significantly (p<0.001) decreased cell numbers in cells transfected with SOD3. (e,f) The BrdU DNA incorporation analysis confirmed significantly (p<0.001) decreased cellular growth caused by SOD3. (g) The 3-D gel invasion assay suggested almost complete lack of invasive capacity of 8505c SOD3 cells. (h,i) The soft agar analysis showed significantly (p<0.001) reduced formation of colonies suggesting decreased anchorage independent growth ability for SOD3 cells as compared to 8505c control vector transfected cells. (j) The growth curve analysis of control and SOD3 cells in the presence of 2.5 mM N-Acetyl-cysteine (NAC) showed growth reduction both in 8505c control and SOD3 expressing cells upon the treatment. (k) CATALASE mRNA expression. The quantitative RT-PCR showed no difference between 8505c control and SOD3 cells in CATALASE expression. Data are expressed as mean ±SD. The p-values (* = p<0.05, ** = p<0.01, *** = p<0.001).
Because SOD3 regulates the availability of superoxide anion and hydrogen peroxide reactive oxygen species, we tested the effect of the general ROS scavenger N-acetyl-cysteine (NAC), which mainly removes superoxide but can also reduce the H2O2 and nitric oxide (NO) concentrations, on cell proliferation. Noteworthy, O2 -, H2O2, and NO derivative peroxynitrate can increase growth by oxidative modification of MEK1/2-ERK1/2 signaling [2,3,14]. The growth curve analytical data showed reduced numbers of 8505c control cells in the presence of 2.5 mM NAC compared with those of the cells grown without NAC. This treatment even decreased the rate of proliferation of 8505c SOD3 over-expressing cells compared with that of untreated 8505c SOD3 cells, emphasizing the role of oxygen and nitrogen free radicals in this process (Fig. 1j). Interestingly, SOD3 over-expression had no impact on catalase expression (Fig. 1k) suggesting differential signal transduction for the enzymes.
SOD3 affected the activation of the cell membrane-associated receptor tyrosine kinases
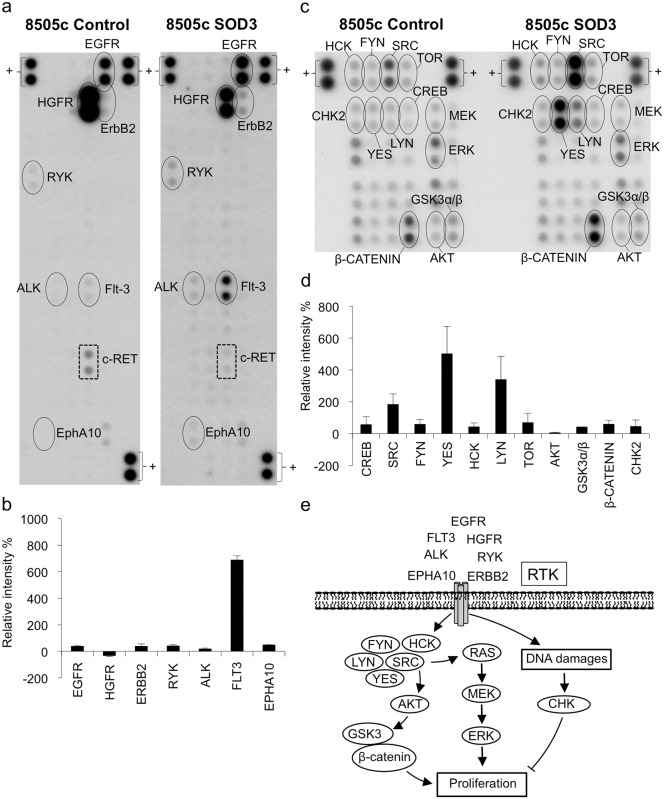
SOD3 is a secreted molecule that interacts reversibly with the cell membrane-associated heparan sulfate proteoglycan molecules, such as collagen [15] and fibulin-5 [16]. To study the effect of high SOD3 expression on the phosphorylation of cell membrane-associated growth-related molecules we analyzed its effect on the activation of receptor tyrosine kinases. The dot intensity analysis of the phospho tyrosine kinase receptor array suggested an increased level of phosphorylation of epithelial growth factor receptor (EGFR), receptor like tyrosine kinase (RYK), anaplastic lymphoma kinase receptor (ALK), fms tyrosine kinase 3 receptor (FLT-3), ephrin A10 receptor (EPHA10), and v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2, HER2) (Fig. 2a and 2b), which are commonly activated in cancer cells [17,18,19,20,21]. In contrast, the level of phosphorylation of HGFR was reduced in the SOD3-positive cells (Fig. 2a and 2b).
Fig 2. The phosphorylation status of receptor tyrosine kinases and cellular kinases in control plasmid and SOD3 transfected cells.
(a,b) A receptor tyrosine kinase phosho-array showed increased levels of phosphorylation of FLT-3, EGFR, ERBB2, RYK, ALK, and EPHA10 in the 8505c SOD3 cells while HGFR activation was downregulated. Note, c-RET is not expressed in 8505c cells and therefore the signal represents unspecific labeling (S1 Fig.). (c,d) Kinase phosphoarray for cytosolic non-receptor kinases and other signaling molecules. SOD3 over-expression increased the activation of CREB, TOR, AKT, GSK3α/β, CHK2, and SCR family members SRC, FYN, YES, HCK, and LYN. Additionally, SOD3 induced β-catenin production. (e) A schematic representation of the main pathways that are affected by robust SOD3 over-expression.
High SOD3 over-expression affected the activation of the AKT/GSK/β-catenin pathway, ERK1/2 pathway, and CHK2 kinase signal transduction pathways
The tyrosine kinase receptors transmit extracellular stimuli to membrane-associated non-receptor tyrosine kinases and small GTPases [22]. To investigate the effect of SOD3 on the activation of intracellular signaling pathways, we evaluated the phosphorylation status of non-receptor tyrosine kinases using a phospho-kinase array. Based on the dot-intensity analysis of an array representing a selection of RTK downstream kinases, robust SOD3 expression induced the increased phosphorylation of SRC (Tyr419), LYN (Tyr397), YES (Tyr426), HCK (Tyr394), and FYN (Tyr420) (Fig. 2c and 2d). SRC kinases are well-characterized proto-oncogenes that mediate the signaling between receptor tyrosine kinases and RAS [23], thereby inducing the activation of the mitogenic pathway, which leads to cellular proliferation. In addition, SRC activation induces the activation of the AKT-GSK3-β-catenin pathway, another major signal transduction pathway that mediates carcinogenic growth. The PI3K/AKT pathway together with the MEK/ERK signaling pathway is a crucial regulator of cell proliferation, survival and invasion. One of the substrates of AKT is the activation of the β-catenin signaling [24]. AKT inactivates the destruction complex by phosphorylating glycogen synthase kinase-3α/β (GSK3α/β) at serines 21 and 9, thereby leading to the accumulation of β-catenin in the cytoplasm [25]. Subsequently, the cytoplasmic β-catenin is translocated into the nucleus, where it interacts with transcription factors to promote cell growth and carcinogenesis [24]. Our results suggest an activation of the AKT-GSK3-β-catenin pathway in SOD3 expressing cells (Fig. 2c, 2d, and 2e). Lastly, consistent with our previous observations [5], the results of the kinase array assay indicated that high SOD3 over-expression led to the increased phosphorylation of CHK2 kinase (Fig. 2c, 2d, and 2e), which is a component of the DNA damage response signal-transduction pathway and is frequently activated due to DNA-strand breaks [26].
