SUMMARY
Regulation of cell expansion is essential to the formation of plant organs. We have characterized 21 mutations, representing six loci, that cause abnormal cell expansion in the root of Arabidopsis thaliana. The phenotype of these mutants is conditional upon the rate of root growth. Calculation of cell volumes indicated that the mutations resulted in defects in either the orientation or the extent of expansion or in both. Analysis of cortical microtubules in the mutants suggested that a shift in the orientation of cell expansion may not be dependent on a change in the orientation of the microtubules. Double mutant combinations resulted in loss of the conditional phenotype suggesting that the genes may act in a similar pathway or encode partially redundant functions.
Keywords: root development, cell expansion, cell polarity, microtubules, core mutants, Arabidopsis
INTRODUCTION
Cell expansion is a key parameter of development in multicellular eucaryotes. Regulation of cell expansion is required for the establishment of cell shape and polarity. In plants, cell expansion is of paramount importance in the determination of organ shape. To the extent that organ shape affects function, the control of cell expansion can be critical to the survival of the plant.
The two principal parameters of cell expansion are orientation and extent. There is evidence that the orientation of cell expansion is controlled by the cellulose microfibrils in the cell wall (Giddings and Staehelin, 1991). These molecules are normally found arranged transverse to the direction of growth and have been likened to hoops around a barrel. Control of the orientation of the cellulose has been attributed to the cortical microtubules of the cytoskeleton. This is based on two types of evidence. First, cortical microtubules are normally found aligned in the same orientation as the cellulose microfibrils. Second, disruption of the microtubules has led to abnormal cellulose microfibril patterns and abnormal cell expansion (Giddings and Staehelin, 1991). A model has been proposed in which microtubules act like tracks between which the cellulose synthesis complex moves (Giddings and Staehelin, 1991). In this way the orientation of the microtubules dictates the orientation of the cellulose microfibrils without any direct contact between the two.
Regulation of the extent of expansion is less well understood. The driving force for expansion is turgor pressure generated by the presence of a high osmotic potential in the vacuoles within the expanding cell. Water movement into the cell is thought to be passive, driven only by the difference in osmotic potential (Cosgrove, 1993). Expansion occurs by addition of cell wall components along the length of the wall. This requires a constant breaking and reforming of bonds within the two co-extensive polymer networks, one made of cellulose and hemicellulose and the other made of pectin (Roberts, 1989). It also requires constant synthesis and transport of structural components to the cell wall. At some point there is a “rigidification” of the wall so that no more bonds are broken and no more wall components are added. At this point the wall resists the turgor pressure. When the wall pressure equals the turgor pressure, expansion ceases (Cosgrove, 1993).
Very little is known about the genetic control of cell expansion. In fact, it has been suggested that plant form is determined not at the cellular level but at the level of the organ. In this theory, expansion of individual cells is subject to a higher regulation that has predetermined the final shape of the organ (Kaplan and Hagemann, 1991). In animal cells, there is also little known about the regulation of cell expansion. Although most animal cells do not have cell walls, current studies of the primary plant cell wall indicate that it resembles the extracellular matrix surrounding animal cells (Roberts, 1989). Therefore, there are likely to be aspects of the regulation of cell expansion that are common to both animals and plants as well as some that are unique to each.
The Arabidopsis root is particularly well suited to studies of cell expansion. Cell expansion in the body of the root is regulated so that the primary axis is normally parallel to the direction of growth. This makes for long cylindrical cells. Polarized expansion in the longitudinal orientation (which we will refer to as “elongation”) occurs primarily in a region at the tip of the root known as the elongation zone (Steeves and Sussex, 1989).
By screening mutagenized plants for roots that have an increased diameter we and others have identified mutants that have deregulated cell expansion (Benfey et al., 1993; Hauser and Benfey, 1993; Baskin et al., 1992). Here, we present a genetic and cellular characterization of 21 mutants representing six loci. Three of these loci have not been previously reported. The phenotype of all 21 mutants was conditional upon the rate of root growth. For this reason these mutants were named Conditional Root Expansion (CORE) mutants. Determination of cellular dimensions suggested that different CORE genes control the orientation and extent of expansion. Cortical microtubule orientation was determined for all mutants. The loss of the conditional phenotype when the CORE mutations were combined indicated that the CORE genes may act in a similar pathway or provide redundant functions.
MATERIALS AND METHODS
Plant strains, growth conditions and genetic analysis
Strains (or ecotypes) Landsberg erecta (Ler), Columbia (Col), Wasselewskija (Ws) of Arabidopsis thaliana (L) were obtained from the Arabidopsis Stock Center (Columbus, OH). The mutants cobra-1 (cob-1), lion’s tail-1 (lit-1) and pom-pom1-1 (pom1-1) were grown as previously described (Benfey et al., 1993; Hauser and Benfey, 1993). Genetic crosses were performed essentially as described by Schiefelbein and Somerville (1990); Benfey et al. (1993). The newly identified CORE mutants, quill (qui), cudgel (cud), and pom-pom2 (pom2) were isolated among 145,000 plants grown from different populations including: pools of T4 T-DNA mutagenized WS lines (Feldmann, 1991), M2 Col seeds mutagenized with ethyl methanesulfonate (EMS), M2 Col seeds mutagenized with X-rays and M2 Col seeds mutagenized with fast neutrons (Lehle Seeds). Plants that exhibited roots with a diameter larger than wild-type were isolated and their progeny were retested in the M3 generation for the conditional phenotype on nutrient agar plates as previously described (Hauser and Benfey, 1993). Pairwise crosses were made between the different CORE mutants to determine complementation groups. Regeneration of roots from calli that had been induced from shoots was according to the regeneration regime described by Valvekens et al. (1988).
Mapping
The chromosomal location of the mutant genes was determined by measuring the recombination frequency between the mutant gene and microsatellite markers (Bell and Ecker, 1994), cleaved amplified polymorphic sequence markers (CAPS) (Konieczny and Ausubel, 1993) and visible markers. The homozygous mutants lit-1 (Col), cob-1 (Col), pom1-1 (Ws), pom1-2 (Col), cud-1 (Col), pom2-1 (Col), qui-1 (Col), qui-2 (Ws) were mapped by crossing them to Landsberg erecta (Ler). pom1-1 was also crossed to Col and lit-1 was also crossed to WS. The resulting F1 plants were selfed to generate F2 seedlings segregating for the mutant phenotype. DNA was isolated from 15-20 independent axenically grown mutant seedlings using a slightly modified method of Konieczny and Ausubel (1993). We modified the microsatellite mapping protocol to separate the fragments on 5% polyacrylamide gels (19:1) in 1× Tris Borate EDTA (TBE), pH 8.3 buffer. Map distances were calculated from recombination frequencies (RF). The recombination frequency of cud-1 which is male gametophytic lethal/sterile was calculated as the number of homozygous plants (both ecotypes) of the total F2 plants exhibiting the mutant phenotype.
Recombination frequencies were as follows, approximate map positions are listed in Table 1:
Table 1. Name and origin of conditional root expansion mutants.
| Mutant/allele | Chromosome number* |
Map position* |
Ecotype | Mutagen | Genetic status |
|---|---|---|---|---|---|
| lit-1 | V | 70 | Col | EMS | recessive |
| cob-1 | V | 90 | Col | EMS | semidominant |
| cob-2 | Col | x-ray | semidominant | ||
| cob-3 | Ws | T-DNA | semidominant | ||
| qui-1 | I | 0 | Col | x-ray | recessive |
| qui-2 | Ws | T-DNA | recessive | ||
| qui-3 | Col | EMS | recessive | ||
| pom1-1 | I | 5 | Ws | T-DNA | recessive |
| pom1-2 | Col | EMS | recessive | ||
| pom1-3 | Ws | T-DNA | recessive | ||
| pom1-4 | Ws | T-DNA | recessive | ||
| pom1-5 | Ws | T-DNA | recessive | ||
| pom1-6 | Ws | T-DNA | recessive | ||
| pom1-7 | Ws | T-DNA | recessive | ||
| pom1-8 | Ws | T-DNA | recessive | ||
| pom1-9 | Col | EMS | recessive | ||
| pom1-10 | Col | EMS | recessive | ||
| pom1-11 | Col | EMS | recessive | ||
| cud-1 | IV | 76 | Col | x-ray | dominant† |
| pom2-1 | II | 60 | Col | fast neutron recessive | |
| pom2-2 | Col | fast neutron recessive | |||
Approximate map positions determined by mapping with CAPS and microsatellite markers (see Materials and Methods).
cudgel is also male-gametophytic lethal. This may be due to a linked mutation resulting perhaps from a deletion induced by the X-ray mutagenesis.