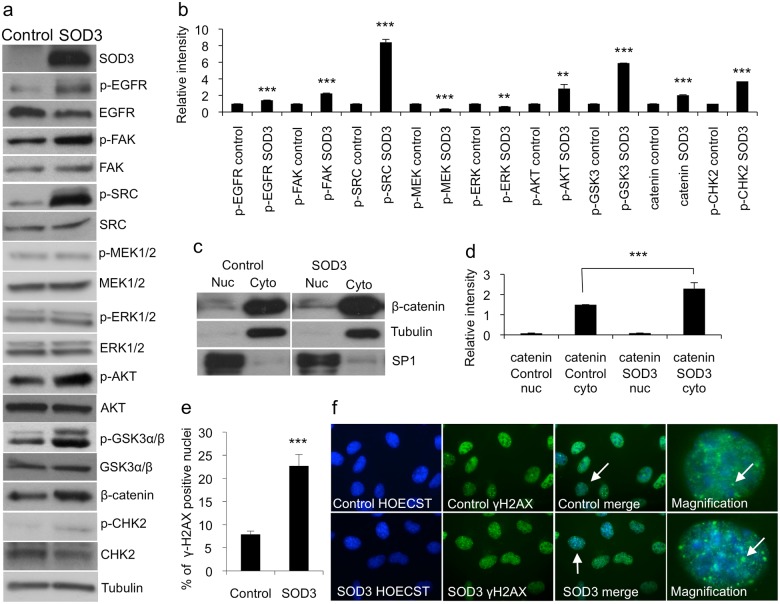
To confirm the SOD3-mediated pathway activation indicated by the results of the protein array assay and to exclude the artifacts caused by nonspecific antibody binding (Fig. 2a and S1 Fig.), we performed Western blotting analysis of SOD3 over-expressing cells and control cells (Fig. 3a and 3b). The analysis confirmed that a robust SOD3 over-expression increased the level of phophorylation of EGFR, FAK, and SRC indicating that it activated cell membrane-associated RTKs and non-receptor tyrosine kinases. Notably, there was no activation of the downstream kinases MEK1/2 and ERK1/2 but rather a modest decrease in the level of their phosphorylation (Fig. 3a and 3b), which was consistent with the reduced levels of cell proliferation (Fig. 1d, 1e, and 1f) and invasive growth (Fig. 1g, 1e, 1h, and 1i) suggesting the existence of a regulatory step in signal transfer between the SRC and MAP kinases.
Fig 3. Western blotting and DNA damage response analysis.
(a,b) Western blotting analyses supported the protein array data showing the increased activation of RTKs and non-receptor kinases in the SOD3-transfected 8505c cells. Note, MEK1/2 and ERK1/2 phosphorylation was moderately downregulated based on the band density analyses. (c,d) Western blotting and the related band intensity analysis for β-catenin nuclear and cytosolic proteins showed significantly (p<0.001) increased β-catenin production in cytosolic compartment. Tubulin was used to normalize cytosolic proteins and SP1 was used to normalize nuclear proteins. (e,f) γH2AX staining for 8505c cells suggested significantly (p<0.001) increased histone H2AX phosphorylation, suggesting DNA damages in 8505c SOD3 cells, which supported the increased CHK2 phosphorylation observed in protoarray and in the Western blotting. The white arrows in the control and the SOD3 merge panels indicate cells that are magnified to detect the nuclear foci. The white arrows in the control and the SOD3 magnification images indicate the nuclear foci. Data are expressed as mean ±SD. The p-values (* = p<0.05, ** = p<0.01, *** = p<0.001).
Our Western blotting data showed the robust activation of the AKT (Thr308) signaling protein in SOD3 over-expressing cells, consistent with our previous observations [8], which led to the phosphorylation of its substrate GSK3α/β at Ser21/9 and the accumulation of β-catenin (Fig. 3a and 3b). Interestingly, the increased amount of cytosolic β-catenin was not accompanied by an increase in the amount of the nuclear fraction (Fig. 3c and 3d), again suggesting the existence of an inhibitory regulation occurring following strong SOD3 over-expression. Lastly, the Western blotting analysis confirmed the increased activation of CHK2 kinase (Fig. 3a and 3b) upon SOD3 over-expression, which was accompanied by increased H2AX histone phosphorylation (γH2AX) (Fig. 3e and 3f).
Robust SOD3 over-expression had a significant impact on proliferation, migration, and invasion related gene expression
To gain a global view of the signaling pathways engaged by SOD3 to cause the reduced growth signaling and consequent reduced rate of cell proliferation, we analyzed the transcriptional alterations that occurred upon SOD3 over-expression by performing a microarray analysis of control plasmid and SOD3 transgene transfected 8505c cells (S2 and S3 Tables).
Analysis of our microarray data revealed that approximately 32% of the SOD3-mediated downregulated and upregulated genes were growth promoters or suppressors (45 of 136 and 20 of 61, respectively), emphasizing the connection between SOD3 and the growth-regulatory genes (Tables 1 and 2). Microarray Gene Ontology (GO) analysis indicated that of the 67 genes involved in the regulation of cell proliferation (GO:0042127), the expression of 24 genes was upregulated and that of 43 genes was downregulated (S4 Table). Of the 33 cell proliferation-related genes (GO:0008283), the expression of 17 genes was upregulated and that of 16 genes was downregulated. In the group of genes involved in the regulation of cell migration (GO:0030334), the expression of 12 of the 27 genes was upregulated, whereas that of the remaining 15 genes was downregulated. Consistent with this result, of the 38 genes in the cell migration group (GO:0016477), the expression of 13 genes was upregulated and that of 25 genes was downregulated. Of the 18 genes in the positive regulation of locomotion group (GO:0040017), the expression of 8 genes was upregulated and that of 10 genes was downregulated. The regulation of cell motility group (GO:2000145) had 25 genes, of which the expression of 11 was upregulated and that of 14 genes was downregulated. Finally, the cell motility group of genes (GO:0048870) included 29 genes, of which 9 showed upregulated expression and 20 showed downregulated expression in the SOD3 over-expressing cells. Thus, the microarray-based functionality analysis indicated that robust SOD3 over-expression mostly downregulated the expression of genes involved in growth and migration.
Table 1. Growth-associated genes showing SOD3-mediated downregulated expression.
| Growth promotors downregulated by SOD3 | ||
|---|---|---|
| Gene | Definition | Reference |
| AKR1C3 | Aldo-keto reductase family 1, member C3 | NM_003739.4 |
| Ang | Angiogenin | NM_001097577.1 |
| ARHGEF16 | Rho guanine exchange factor (GEF) 16 | NM_014448.2 |
| CCNA1 | Cyclin A1 | NM_003914.2 |
| CSF2 | Colony stimulating factor 2 (granulocyte-macrophage) | NM_001781.1 |
| ELF3 | E74-like factor 3 | NM_004433.3 |
| DDIT4/REDD1 | DNA-damage-inducible transcript 4 | NM_019058.2 |
| ERBB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 | NM_001982.2 |
| FOXO1 | Forkhead box O1 | NM_002015.3 |
| GCHFR | GTP cyclohydrolase I feedback regulator | NM_005258.2 |
| HAS3 | Hyaluronan synthase 3 | NM_005329.2 |
| IL1β | Interleukine 1β | NM_000576.2 |
| LTB | Lymphotoxin beta | NM_002341.1 |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | NM_005204.2 |
| MSLN | Mesothelin | NM_013404.3 |
| NUAK2 | SNF1-like kinase, 2 | NM_030952.1 |
| NOX5 | NADPH oxidase 5 | NM_024505.2 |
| PDGFRL | Platelet-derived growth factor receptor-like | NM_006207.1 |
| RGL1 | Ral guanine nucleotide dissociation stimulator-like 1 | NM_015149.3 |
| PLAT | Plasminogen activator, tissue | NM_000930.2 |
| SCD | Stearoyl-CoA desaturase | NM_005063.4 |
| PRSS7 | Protease serine 7 (enterokinase) | NM_002772.1 |
| RIPK4 | Receptor-interacting serine-threonine kinase 4 | NM_020639.2 |
| RRAD | Ras-related associated with diabetes | NM_004165.1 |
| VEGFA | Vascular endothelial growth factor A | NM_001025367.1 |
| VCAM1 | Vascular cell adhesion molecule 1, CD106 | NM_001078.2 |
| WNT7A | Wingless-type MMTV integration site family, member 7A | NM_004625.3 |
Table 2. Growth-associated genes showing SOD3-mediated upregulated expression.
| Growth promotors upregulated by SOD3 | ||
|---|---|---|
| Gene | Definition | Reference |
| ABCE1 | ATP-binding cassette, sub-family E,1 | NM_001040876.1 |
| CCND2 | Cyclin D2 | NM_018404.2 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | NM_001554.3 |
| CTPS | CTP synthase | NM_001905.1 |
| EEF1A2 | Elongation factor 1 alpha 2 | NM_001958.2 |
| ID1 | Inhibitor of DNA binding 1 | NM_181353.1 |
| ID3 | Inhibitor of DNA binding 3 | NM_002167.2 |
| NMU | Neuromedin U | NM_006681.1 |
| PLAC8 | Placenta-specific 8 | NM_016619.1 |
To validate the microarray data we selected 18 genes involved in growth, migration, and invasion (S2 Fig.) and confirmed using real-time RT-PCR the SOD3-mediated alteration of gene expression. To cluster the genes involved in signal transduction pathways, we applied the categories of the Kyoto Encyclopedia of Genes and Genomes (KEGG) finding 254 genes related to mitogen activated protein kinase (MAPK) signal transduction among the SOD3-regulated transcripts (Table 3). Accordingly, the expression of 312 genes related to oncogenic pathways was affected upon SOD3 over-expression. Consistent with the results of the KEGG analysis, the stimulated growth-related signaling cascades in SOD3 over-expressing cells that were identified by searching the Gene Ontology project (GO) indicated that cell proliferation and mitogen regulated pathways play a central role in the response to SOD3 over-expression (Table 3). Finally, Gene Ontology (GO) analysis of the microarray data revealed the SOD3-mediated increased expression of the WWTR1 and SNAI2 genes, which are responsible for β-catenin cytoplasmic arrest [26] and binding [27], respectively, in addition to the SOD3-mediated decreased expression of AXIN2, which drives the degradation of cytoplasmic β-catenin [28] (Table 3). These data supported the results of the protein array and Western blotting analyses, which showed an increased level of β-catenin protein in the cytoplasm of SOD3 over-expressing cells (Fig. 3c and 3d).
Table 3. The results of the KEGG, GO, and GOrilla functional analysis.
| KEGG signaling pathway analysis | |||
|---|---|---|---|
| Pathway | p-value | Genes | Pathway description |
| 4010 | 0.02 | 254 | MAPK signaling pathway |
| 5200 | 0.023 | 312 | Pathways in cancer |
| 5222 | 0.026 | 78 | Small lung cancer |
| 5215 | 0.027 | 83 | Prostate cancer |
| 5219 | 0.029 | 39 | Bladder cancer |
| 4012 | 0.047 | 83 | ErbB signaling pathway |
| 5210 | 0.048 | 81 | Colorectal cancer |
The data indicated that SOD3 played a role in the regulation of the expression of genes involved in cell-proliferative pathways.