For lit, 18.8% with DFR (CAPS), 20% with LFY (CAPS). For cob, O.8% with LFY. For qui, 8% with PVV4 (CAPS), 10% with nga59 (microsatellite). For pom1, 4.7% with nga59, 1.2% with nga63 (microsatellite). For pom2, 4% with GPA1 (CAPS), 12% with m429 (CAPS). For cud, 16% with DHSI (CAPS), 26% with AG (CAPS).
Confocal microscopy, immunolocalization of microtubules, nuclear staining and image analysis
Seedlings were stained with acriflavine as described (Aeschbacher et al., 1995). Hypocotyls were stained with two additional 30 minute steps in 95% ethanol after fixation, to reduce the chlorophyll content. Microtubule immunolocalization was a modification of the method of Colasanti et al. (1993); Doonan et al. (1993) and performed as follows. Seedlings were fixed in a solution of 4% paraformaldehyde, 5% DMSO, 0.1% glutaraldehyde, 50 mM PIPES, 5 mM EGTA, 5 mM MgSO4, pH 7.0 by vacuum infiltration at 4°C for 1 hour. Seedlings were then washed three times in wash buffer (50 mM PIPES, 5 mM EGTA, 5 mM MgSO4, pH 7.0) for 30 minutes. Seedlings were then transferred to a solution of 2% driselase (Sigma), which was vacuum infiltrated for 1 hour at room temperature. The seedlings were washed twice followed by a 10 minute incubation with 100% methanol at −20°C. Seedlings were washed twice then infiltrated for 15 minutes with 0.5% Nonidet P-40 in wash buffer. Seedlings were washed twice then labelled for at least 1 hour at room temperature with 300 μl washing buffer containing 3% BSA (Sigma) and a 1:75 dilution of the rat monoclonal antibody YOL1/34 anti-yeast-α-tubulin subunit antibody (Sera Labs). The antibody recognizes most eucaryotic α-tubulins including yeast, mammal, and plant. The antibody has a lower affinity to native microtubules and a higher affinity to fixed microtubules. Seedlings were washed three times then incubated for 1 hour at room temperature in 300 μl wash buffer containing 3% BSA (Sigma) and a 1:100 dilution of a tetramethylrhodamine-5-isothiocyanate-(TRITC) labelled secondary antibody (anti-rat IgG, Sigma). Seedlings were rinsed in a wash buffer/glycerin series (30%, 50%, 70%) and the roots were mounted on a microscope slide with Citifluor (Ted Pella). Confocal laser scanning microscopy (CLSM) was performed with a Zeiss confocal microscope with a helium line at 543 nm.
For root tip staining, young seedlings were fixed in 3.7% formaldehyde in PBT buffer (130 mM NaCl, 10 mM NaHPO4 buffer, pH 7.0, 0.1% Tween 20) for at least 4 hours at room temperature, washed three times with PBT, and stained with 2 μM 4,6-diamindino-2-phenylindole (DAPI, Sigma) in PBT buffer for 10 minutes. Excess stain was removed by washing with PBT, then seedlings were washed in 50% and 70% glycerin in PBT and mounted with Citifluor (Ted Pella). The root tips were analyzed with a Zeiss confocal microscope with the Argon/Krypton laser line of 488 nm. Images were stored as TIFF files, manipulated in Adobe Photoshop and printed on a Kodak XL7700 dye sublimation printer.
Cell dimensions, cell volume calculations and growth condition shift experiments
Cellular dimensions were determined from CLSM pictures taken in the differentiation zone of wild-type and CORE mutant roots. Between 150 and 200 cells were measured from four to 14 different roots for wild type and each mutant. For hypocotyls, approximately 80 cells were measured from five different plants. Cell volumes were calculated assuming that epidermis and cortex cells were approximately cylindrical for both wild type and mutants. This was based on observations of numerous longitudinal and fresh transverse sections. In transverse sections, the cross-sectional areas of endodermis cells had a rectangular shape. For these cells, the circumferential wall was taken as the length and the radial wall was used for the width so that volume = circumferential length × radial width × cell length. As controls for these measurements, cross-sectional areas were obtained directly from digitized images of fresh sections as described previously (Benfey et al., 1993). Areas obtained in this way were approximately equivalent to areas calculated from diameters measured from optical sections.
For the growth condition shift experiments, seeds were planted on nutrient agar medium supplemented with 0.1% sucrose. After 8 days growth, root epidermal cells were marked with carbon grains as described by Okada and Shimura (1990). Seedlings were then transferred to either nutrient agar medium supplemented with 4.5% sucrose or 0.1% sucrose. At intervals of 4, 8, 12, 24 and 36 hours root tips were photographed under a Nikon SMZ-U stereomicroscope. Tracings of root tips and the location of carbon grains were made from projections of 35 mm slides.
RESULTS
Phenotypic and genetic analysis of root expansion mutants with conditional phenotypes
To investigate the genetic basis of root morphogenesis we screened progeny of mutagenized seeds for abnormal root expansion. We isolated 21 mutants that had reduced root length and exhibited a large variability in root diameter along the length of the root. Reciprocal crosses among the mutants revealed six complementation groups. Three loci, cobra (cob), lion’s tail (lit) and pom-pom1 (pom1) have been described previously (Benfey et al., 1993; Hauser and Benfey, 1993). Here we describe the isolation and characterization of new alleles of these mutants as well as three new loci, quill (qui), pom-pom2 (pom2) and cudgel (cud) (Table 1). All mutations except for cob and cud-1 were found to be recessive. cob is semidominant (Benfey et al., 1993). cud-1 was found to be dominant and male-sterile as evidenced by 100% wild-type progeny when pollinating wild type and 50% mutant progeny when receiving pollen from wild type. The segregation of the progeny of cud-1 mutant plants was 1:1 mutant to wild type. The chromosomal locations of the loci were determined by use of molecular markers (Table 1).