Robust SOD3 over-expression modified the expression of the small GTPase regulatory GEF, GAP and GDI mRNAs
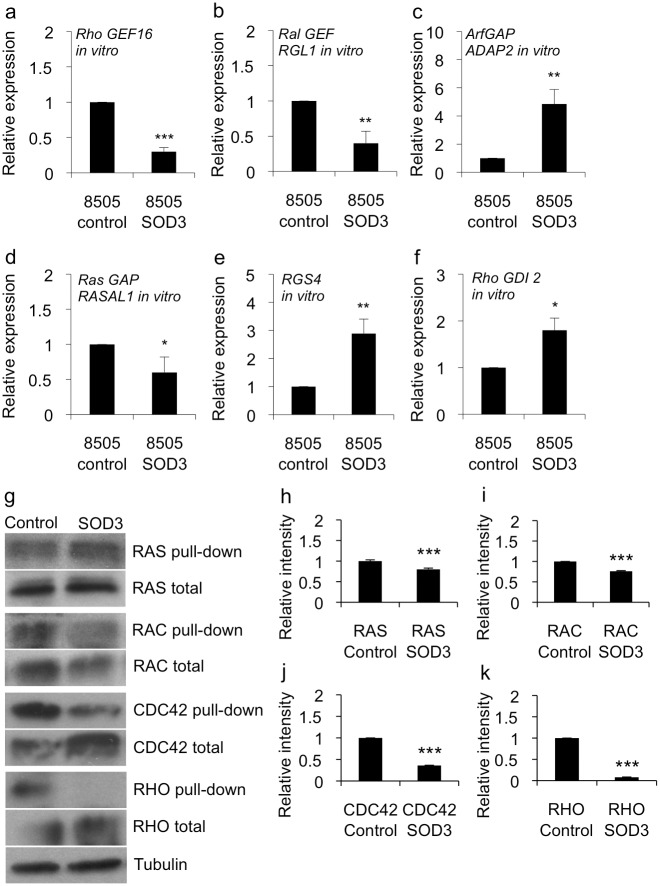
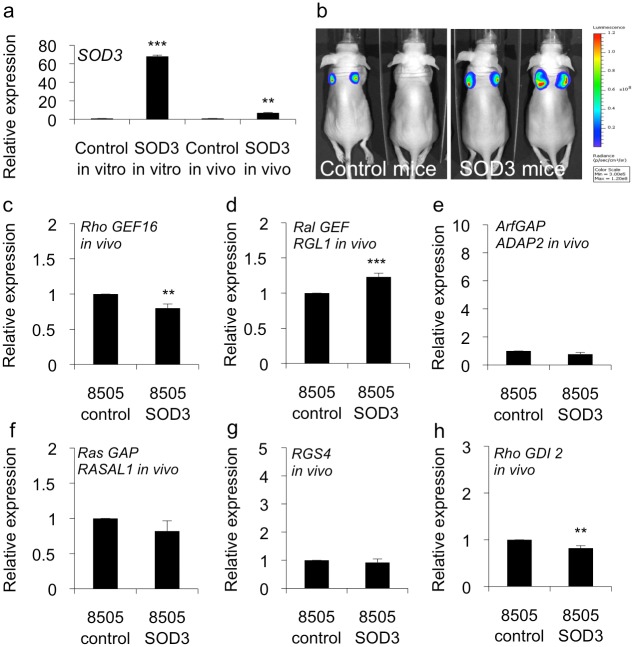
Interestingly, our microarray data revealed changes in the levels of expression of guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs), and a Rho guanine nucleotide dissociation inhibitor (GDI) that were affected by SOD3 over-expression (Fig. 4a, 4b, 4c, 4e, and 4f). GEFs are involved in the exchange of GDP for GTP, which activates members of the small G-protein superfamily of GTPases, whereas GAPs terminate the active state by inducing GTP hydrolysis [29,30]. GDIs in turn inhibit the dissociation of GDP from small G-protein GTPases thereby maintaining these molecules in their inactive form [31]. Robust in vitro SOD3 expression downregulated the level of transcription of Rho guanine exchange factor 16 (RHO GEF16), Ral guanine nucleotide dissociation stimulator-like 1 (RGL1), and Ras protein activator like 1 (GAP1 like) (RASAL1) genes. Among the genes with upregulated mRNA expression in the SOD3 over-expressing cells, the most relevant GTPase-associated genes were RHO GDP dissociation inhibitor beta (RHO GDI2), ArfGAP with dual PH domains 2 (ADAP2), and Regulator of G-protein signaling 4 (RGS4), the latter one having also GAP activity (Fig. 4a, 4b, 4c, 4d, 4e, and 4f).
Fig 4. SOD3 regulated the expression of small GTPase regulatory genes and affected the activation of small GTPases.
(a-f) Quantitative RT-PCR for GTPase superfamily regulatory genes. The panels show the expression levels of the regulatory genes in vitro cultured cells containing high-level SOD3 mRNA and reduced growth abilities. (g-k) Pull-down assay for small GTPase proteins RAS, RAC, CDC42, and RHO. The assay suggested significantly (p<0.001) reduced GTP loading to each tested GTPase therefore being in line with reduced cell proliferation, migration, and growth related signal transduction. Data are expressed as mean ±SD. The p-values (* = p<0.05, ** = p<0.01, *** = p<0.001).
To link these data to the activation of small G proteins, we performed pull-down assays to evaluate the status of the small GTPase RAS, RAC, CDC42, and RHO, which showed a reduced level of small GTPase activation in the SOD3 over-expressing cells (Fig. 4g, 4h, 4i, 4j, and 4k) supporting the observed decreased activation of the MEK1/2 and ERK1/2 mitogen activated signaling pathway and the decreased rate of cellular proliferation in these cells. These data hence indicated the existence of a major SOD3-related regulatory mechanism that affected the activation of the membrane-associated small GTPase molecule.
SOD3 over-expression regulates small GTPase regulatory gene mRNA production in a dose dependent manner
We previously demonstrated that the effect of SOD3 on the expression of growth-related genes correlated with the level of expression of this enzyme in vitro and in vivo. A robust SOD3 expression caused growth arrest, whereas a moderately increased SOD3 production supported cell survival, growth and tumorigenesis [5,6,8,32]. In the current study, we evaluated mice that had been previously transplanted with 8505c cells transfected with the luciferase expression control vector or SOD3 expression vector [5] to study the effect of low-dose growth promoting SOD3 synthesis levels on the expression of GEFs, GAPs, and GDIs (Fig. 5a and 5b). While the in vitro expression of GTPase regulatory genes favored the reduced growth rate and the inactivation of small GTPases (Fig. 4a, 4b, 4c, 4d, 4e, and 4f), the level of mRNA expression of GEF, GAP, and GDI genes in vivo isolated tumors was reversed (Fig. 5c, 5d, 5e, 5f, 5g, and 5h) supporting the observation of low SOD3 level induced increased rate of tumor growth (Fig. 5b) [5]. Although a significantly decreased expression of the RHO GEF16 and RAL GEF RGL1 genes was observed in vitro in the 8505c SOD3 over-expressing cells (Fig. 4a and 4b), the level of expression of these genes was increased in tumors that showed a moderate expression level of SOD3 mRNA (Fig. 5c and 5d). Similarly, the significantly increased expression of the GTPase activator proteins Arf GAP ADAP2 and RGS4 was correlated with a high level of SOD3 expression in vitro (Fig. 4c and 4e) and was markedly decreased in the in vivo tumors (Fig. 5e and 5g). Finally, the expression of RHO GDI 2, of which increased mRNA levels were found in vitro (Fig. 4f), was downregulated in vivo (Fig. 5h).
Fig 5. In vivo transplantation of 8505c SOD3 cells results in tumorigenesis.
(a) There was a robust over-production of SOD3 mRNA in 8505c SOD3 stable cells while the tumors derived from 8505c SOD3 stable cells contained moderately increased SOD3 mRNA consistent with our previously published data. (b) Bioluminescence imaging showing the increased tumor size in 8505c SOD3 transplanted mice as compared to control cell transplanted animals. (c-h) GEF, GAP, and GDI mRNA expression in vivo. The mice showing a moderately increased SOD3 expression in transplanted 8505c cells had reversed small GTPase regulatory gene expression as compared to in vitro situation. Data are expressed as mean ±SD. The p-values (* = p<0.05, ** = p<0.01, *** = p<0.001).
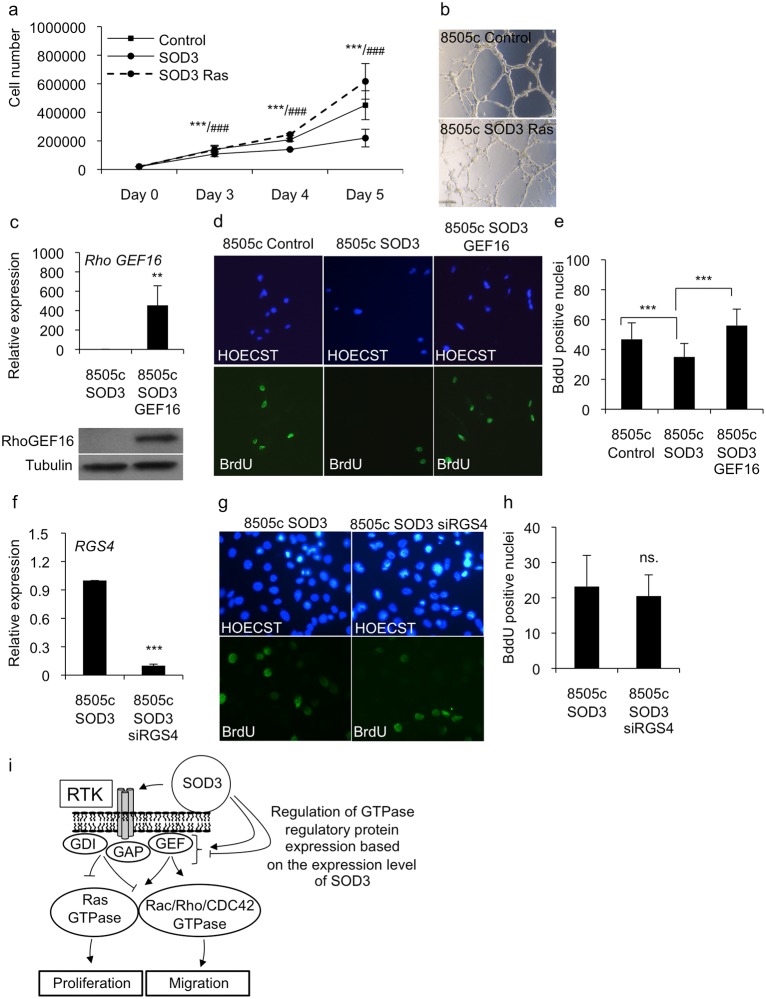
To dissect the effects of SOD3 on the expression of the small GTPase regulatory genes, we investigated the functional effects of RAS and RHO GEF16 over-expression in SOD3 transfected cells. Our data demonstrated that the stable expression of the H-RasV12 oncogene in 8505c SOD3 cells reverted the SOD3-mediated growth inhibitory effect, confirming the negative regulation of RAS by SOD3 (Fig. 4g and 4h) and suggesting a role for RAS as a mediator of SOD3 function, as previously described (Fig. 6a) [6]. Analysis of the invasive capability as determined using 3D Matrigel assay confirmed the ability of H-RasV12 to revert the growth inhibition of SOD3 cells to the levels of 8505c control cells (Fig. 6b). Similarly to the effect of H-RasV12 expression, the over-expression of the RHO GEF16 gene in 8505c SOD3 cells caused an increased level of cell proliferation compared with that of the 8505c control and 8505c SOD3 cells (Fig. 6c, 6d, and 6e). The data suggests that SOD3-mediated inhibition of RHO GEF16 expression is an important step in the SOD3-mediated regulation of cell growth, which supports previous observations concerning the oncogenic character of RHO GEF16 [33]. In contrast, RNAi of RGS4 expression had no effect on the growth potential of SOD3 expressing cells, suggesting that modulation of a single gene encoding a protein with GAP activity was insufficient to inhibit the effects of SOD3 over-expression (Fig. 6f, 6g, and 6h).
Fig 6. Reversed growth characteristics by H-RasV12 and RHO GEF16 over-expression in SOD3 transfected cells.
(a,b) The 8505c SOD3 cells were stable transfected with H-RasV12 oncogene and analyzed by growth curve and 3-D invasion analysis. Both analysis suggested marked increase in the growth of SOD3 cells after H-RasV12 transfection. (c) The 8505c SOD3 cells were stably transfected with RHO GEF16. The real time RT-PCR suggested significant (p<0.01) increase in RHO GEF16 mRNA production that was confirmed by Western blotting (lower panel). (d,e) The BrdU DNA incorporation analysis showed significantly (p<0.001) increased nuclear proliferation after RHO GEF16 over-expression as compared to 8505c SOD3 cells. (f) The RNAi of RGS4 in 8505c SOD3 cells showed significant downregulation of RGS4 mRNA production. (g,h) However, the BrdU DNA incorporation analysis failed to show differences between 8505c control and 8505c SOD3/siRGS4 cells. (i) A schematic drawing hypothesizing the SOD3 action in the cells. SOD3 over-expression activates cell membrane RTKs and membrane associated non-receptor tyrosine kinases. Depending on the expression level of SOD3 the proliferative and migratory signal is either halted or promoted at the level of small GTPase regulatory genes and small GTPases.
The 8505c control, 8505c SOD3, 8505c SOD3 Ras, and in 8505c SOD3 GEF16 cells were stained with dihydroethidium (DHE) and with MitoTracker RED CMX. DHE staining of live cells suggested no difference between 8505c control and 8505c SOD3 cells, a modest increase in cytoplasmic staining in 8505c SOD3 Ras cells as compared to control cells and a marked increased stainining in 8505c SOD3 GEF16 cells (S3 Fig.). MitoTracker RED CMX staining suggested similar mitochondrial staining in the all cells (S3 Fig.).
In summary, our data revealed a dose-dependent effect of SOD3 on the activity of small GTPase regulatory genes. These data clarified the correlation between high SOD3 expression levels and reduced cell proliferation, colony formation, and cell migration. These effects are regulated by a large set of genes that appear to converge on the small GTPase regulatory genes (Fig. 6i), β-catenin, and growth promoting/suppressing genes.