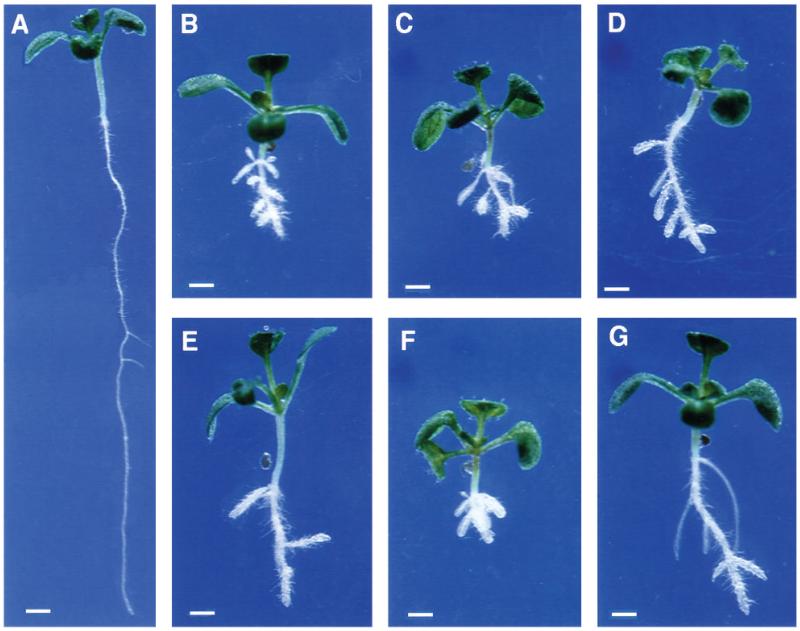
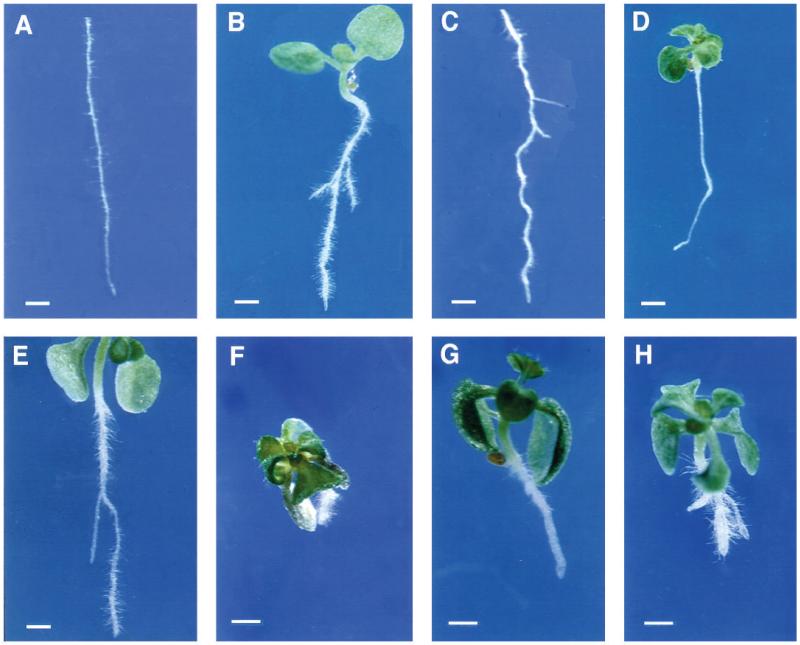
The phenotype of all 21 mutants was conditional upon the roots growing at a maximum rate. As noted above, we have named this class of mutants, Conditional Root Expansion (CORE) mutants. We have shown that wild-type root growth rate is dependent on various factors. These include the concentration of sucrose in nutrient agar media as well as light and temperature conditions (Benfey et al., 1993; Hauser and Benfey, 1993). When the 21 root expansion mutants were grown under conditions that provide maximal wild-type root growth they exhibited the expanded phenotype (Fig. 1). Wild-type growth rates can be diminished by growing the plants on medium with a lower concentration of sucrose or by reducing temperature and light. Under these conditions the mutant roots had relatively normal diameters (Fig. 2). Optical sections revealed that the cellular morphology of the mutants was also relatively normal under these permissive conditions (Fig. 3). In particular, the cellular morphology of cob-1 grown on media containing 0.5% sucrose was difficult to distinguish from wildtype (Fig. 3B) (Benfey et al., 1993). This was also true of pom2 (data not shown). Under these conditions cob-1, pom1-8 (Fig. 2B), pom1-7 (Fig. 3D), qui-2 (Fig. 2C) and cud-1 had reduced root length but no apparent radial cell expansion. The only mutant with any detectable radial cell expansion when grown under these conditions was lit-1 (Figs 2D, 3E and 3F). However, this was dramatically reduced from the extent of expansion when grown on higher sucrose levels (compare Figs 2D and 1B).
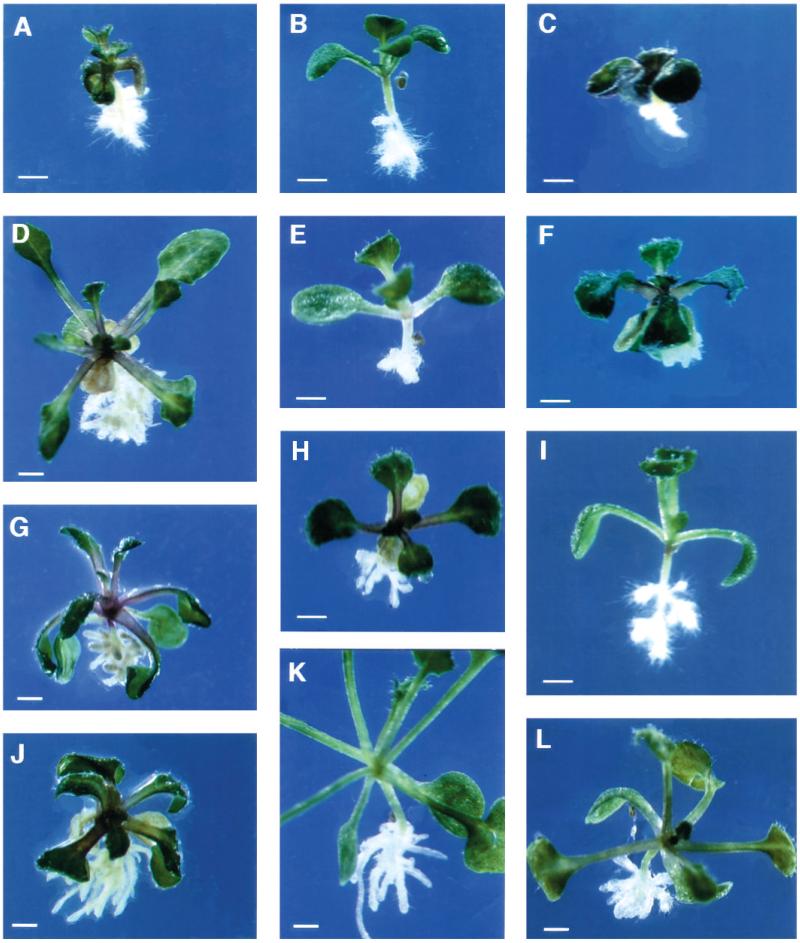
Fig. 1.
Wildtype and CORE seedlings on medium supplemented with 4.5% sucrose, 11 days after germination (DAG). (A) Wild-type WS, (B) lion’s tail-1, (C) cobra-1, (D) quill-1, (E) pom-pom1-1, (F) cudgel-1, (G) pom-pom2-1. Bar, 1 mm.
Fig. 2.
Wildtype and CORE single and double mutants grown on medium supplemented with 0.5% sucrose. (A) Wild-type Col, (B) pom-pom1-8, (C) quill-1, (D) lion’s tail-1. Double mutants between (E) pom-pom2-1 and cobra-1, (F) cudgel-1 and pom-pom1-1, (G) cudgel-1 and quill-1, (H) pom-pom2-1 and lion’s tail-1. Bar, 1 mm.
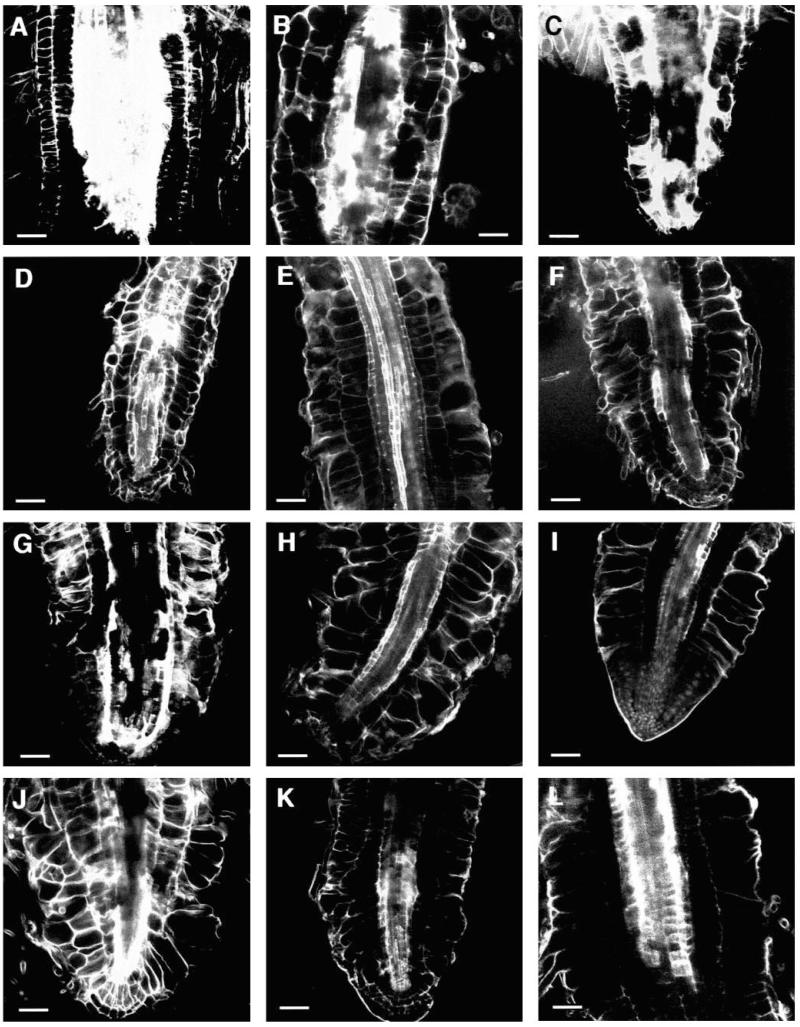
Fig. 3.
Optical sections through the roots of seedlings grown on medium supplemented with 0.5% sucrose. Median longitudinal sections through the differentiation zone of (A) wild-type Ws, (B) cobra-3, (C) quill-2, (D) pom-pom1-7, (E) half of the expanded part of the root of lion’s tail-1, (F) unexpanded part of same lion’s tail-1 root, (G) double mutant of cobra-1 and pom-pom2-1, (H) double mutant of cudgel-1 and quill-2, (I) double mutant of cudgel-1 and pom-pom1-1, (J) double mutant of lion’s tail-1 and pom-pom2-1. Bar, 50 μm.
To test whether the expansion at higher sucrose levels was influenced by the increased osmotic potential, we grew the CORE mutants on two different concentrations of mannitol, a non-metabolized carbohydrate. We found that there was no difference in root expansion in any of the CORE mutants when grown on low (0.25%) or high (2.25%) mannitol-containing medium. The root phenotype of the CORE mutants was similar to the phenotype on normal growth medium supplemented with 0.5% sucrose.
There was no apparent abnormality in the aerial phenotype of CORE mutant seedlings when grown under restrictive or permissive conditions (Figs 1 and 2). When grown on soil, the mutants lit and pom1 exhibited reduced growth and smaller leaves. The flower phenotypes of the CORE mutants were normal with the exception of pom2 which had reduced filament elongation as well as thickening of the style (data not shown). This flower phenotype was probably the reason for the extremely low fertility of the pom2 mutants.
To determine whether the mutant phenotypes were specific for the developmental context of the seedling, we examined roots induced from callus culture. Callus-induced roots from lit-1, cob-1, cud-1, pom1-1, qui-1 and pom2-1 displayed the mutant phenotype when they grew under restrictive conditions (data not shown). All the mutants responded to a gravitropic stimulus in a manner similar to wildtype although the change of the direction of growth was slower than wildtype.
Root morphology and cell volume of CORE mutants
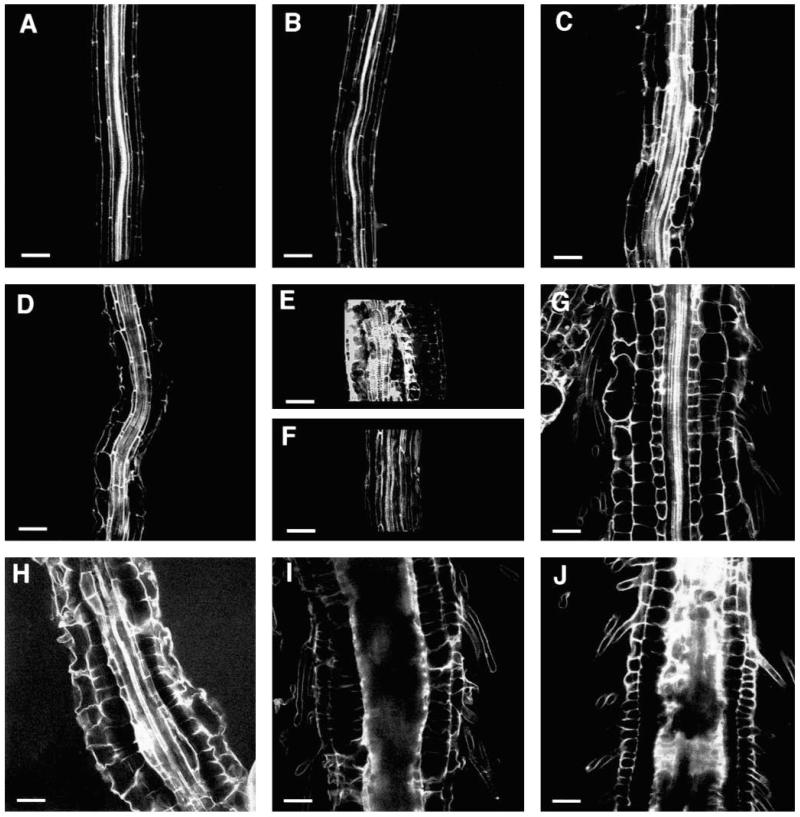
The structure of the expanded root of the CORE mutants was analyzed by confocal laser scanning microscopy. Optical sections through the differentiation zone revealed differences in cell shape among the different mutants (Fig. 4B-G). To determine how the shape changes affected cell volumes we measured cellular dimensions for the epidermis, cortex and endodermis (Table 2). The cell walls in the stele were hard to distinguish in the optical sections due to high autofluorescence (Fig. 4) and therefore the cellular dimensions could not be determined for these tissues.
Fig. 4.
Optical sections of wildtype and CORE mutant roots of plants grown on medium supplemented with 4.5% sucrose. Median longitudinal sections through the differentiation zone of (A) wild-type Col, (B) lion’s tail-1, (C) cobra-1, (D) quill-1, (E) pom-pom1-1, (F) cudgel-1, (G) pom-pom2-1. Median longitudinal sections through the root tips of (H) wild-type Col, (I) lion’s tail-1, (J) cobra-1, (K) pom-pom1-1 and (L) pom-pom2-1. Bar, 50 μm.
Table 2. Cellular dimensions of fully expanded roots of wild type and CORE mutants.
| Genotype | Endodermis length/diameter |
Cortex length/diameter |
Endodermis length/radial width circumferential length |
Stele diameter | Root diameter |
|---|---|---|---|---|---|
| wild type | 174±38.5 13.2±1.8 |
156±3.5 18.0±2.8 |
112.8±29 6.2±1.4 13.5±2.6 |
38.0±4.7 | 120.5±29 |
| lit-1 | 20.0±9.0 23.6±7.6 |
20.4±7.6 32.7±4.6 |
18.4±6.6 13.9±1.9 20.0±2.5 |
82.9±13.8 | 229.6±17 |
| cob-1 | 21.7±8.8 68.9±11.2 |
20.1±6.6 30.8±10.0 |
15.4±5.0 15.2±3.2 21.8±8.8 |
52.5±8.9 | 289.7±39.4 |
| qui-1 | 33.7±11.7 65.4±27.8 |
26.5±8.9 26.3±9.7 |
21.9±7.0 8.4±1.1 12.7±2.4 |
44.8±5.1 | 259.6±56.5 |
| pom1-1 | 51.7±21.5 44.1±9.7 |
38.8±16.9 45.2±10.7 |
27.5±13.0 10.1±3.1 17.9±4.9 |
58.2±22 | 259.8±55.8 |
| cud-1 | 30.2±11.5 64.9±18.2 |
25.1±6.5 35.5±6.2 |
19.4±6.5 14.9±5.9 19.9±2.6 |
70.3±15.3 | 319.8±79.2 |
| pom2-1 | 59.0±21.5 34.9±8.0 |
47.1±16.1 41.5±5.3 |
32.8±10.5 11.5±1.9 16.1±4.2 |
54.7±13.5 | 238.2±33.5 |
All measurements are in micrometers.