Discussion
Reactive oxygen species O2 .- and H2O2, which were originally thought to be harmful free radicals, are important second messengers in cellular signaling. The primary reactive oxygen species O2 .- is a precursor to a number of secondary radicals, such as H2O2, which can induce DNA damage, lipid peroxidation, and protein oxidation [3]. Noteworthy, at a low physiological concentration, H2O2 functions as a growth stimulator that increases the rate of cell proliferation whereas at high concentrations, H2O2 has toxic effects on cellular functions [3]. Superoxide dismutases (SODs) have been traditionally classified as antioxidative enzymes that remove harmful O2 .- radicals, producing more stable, although still reactive, H2O2 molecules. Thus SODs regulate the availability of O2 .- and H2O2 and should be considered intermediary molecules between oxidative free radical-forming enzymes and antioxidative free radical removing enzymes.
A recent report demonstrated that SOD3 acted as a growth regulator suggesting its ability to affect RAS activation and to stimulate mitogenic growth signaling [6]. However, the mechanism for SOD3—derived RAS activation is poorly characterized. In the present work we analyzed the effect of high SOD3 expression on the phosphorylation status of the cell membrane-associated RTKs and the cytoplasmic non-receptor kinases as well as on the transcriptome to achieve a global view of the transcriptional changes occurring during SOD3 expression. Consistent with the results of previous studies, our functional analysis showed that robust SOD3 expression reduced cellular growth (Fig. 1). Interestingly, high-level SOD3 expression markedly increased the activation of the cell membrane-associated RTKs and the downstream signaling molecules, such as the SRC protein family members SRC, FYN, YES, HCK, and LYN (Fig. 2) all of which function upstream of the small GTPase proteins, such as RAS [34]. More interestingly, the greatly increased level of SRC family signaling in 8505c SOD3 cells did not increase the activation status of small GTPase proteins as compared to control cells (Fig. 4); in contrast, the levels of activation of these proteins in the former cells were moderately decreased thus confirming our earlier observations that SOD3 regulates mitogen signaling at the level of RAS GTPase [6]. The neutralization of the activity of the mitogen signal transduction pathway (Fig. 3a and 3b) was associated with the modification of β-catenin signaling, suggesting that the reduced nuclear localization of β-catenin in SOD3 over-expressing cells (Fig. 3a, 3b, 3c, and 3d) and the simultaneous DDR activation (Fig. 3a, 3b, 3e, and 3f) enhanced the resultant decrease in the rate of cell proliferation observed in the cell proliferation and cell migration experiments (Fig. 1).
The involvement of SOD3 in the growth-regulated signal transduction was confirmed by the microarray screening combined with KEGG, GO, and GOrilla functional analyses (Table 3). The microarray results further demonstrated the SOD3–driven regulation of the genes responsible for the activation of small GTPases. Most importantly, the screening indicated that SOD3 regulated the expression of six GEFs, GAPs, and GDIs (Figs. 4 and 5) that together with the downregulated growth promoters, and the upregulated growth suppressors (Tables 1, 2, and 3) could have caused decreased mitogen signaling despite the presence of a strong upstream mitogen stimulus. Noteworthy, the simultaneous expression of SOD3 and RAS or SOD3 and RHO GEF16 increased the rate of cellular growth to the levels of the control cells strengthening the hypothesis that RAS and RHO GEF16 are mediators of the SOD3 effect on growth and migration (Fig. 6).
Small GTPases play pivotal roles in the regulation of the cytoskeletal rearrangements and cell motility, but are also involved in cell proliferation, transformation and differentiation, and participate in many steps for carcinogenesis initiation and the progression of cancers. Once bound to the cell membrane, the GTPase superfamily members interact with the upstream stimulators, such as SRC family proteins, and the cytoplasmic molecules that tightly are regulating their activity [34]. The activation of small GTPases requires GDP-GTP exchange, which is catalyzed by various guanine nucleotide exchange factors (GEFs) [35,36]. In contrast, GTPase-activating proteins (GAPs) inactivate small GTPases by stimulating the hydrolysis of GTP [35,36]. Inactive GTPases are sequestered in the cytosol by the guanine nucleotide dissociation inhibitors (GDI) [35,36,37]. Due to their ability to increase the activity of small GTPase superfamily, GEFs are potential oncogenes that affect tumorigenesis, e.g. RAL GEFs have been shown to increase the anchorage independent growth in vitro and the transformation activity after their membrane translocation [38].
GAPs activate the intrinsic GTPase activity of Ras superfamily GTPase members, which hydrolyze GTP and return the GTPase to the inactive GDP bound form. Interestingly, GAPs can hydrolyze only the wild-type RAS to terminate its signaling, whereas these molecules have no effect on mutant RAS activity, therefore being less attractive drug targets [39,40]. However, several studies have shown the tumor suppressive properties of GAPs [41] hence suggesting that they play a role in early stages of tumorigenesis.
The third group of regulators is GDIs, which maintain the GTPases in GDP-bound inactive form in the cytosolic compartment and control their GTPase membrane-binding capacity. RHO GDI2 has been shown to be a metastasis suppressor in bladder and breast cancers [42,43]. In line, the level of RHO GDI2 expression was increased during the early phase of breast epithelial hyperplasia and was significantly decreased in transformed metastatic cells. Indeed, it has been shown that RHO GDI2 reduced the extend of cell migration and served as a metastasis inhibitor rather than a cell proliferation suppressor [44].
We have recently demonstrated that SOD3 has a dose-dependent effect on cellular growth in vitro and in vivo [5]. High-level SOD3 expression decreased the growth rate by inducing apoptosis in p53 responsive cancer cells whereas a moderately upregulated level of SOD3 production increased the proliferation of primary and cancer cells, and promoted cellular immortalization and neoplastic transformation. Noteworthy, the induction of p53 activation and the resultant growth arrest were observed in both in primary cells and in cancer cells containing functional p53 response. Furthermore, the p53 induction occurred at both at low and high SOD3 expression levels: the response induced by high-level SOD3 expression was the immediate death of most of the cells within 24 hours, whereas low-level SOD3 expression first activated a proliferative burst that was followed by DNA strand break stimulated p53-p21 growth arrest and a senescence phase in a fashion similar to that caused by the H-RasV12 oncogene [5]. However, the growth arrest pathway activation does not solely explain this reduced cellular growth. A number of in vivo SOD3 mouse model studies have demonstrated the decreased level of tumorigenesis under both p53-responsive and p53-inactive conditions, supporting our recent in vitro data [45,46,47,48] and indicating the existence of additional growth-signaling-related regulatory systems. Therefore, the current data showing decreased GEF expression and increased GAP and GDI production, which paralleled with a reduced activation of small GTPases at high SOD3 levels, together with the observed effects on β-catenin signaling supported the previous observations and may explain how robust SOD3 expression reduces cellular growth. In turn, a moderately increased SOD3 mRNA expression reversed the GEF, GAP, and GDI synthesis suggesting the regulatory characteristics of SOD3 in controlling the activation the status of small GTPases.