The cell length measurements indicated that all mutants had dramatically reduced cell elongation (Table 2). The mutants varied greatly in the extent of radial expansion. The ratio of cell length to diameter provided an indication of the polarity of cell expansion. In wild-type epidermal cells the length was significantly greater than the diameter (Table 2). However, in cob-1, qui-1, and cud-1 the reverse was true, suggesting that there was a change in polarity of cell expansion in the epidermal cells (Table 2). For lit-1, pom1-1 and pom2-1 the length and diameter of epidermal cells were approximately equivalent (Table 2). For wild-type cortex and endodermal cells the length was also significantly greater than the diameter (Table 2). For all mutants the cortex and endodermis cells had approximately equivalent cell lengths and diameters (Table 2).
Cell volumes were calculated from the cellular dimensions (See Materials and Methods). For the mutants cob-1, qui-1 and cud-1 the volume calculation was based on the shift in polarity described above so that the cell length was now the diameter and vice versa. From the volume calculations, the mutants could be divided into three groups. The first class had volumes of epidermis, cortex and endodermis that were significantly smaller than wildtype. The mutant lit-1 was the only member of this class. Cell volumes in lit-1 roots were approximately one quarter those of wildtype (Table 3). This indicates that mutation of the lit gene results in defective control of the extent of cell expansion so that normal cell volumes are never obtained. In the second class, represented by cob-1, the epidermal cell volume was approximately equal to wildtype (Table 3). This indicates that the cob mutation does not have a dramatic effect on the extent of expansion since volume (at least of the epidermis) is conserved. It suggests that the primary defect in cob could be a shift in the polarity of cell expansion. The third class of mutants had cell volumes that were significantly greater than wildtype for at least one tissue. In qui-1 and cud-1 roots, epidermal tissue had a greater volume than wildtype (Table 3). In both pom1-1 and pom2-1 there was increased volume in epidermis as well as cortex. This suggests that these mutations cause a loss of regulation of the extent of expansion.
Table 3. Mean and range* of calculated volumes of fully expanded root cells.
| Genotype | Epidermis mean (max-min) |
Cortex mean (max-min) |
Endodermis mean (max-min) |
|---|---|---|---|
| wildtype | 24 (34-14) | 40 (57-22) | 9 (14-4) |
| lit-1 | 6 (10-2) | 17 (26-8) | 6 (8-3) |
| cob-1 | 26 (44-8) | 15 (25-5) | 6 (9-2) |
| qui-1 | 58 (102-14) | 15 (15-4) | 3 (4-1) |
| pom1-1 | 79 (130-28) | 63 (104-21) | 5 (9-1) |
| cud-1 | 47 (81-13) | 25 (37-13) | 6 (10-2) |
| pom2-1 | 56 (92-21) | 64 (96-32) | 6 (9-3) |
Calculation of mean used mean values for cell diameters and lengths from Table 2. Lower values for range calculated using mean minus standard deviation for both diameter and length, upper values used mean plus standard deviation for both diameter and length.
All measurements are in picoliters.
The cell volume of the endodermis was reduced in all mutants. qui-1 showed the most dramatic reduction. This may be due to positional constraints of these cells between the cortex and the stele. It is important to note that the range of calculated cell volumes was much larger in the mutants than in wild type (Table 3). This may due to the conditional nature of the mutations which causes a variability in the diameter of the root. An alternative explanation is that the tight regulation of volume and shape has been lost in the different tissues of all the mutants.
To determine if the mutant phenotypes were specific for the root we examined the dimensions of hypocotyl cells. The CORE mutants differed little from wildtype in the cell lengths of the epidermis, cortex and endodermis (Table 4). The large standard deviation of the cell length data results from the gradual increase of cell length along the hypocotyl. The diameter of the hypocotyl was similar to wildtype for lit-1, cob-1 and qui-1. However, for pom1-1, cud-1 and pom2-1 there was a small increase in hypocotyl diameter. This increase was due primarily to a slight increase in the radial expansion of the two cortex layers. The radial expansion of the hypocotyl in these three mutants (approximately 40%) was still significantly smaller than the radial expansion of the roots (200-260% for these three mutants). When grown in the dark to stimulate more rapid elongation of hypocotyl cells there was still no major increase in the hypocotyl diameter (data not shown). We conclude that the radial expansion phenotype of the CORE mutants affects primarily root cells.
Table 4. Hypocotyl cell dimensions.
| Length (mm) |
Diameter (μm) |
Epidermis length (μm) |
Cortex length (μm) |
Endodermis length (μm) |
|
|---|---|---|---|---|---|
| wt | 1.3±0.4 | 252±32 | 142±60 | 106±30 | 97.0±31 |
| lit-1 | 0.9±0.2 | 297±39 | 116±38 | 75.9±20 | 62.1±17 |
| cob-1 | 1.5±0.4 | 292±27 | 152±47 | 84.4±28 | 63.6±19 |
| qui-1 | 0.9±0.2 | 263±33 | 109±65 | 82.8±36 | 70.4±24 |
| pom1-1 | 1.3±0.4 | 319±57 | 93.4±52 | 71.5±47 | 61.7±30 |
| cud-1 | 0.8±0.3 | 311±20 | 96.5±27 | 76.5±17 | 65.5±16 |
| pom2-1 | n.d. | 363±49 | 113.6±38 | 80.2±20 | 68.0±23 |
Measurements were made 4 days after germination.
Apparent cell division rate of mutant roots
Under conditions that favor higher growth rates for wild-type roots, the CORE mutant roots are expanded and very short. We wished to determine whether the reduction in cell elongation was sufficient to account for the reduced growth rate of the mutants. Root growth was determined by measuring the root length differences between days 4 and 6 after germination (Table 5). As would be expected from the severe defects in cell elongation, root growth (as defined by length increase) was severely reduced in all the CORE mutants. Assuming constant cell cycle time, we calculated the cell division rate by dividing the root growth per day by the cell length (plus or minus the standard deviation) (Table 5). For the mutants qui-1 and pom2-1 the reduction in root growth appeared to be due primarily to the reduction of cell elongation. For the other mutants the shorter root length and the slower root growth appeared to be a combination of reduced cell elongation and an apparent reduction in the division rate. The most dramatic apparent reduction in cell division rate was observed in lit-1 and cud-1.
Table 5. Comparison of root growth and apparent cell division rate under maximal growth rate conditions.
| Root growth (mm/day) |
Division rate (cell number increase/day) |
|||
|---|---|---|---|---|
| epidermis | cortex | endodermis | ||
| WT | 6.0 | 28-44 | 32-49 | 43-72 |
| lit-1 | 0.15 | 5-14 | 5-13 | 6-14 |
| cob-1 | 0.3 | 10-23 | 11-23 | 15-30 |
| qui-1 | 1.0 | 22-46 | 27-55 | 34-66 |
| pom1-1 | 0.65 | 9-21 | 12-30 | 16-43 |
| cud-1 | 0.1 | 2-5 | 3-6 | 4-8 |
| pom2-1 | 1.3 | 16-35 | 21-42 | 30-57 |
Root growth was determined by measuring the root length differences between day 4 and 6 after germination. The apparent cell division rate was calculated by dividing the root growth per day by the cell length (± the standard deviation; see Table 2).
From optical sections of root tips (Fig. 4H-L) we calculated the cell number in the cortex tissue file between the initial cells and the first cell that exhibited polar or increased cell expansion. This is approximately equivalent to the region that has been termed the post-mitotic isodiametric growth (PIG) zone (Baluska et al., 1993). In wildtype this region consists of about 20 cells. For the mutants cob-1, lit-1, and cud-1 which have the greatest apparent reduction in cell division rate there were only about 4 cells in this region. The mutants qui-1, pom1-1 and pom2-1 have a cortex cell number of about 9, 7 and 10 respectively in this region. These data suggest that the timing of cell expansion and differentiation is not necessarily linked to a specific number of cell divisions.
Cell expansion upon shifting to restrictive conditions
To determine if all growing cells were sensitive to the effects of the cobra mutation we performed experiments in which plants were shifted from permissive to restrictive conditions. Seedlings were grown for 8 days on nutrient medium supplemented with 0.1% sucrose. Epidermal cells at the tip of the root were marked with carbon grains (Okada and Shimura, 1990). Plants were then transferred to nutrient medium supplemented with either 4.5% sucrose or with 0.1% sucrose as a control. Images of roots taken at intervals were then analyzed for the relative movement of the carbon grains (Fig. 5). Estimates of elongation rates were determined from distance increases between adjacent carbon grains during a particular time interval. Increases in root diameter were used as estimates of radial expansion for the same time interval.
Fig. 5.
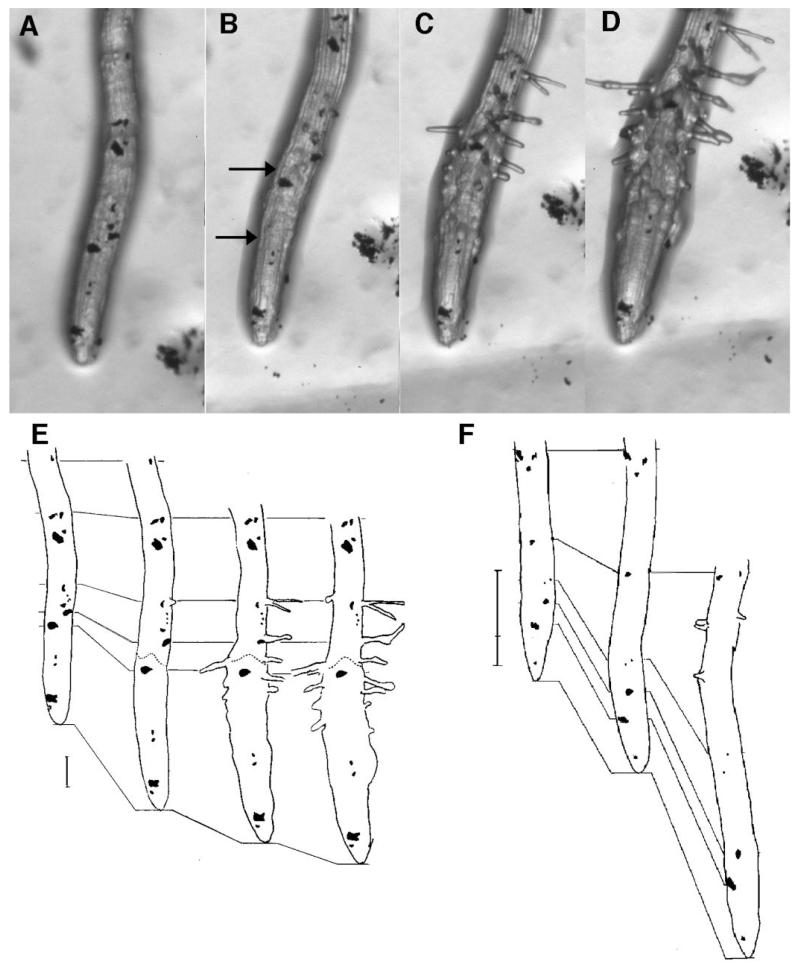
The effect on cell expansion of a shift from permissive to restrictive conditions. cobra was grown for 8 days on medium supplemented with 0.1% sucrose and root epidermal cells were marked with carbon grains. Seedlings were then transferred to media supplemented with 4.5% sucrose or 0.1% sucrose. (A) cob-1 root tip at 0 time after transfer to 4.5% sucrose. (B) cob-1 at 4 hours after transfer to 4.5% sucrose. (C) cob-1 at 8 hours after transfer to 4.5% sucrose. (D) cob-1 at 12 hours after transfer to 4.5% sucrose. (E) Tracings of root tips shown in A-D with location of carbon grains indicated. Lines connect equivalent regions of the root as evidenced by the presence of distinct carbon grains. Dotted line indicates point above which there is very little abnormal radial expansion. Bar, 100 μm. (F) Tracings of root tips of cob-1 seedlings transferred from 0.1% sucrose to 0.1% sucrose. From left to right, times are 0, 4 and 8 hours after transfer. Bar, 300 μm. Note the carbon grains at the very tip of both sets of roots are on root cap cells and therefore show little movement relative to the tip.
Abnormal radial expansion was already observable 4 hours after the shift to restrictive conditions (Fig. 5B). Abnormal radial expansion appeared to be restricted to a subset of growing cells (Fig. 5B,E). Above this region there were cells in which elongation occurred without any apparent abnormal radial expansion. In particular, the relative movement of carbon grains located approximately 350 μm from the root tip indicated continued elongation of these cells while there was little increase in the root diameter at this point (Fig. 5E). The meristematic zone also exhibited little or no change in shape during the first 8-12 hours after the shift to restrictive conditions (Fig. 5B-D). Estimates of relative expansion rates suggested that in the region of abnormal radial expansion, the rate of radial expansion exceeded that of elongation. This provides additional evidence that the cobra mutation results in a shift in the orientation of polar expansion.
Orientation of cortical microtubules
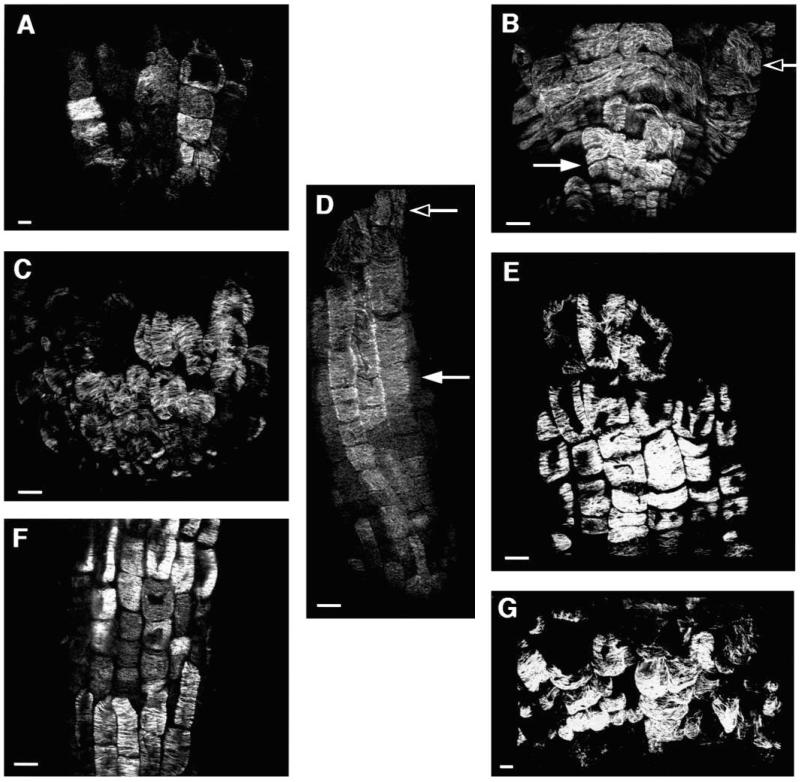
All the CORE mutants have abnormal polarity as evidenced by the relative dimensions of the wall lengths of their epidermal cells. Particularly striking is the shift in polarity of the epidermal cells in cob. The prevailing model of expansion regulation predicts that there should be a shift in the orientation of the microtubules when there is a change in the polarity of expansion (Giddings and Staehelin, 1991) (see Discussion). To examine whether such a shift had occurred in the CORE mutants, we analyzed microtubule orientation by in situ localization with an anti-tubulin antibody.
In wild-type roots, cortical microtubules in the elongation zone were aligned transversely to the axis of growth as predicted by the model (Fig. 6D). In cells that had entered the differentiation zone the microtubule orientation was more random (Fig. 6D). In cob-1 there did not appear to be a dramatic shift in the orientation of microtubules (Fig. 6B). From the staining pattern it appeared that the orientation of the microtubules may be somewhat more random and the microtubule bundles appeared somewhat thicker. As with wildtype, outside the elongation zone the cob-1 microtubule orientation appeared much more random (Fig. 6B). The same was found for qui-1 and cud-1 in which epidermal polarity appeared shifted (Fig. 6C,G). Cortical microtubules of lit-1, pom1-1 and pom2-1 were also found to be predominantly in the same orientation as wildtype (Fig. 6A,E,F).