Our current and previous data thus suggest that SOD3 has a dose-dependent effect on growth signaling. A moderate level of SOD3 expression induced proliferative signal transduction through phophorylation of the RTKs and cytosolic non-receptor kinases, leading to RAS and ERK activation. In contrast, high SOD3 levels reduced the rates of cell proliferation and cell migration by affecting the expression of a family of small GPTase regulatory genes that negatively regulate the activity of small GTPases and ERK kinases. These data revealed a new scenario, in which SOD3 acts in a dose-dependent manner to simultaneously activate distinct pathways that have conflicting effects. Because SOD3 itself is a drug target of great interest, the current study might indicate molecules that mediate the effects of SOD3 on cell proliferation and hence are potential druggable targets in the early phase of tumorigenesis.
Supporting Information
RET and phospho-RET Western blotting from 8505c cells showed no RET expression. The bands observed in the p-RET lane are the result of nonspecific labeling.
(TIF)
(TIF)
(a) The images show Hoechst nuclear staining and DHE ROS staining. DHE staining of the cells shows minor differences in ROS staining in 8505c SOD3 as compared to 8505c control cells. A moderate increase was observed in 8505c SOD3 Ras cells and a marked increased in 8505c SOD3 GEF16 cells. (b) Mitochondrial ROS staining showed no differences between cell lines. Images are shown in grey scale (left side panes) and with red fluorescence (right side panels).
(TIF)
(XLSX)
(XLS)
(XLS)
(XLS)
Data Availability
Data is available in NCBI GEO (accession number GSE56546): Extracellular Superoxide Dismutase Regulates the Expression of GTPase Guanine Nucleotide Exchange Factors (GEFs), GTPase-activating proteins (GAPs), and Rho Guanine nucleotide disassociation inhibitor (GDI).
Funding Statement
This work was supported by Fondazione SDN (decision number RC2010-M-0001, http://www.sdn-napoli.it/).
References
- 1. Marklund SL. Human copper-containing superoxide dismutase of high molecular weight. Proc Natl Acad Sci U S A. 1982;79:7634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Irani K, Goldschmidt-Clermont PJ. Ras, superoxide and signal transduction. Biochem Pharmacol. 1998;55:1339–46. [DOI] [PubMed] [Google Scholar]
- 3. Song Y, Driessens N, Costa M, De Deken X, Detours V, Corvilain C, et al. Roles of hydrogen peroxide in thyroid physiology and disease. J Clin Endocrinol Metab. 2007;92:3764–73. [DOI] [PubMed] [Google Scholar]
- 4. Laurila JP, Laatikainen LE, Castellone MD, Laukkanen MO. SOD3 reduces inflammatory cell migration by regulating adhesion molecule and cytokine expression. PLoS One. 2009;4:e5786 10.1371/journal.pone.0005786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castellone MD, Langella A, Cantara S, Laurila JP, Laatikainen LE, Bellelli R, et al. Extracellular Superoxide Dismutase Induces Mouse Embryonic Fibroblast Proliferative Burst, Growth Arrest, Immortalization, and Consequent In Vivo Tumorigenesis. Antioxid Redox Signal. 2014; 21:1460–74. 10.1089/ars.2013.5475 [DOI] [PubMed] [Google Scholar]
- 6. Laurila JP, Castellone MD, Curcio A, Laatikainen LE, Haaparanta-Solin M, Gronroos TJ, et al. Extracellular superoxide dismutase is a growth regulatory mediator of tissue injury recovery. Mol Ther. 2009;17:448–54. 10.1038/mt.2008.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sibenaller ZA, Welsh JL, Du C, Witmer JR, Schrock HE, Du J, et al. Extracellular superoxide dismutase suppresses hypoxia-inducible factor-1alpha in pancreatic cancer. Free Radic Biol Med. 2014;69:357–66. 10.1016/j.freeradbiomed.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laatikainen LE, Incoronato M, Castellone MD, Laurila JP, Santoro M, Laukkanen MO. SOD3 decreases ischemic injury derived apoptosis through phosphorylation of Erk1/2, Akt, and FoxO3a. PLoS One. 2011;6:e24456 10.1371/journal.pone.0024456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwon MJ, Han J, Kim BH, Lee YS, Kim TY. Superoxide dismutase 3 suppresses hyaluronic acid fragments mediated skin inflammation by inhibition of toll-like receptor 4 signaling pathway: superoxide dismutase 3 inhibits reactive oxygen species-induced trafficking of toll-like receptor 4 to lipid rafts. Antioxid Redox Signal. 2012;16:297–313. 10.1089/ars.2011.4066 [DOI] [PubMed] [Google Scholar]
- 10. Nozik-Grayck E, Suliman HB, Majka S, Albietz J, Van Rheen Z, Roush K, et al. Lung EC-SOD overexpression attenuates hypoxic induction of Egr-1 and chronic hypoxic pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2008;295:L422–30. 10.1152/ajplung.90293.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teoh ML, Fitzgerald MP, Oberley LW, Domann FE. Overexpression of extracellular superoxide dismutase attenuates heparanase expression and inhibits breast carcinoma cell growth and invasion. Cancer Res. 2009;69:6355–63. 10.1158/0008-5472.CAN-09-1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laatikainen LE, Castellone MD, Hebrant A, Hoste C, Cantisani MC, Laurila JP, et al. Extracellular superoxide dismutase is a thyroid differentiation marker down-regulated in cancer. Endocr Relat Cancer. 2010;17:785–96. 10.1677/ERC-10-0021 [DOI] [PubMed] [Google Scholar]
- 13. Svensk AM, Soini Y, Paakko P, Hiravikoski P, Kinnula VL. Differential expression of superoxide dismutases in lung cancer. Am J Clin Pathol. 2004;122:395–404. [DOI] [PubMed] [Google Scholar]
- 14. Pesse B, Levrand S, Feihl F, Waeber B, Gavillet B, Pacher P, et al. (2005) Peroxynitrite activates ERK via Raf-1 and MEK, independently from EGF receptor and p21Ras in H9C2 cardiomyocytes. J Mol Cell Cardiol. 2005;38:765–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersen SV, Oury TD, Ostergaard L, Valnickova Z, Wegrzyn J, Thogersen IB, et al. Extracellular superoxide dismutase (EC-SOD) binds to type i collagen and protects against oxidative fragmentation. J Biol Chem. 2004;279:13705–10. [DOI] [PubMed] [Google Scholar]
- 16. Nguyen AD, Itoh S, Jeney V, Yanagisawa H, Fujimoto M, Ushio-Fukai M, et al. Fibulin-5 is a novel binding protein for extracellular superoxide dismutase. Circ Res. 2004;95:1067–74. [DOI] [PubMed] [Google Scholar]