Fig. 6.
Optical sections of the root epidermis of seedlings grown on medium supplemented with 4.5% sucrose after immunolocalization of microtubules (A) lion’s tail-1, (B) cobra-1, (C) quill-1, (D) wild-type Col, (E) pom-pom1-1, (F) pom-pom2-1, (G) cudgel-1. Bar, 10 μm.
Analysis of CORE mutant interactions
To examine the genetic interactions of the CORE mutants we generated double mutant combinations (Table 6). Combinations of lit-1 and the other CORE mutants tested had phenotypes similar to lit-1 for the root expansion character. The double mutants had reduced cell volume of the epidermis, cortex and endodermis (Fig. 7A-D). However, the root length of these double mutants was dramatically shorter than either parent. The double mutants also had an aerial phenotype that was severely stunted (Fig. 8A-D). This was particularly evident in the combinations of lit-1 with cud-1 and with pom1-1 (Fig. 8A,D).
Table 6. Analysis of CORE double mutants.
| Seedling phenotypes |
||||||
|---|---|---|---|---|---|---|
| Parental genotype mut1 × mut2 |
Total | Wild-type Obs./Exp. |
Mut1 Obs./Exp. |
Mut2 Obs./Exp. |
Double Obs./Exp. |
χ 2 |
| cob-1 × lit-1 | (Benfey et al., 1993) | |||||
| qui × lit | n.d. | |||||
| pom1-1 × lit-1 | 228 | 133/128 | 44/42.8 | 38/42.8 | 13/14.3 | 0.90 |
| cud-1 × lit-1 | 170 | 59/63.8 | 64/63.8 | 21/21.3 | 26/21.3* | 1.4 |
| pom2-1 × lit-1 | 218 | 121/122.6 | 46/40.9 | 39/40.9 | 12/13.6 | 0.93 |
| qui-2 × cob-1 | 347 | 64/65.1 | 283/281.9 | (39/21.7)†,§ | 0.02 | |
| pom1-10 × cob-1 | 750 | 430/421.9 | 256/281.3 | 64/46.9† | 8.7** | |
| 430/421.9 | 320/328.2 | 0.36 | ||||
| cud-1 × cob-1 | 213 | 77/79.9 | 106/106.5 | 30/26.6*,† | 0.8 | |
| pom2-1 × cob-1 | 148 | 82/83.3 | 18/27.8 | 32/27.8 | 16/9.3† | 8.9** |
| 82/83.3 | 66/64.9 | 0.04 | ||||
| pom1-1 × qui-1 | 564 | 334/328 | 212/219 | 38/36.5‡ | 0.4 | |
| cud-1 × qui-2 | 129 | 61/48.4 | 38/48.4 | 15/16.2 | 15/16.1* | 5.67 |
| pom2-1 × qui-1 | 155 | 76/87.2 | 69/58.1 | 10/9.69‡ | 3.48 | |
| cud-1 × pom1-1 | 249 | 105/93.4 | 116/124.5 | 28/31.1 | 2.33 | |
| pom2-1 × pom1-1 | 292 | 149/164.3 | 122/109.5 | 21/18.3‡ | 3.25 | |
| cud × pom2 | n.d. | |||||
cud is dominant and male gametophytic lethal and therefore the dihybrid cross segregates 3: 3: 1: 1 = wt: cud: mut2: dbl
cob is semidominant and therefore we observed a gradient of slightly different phenotypes which we scored together. They include the genotypes cob/cob +/+, cob/cob mut2/+, cob/+ mut2/mut2, +/+ mut2/mut2 and in case of the cross of cob with qui probably also cob/+ qui/+ and cob/+ +/+. In a cob/+, qui/+ trans-heterozygote we observed enhancement of the cob semidominant phenotype.
Due to very similar phenotype it was not possible to distinguish pom1, pom2 and qui as single mutants. The double mutant phenotype was easily scored.
Some of the plants scored as double mutants survived to give F3 seeds. The analysis of these F3 as well as testcross results revealed that the surviving putative double mutants had the genotype cob/cob.qui/+. Therefore we placed this number in parentheses.
The χ2 value for these gave a P value smaller than 0.05. This is probably due to the semidominant cob phenotype which made it difficult to distinguish double mutant homozygotes from the double mutant combination with a heterozygous cobra allele. Abbreviations: mut., mutant; obs, observed; exp, expected, n.d., not determined.
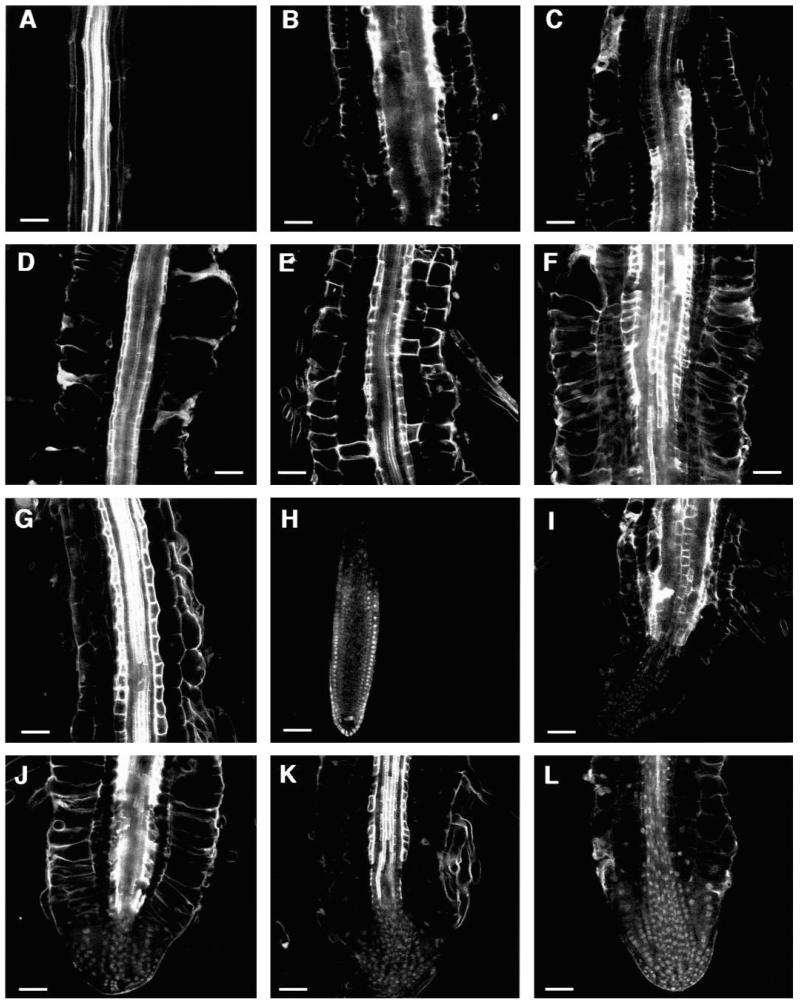
Fig. 7.
Optical sections of CORE double mutants grown on medium supplemented with 4.5% sucrose. Median longitudinal sections through the differentiation zone of double mutants of (A) cudgel-1 and lion’s tail-1, (B) cobra-1 and lion’s tail-1, (C) pom-pom1-1 and lion’s tail-1, (D) pom-pom2-1 and lion’s tail-1, (E) cudgel-1 and quill-2, (F) cudgel-1 and pom-pom1-1. Median longitudinal sections through root tips of double mutants of (G) cudgel-1 and cobra-1, (H) quill-1 and pom-pom1-1, (I) cobra-1 and quill-2, (J) cobra-1 and pom-pom1-10, (K) pom-pom1-1 and pom-pom2-1, (L) cobra-1 and pom-pom2-1. Bar, 50 μm.
Fig. 8.