- 17. Ciardiello F, Kim N, Saeki T, Dono R, Persico MG, Plowman GD, et al. Differential expression of epidermal growth factor-related proteins in human colorectal tumors. Proc Natl Acad Sci U S A. 1991;88:7792–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia R, Mayer BJ, Hanafusa T, Hanafusa H. A novel oncogene, v-ryk, encoding a truncated receptor tyrosine kinase is transduced into the RPL30 virus without loss of viral sequences. J Virol. 1992;66:5975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–4. [DOI] [PubMed] [Google Scholar]
- 20. Timeus F, Ricotti E, Crescenzio N, Garelli E, Doria A, Spinelli M, et al. Flt-3 and its ligand are expressed in neural crest-derived tumors and promote survival and proliferation of their cell lines. Lab Invest. 2001;81:1025–37. [DOI] [PubMed] [Google Scholar]
- 21. Fox BP, Tabone CJ, Kandpal RP. Potential clinical relevance of Eph receptors and ephrin ligands expressed in prostate carcinoma cell lines. Biochem Biophys Res Commun. 2006;342:1263–72. [DOI] [PubMed] [Google Scholar]
- 22. Sibley DR, Benovic JL, Caron MG, Lefkowitz RJ. Regulation of transmembrane signaling by receptor phosphorylation. Cell. 1987;48:913–22. [DOI] [PubMed] [Google Scholar]
- 23. Sen B, Johnson FM Regulation of SRC family kinases in human cancers. J Signal Transduct. 1987;2011:865819 10.1155/2011/865819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen RH, Ding WV, McCormick F. Wnt signaling to beta-catenin involves two interactive components. Glycogen synthase kinase-3beta inhibition and activation of protein kinase C. J Biol Chem. 2000;275:17894–9. [DOI] [PubMed] [Google Scholar]
- 25. Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–9. [DOI] [PubMed] [Google Scholar]
- 26. Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7. [DOI] [PubMed] [Google Scholar]
- 27. D'Uva G, Bertoni S, Lauriola M, De Carolis S, Pacilli A, D'Anello L, et al. Beta-catenin/HuR post-transcriptional machinery governs cancer stem cell features in response to hypoxia. PLoS One. 2013;8:e80742 10.1371/journal.pone.0080742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, et al. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J Biol Chem. 2002;277:21657–65. [DOI] [PubMed] [Google Scholar]
- 29. Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. [DOI] [PubMed] [Google Scholar]
- 30. Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. [DOI] [PubMed] [Google Scholar]
- 31. DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–63. [DOI] [PubMed] [Google Scholar]
- 32. Laukkanen MO, Kivela A, Rissanen T, Rutanen J, Karkkainen MK, Leppanen O, et al. Adenovirus-mediated extracellular superoxide dismutase gene therapy reduces neointima formation in balloon-denuded rabbit aorta. Circulation. 2002;106:1999–2003. [DOI] [PubMed] [Google Scholar]
- 33. Oliver AW, He X, Borthwick K, Donne AJ, Hampson L, Hampson IN, et al. The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. Br J Cancer. 2011;104:324–31. 10.1038/sj.bjc.6606026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–95. 10.1038/nrclinonc.2009.129 [DOI] [PubMed] [Google Scholar]
- 35. Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–54. [DOI] [PubMed] [Google Scholar]
- 36. Pechlivanis M, Kuhlmann J. Hydrophobic modifications of Ras proteins by isoprenoid groups and fatty acids—More than just membrane anchoring. Biochim Biophys Acta. 2006;1764:1914–31. [DOI] [PubMed] [Google Scholar]
- 37. Luttrell DK, Luttrell LM. Not so strange bedfellows: G-protein-coupled receptors and Src family kinases. Oncogene. 2004;23:7969–78. [DOI] [PubMed] [Google Scholar]
- 38. Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ, et al. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem. 2010;285:34729–40. 10.1074/jbc.M110.116756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–57. 10.1038/nrc2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal. 2011;23:969–79. 10.1016/j.cellsig.2010.10.022 [DOI] [PubMed] [Google Scholar]
- 41. Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. [DOI] [PubMed] [Google Scholar]
- 42. Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF Jr, et al. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res. 2004;10:3800–6. [DOI] [PubMed] [Google Scholar]
- 43. Hu LD, Zou HF, Zhan SX, Cao KM. Biphasic expression of RhoGDI2 in the progression of breast cancer and its negative relation with lymph node metastasis. Oncol Rep. 2007;17:1383–9. [PubMed] [Google Scholar]
- 44. Harding MA, Theodorescu D. RhoGDI signaling provides targets for cancer therapy. Eur J Cancer. 2010;46:1252–9. 10.1016/j.ejca.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanaka M, Kogawa K, Nakamura K, Nishihori Y, Kuribayashi K, Hagiwara S, et al. Anti-metastatic gene therapy utilizing subcutaneous inoculation of EC-SOD gene transduced autologous fibroblast suppressed lung metastasis of Meth-A cells and 3LL cells in mice. Gene Ther. 2001;8:149–56. [DOI] [PubMed] [Google Scholar]
- 46. Wheeler MD, Smutney OM, Samulski RJ. Secretion of extracellular superoxide dismutase from muscle transduced with recombinant adenovirus inhibits the growth of B16 melanomas in mice. Mol Cancer Res. 2003;1:871–81. [PubMed] [Google Scholar]
- 47. Kim SH, Kim MO, Gao P, Youm CA, Park HR, Lee TS, et al. Overexpression of extracellular superoxide dismutase (EC-SOD) in mouse skin plays a protective role in DMBA/TPA-induced tumor formation. Oncol Res. 2005;15:333–41. [DOI] [PubMed] [Google Scholar]
- 48. Teoh ML, Sun W, Smith BJ, Oberley LW, Cullen JJ. Modulation of reactive oxygen species in pancreatic cancer. Clin Cancer Res. 2007;13:7441–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RET and phospho-RET Western blotting from 8505c cells showed no RET expression. The bands observed in the p-RET lane are the result of nonspecific labeling.
(TIF)
(TIF)
(a) The images show Hoechst nuclear staining and DHE ROS staining. DHE staining of the cells shows minor differences in ROS staining in 8505c SOD3 as compared to 8505c control cells. A moderate increase was observed in 8505c SOD3 Ras cells and a marked increased in 8505c SOD3 GEF16 cells. (b) Mitochondrial ROS staining showed no differences between cell lines. Images are shown in grey scale (left side panes) and with red fluorescence (right side panels).
(TIF)
(XLSX)
(XLS)
(XLS)
(XLS)
Data Availability Statement
Data is available in NCBI GEO (accession number GSE56546): Extracellular Superoxide Dismutase Regulates the Expression of GTPase Guanine Nucleotide Exchange Factors (GEFs), GTPase-activating proteins (GAPs), and Rho Guanine nucleotide disassociation inhibitor (GDI).