Seedlings of CORE double mutants grown on medium supplemented with 4.5% sucrose approximately 17 DAG. Double mutant of (A) cudgel-1 and lion’s tail-1, (B) quill-3 and lion’s tail-1, (C) pom-pom1-1 and lion’s tail-1, (D) pom-pom2-1 and lion’s tail-1, (E) cudgel-1 and quill-2, (F) cudgel-1 and pom-pom1-1, (G) cudgel-1 and cobra-1, (H) quill-1 and pom-pom1-1, (I) cobra-1 and quill-1, (J) cobra-1 and pom-pom1-10, (K) pom-pom1-1 and pom-pom2-1, (L) cobra-1 and pom-pom2-1. Bar, 1 mm.
All cud-1 double mutant combinations had root expansion characteristics reminiscent of cud-1 single mutants. The phenotype of the double mutants included dramatically expanded stele tissue as well as radially expanded epidermal and cortex tissue (Fig. 7E-G). In addition, double mutants of both lit-1 and cud-1 had extremely reduced elongation zones that resulted in roots with cells that were expanded in the zone where cell division normally takes place (Fig. 7C,D,F,G). Severely stunted aerial phenotypes similar to those of the lit-1 double mutants were observed in the double mutants of cud-1 with the other CORE mutants (Fig. 8E-G).
Double mutant combinations of cob-1 with pom1-10 or pom2-1 appeared to have an additive phenotype for expansion in the epidermis and cortex (Fig. 7J,L). The dramatic radial expansion of the epidermis which is characteristic of cob-1 was apparent in these double mutant combinations. However, in combination with cud-1 and qui-2, cob-1 did not appear to provide any additional radial expansion of the epidermis beyond that which is characteristic of these two mutants (Fig. 7G,I). In a similar fashion, double mutants with qui-2 did not have increased epidermal cell expansion (Fig. 7E,I). We note though, that double mutants with any of the alleles of qui were more affected in the aerial part of the plant (compare Fig. 8E and 8G, or 8H and 8J). The pom2-1 mutation seemed to enhance slightly all other mutant phenotypes when in a double mutant combination (Fig. 7K,L). Double mutant combinations with pom2-1 exhibited a nearly wild-type aerial part, but due to the extremely reduced fertility it was not possible to get F3 seeds. Double mutant combinations with either pom1-1 or pom1-10 were characterized by shorter roots that had differentiated and expanded cells in what would normally be the meristematic zone (Fig 7C,F,H,J,K).
To test if the conditional phenotype was maintained in the double mutants we analyzed root growth under normally permissive growth conditions. All of the double mutant combinations tested had expanded roots when grown under conditions in which the single mutants do not have detectable expansion (Figs 2E-H, 3G-J). This suggests that these genes may have at least partially redundant functions or act in a similar pathway (see Discussion). Most of these double mutants had extremely low fertility precluding further genetic testing.
DISCUSSION
The regulation of cell expansion is of paramount importance to the determination of organ shape in higher plants. The two principal parameters of cell expansion are orientation and extent. We have identified six genetic loci that when mutated, result in deregulation of cell expansion. Three of the loci were newly isolated and additional alleles of previously identified loci have been characterized. Common features of these CORE mutations include the dramatically reduced elongation of the root, the variability in the degree of expansion along the length of the root and the root-specificity of the expansion defects. From calculations of cell volumes we have been able to place the mutants into three classes: those with volumes less than, equal to, and greater than wildtype. This indicates that the mutations result in abnormal regulation of the extent or the orientation of expansion or of both.
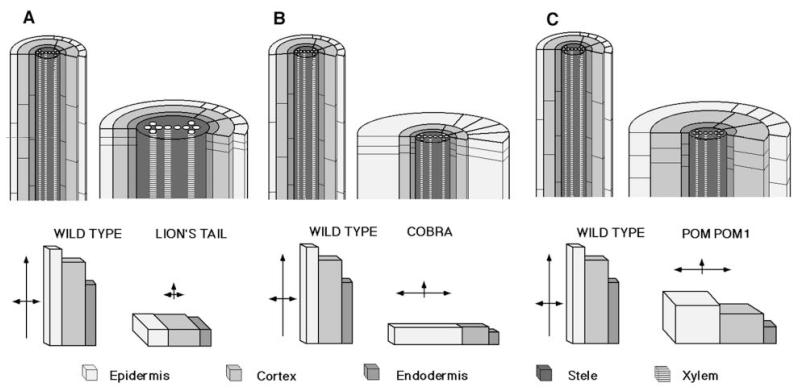
The volume calculations indicate that the primary defect in lit may be an inability to achieve correct elongation with little compensating radial expansion (Fig. 9A). This suggests that this gene regulates the cell’s ability to attain its proper volume. For cobra we have determined that epidermal cell volume of the mutant is approximately equal to wildtype (Fig. 9B). This suggests that the primary defect may be in the control of the orientation of cell expansion. The principal axis of expansion in these mutants may be in the radial rather than the longitudinal direction. The third class, which includes quill, cudgel, pom-pom1 and pom-pom2, has cell volumes that are significantly greater than wildtype. This indicates that these genes play a role in controlling the extent of expansion (Fig. 9C).
Fig. 9.
Schematic representation of the three classes of CORE mutants. (A) In lion’s tail cell volume of the outer three layers is reduced as compared to wildtype. (B) In cobra there is approximate conservation of cell volume. Therefore, radial expansion compensates for the reduction in elongation. This leads to a shift in the polarity of expansion. (C) In the other four mutants, represented here by pom-pom1 there is a defect in volume regulation so that the cells attain a greater volume than wildtype. This results from a major increase in radial and circumferential expansion that overcompensates for the reduction in elongation.
The role of microtubules in determining the orientation of expansion
To analyze further the effect of the cobra mutation on cell expansion we shifted plants from permissive to restrictive conditions. These experiments suggested that the cobra mutation affected cells only for a limited period during their transit through the elongation zone. Preliminary measurements also suggested that the mutation caused a change from anisotropic growth in the longitudinal direction to anisotropic growth in the radial orientation. If a true shift in the polarity of expansion occurs in this mutant it could be used to test theories concerning the role of different macromolecules in the control of cell expansion. It was expected that the cortical microtubules in the epidermal cells of cobra would exhibit a dramatic shift in polarity. This was found not to be the case. It is possible that this is a situation in which a dramatic shift in microtubule orientation would not be required to obtain the observed cell shapes. One possibility is that elongation is arrested while “normal” radial expansion is unimpaired by the mutation. In this case, the mutation must allow radial expansion to continue beyond the limit found in wildtype. Another possibility is that the CORE mutations result in a change from anisotropic to isotropic expansion. If this were true then a dramatic shift in microtubule orientation would not be observed. Rough estimates of relative expansion rates suggest that at least in cobra there has been a shift from anisotropic expansion in the longitudinal direction to anisotropic expansion in the radial direction. However, this can only be resolved by accurate measurements of cellular expansion rates. It should also be noted that although the microtubules do not shift their orientation, the microtubule bundles appear to be more aggregated and slightly more disorganized than wildtype. A key question for future investigations is whether the cellulose microfibrils are still in the same orientation as the microtubules or whether they have shifted their orientation.
These observations raise the possibility that microtubules may not play a critical role in regulating the orientation of cell expansion in Arabidopsis root cells. Further evidence for this comes from studies of ethylene treated roots and of roots treated with microtubule stabilizing and destabilizing agents (Baskin and Williamson, 1992; Baskin et al., 1994).
Several of the CORE mutants appeared to have reduced cell division rates. The calculations of cell division rates were based on the assumption that cells were dividing at constant rates. Because we do not have direct evidence for this steady state situation these observations should be interpreted with caution. We note, however, apparent division rates were particularly low for the mutants lit, cud, and cob that have a large amount of expansion in the stele (see Table 2). It is possible that a lower cell division rate is caused by defects in the normal functioning of nutrient transport of the root vascular tissue. We do not believe that abnormal cell expansion is a secondary effect of reduced cell division because we have analyzed several mutants in which cell division slows down and then ceases entirely (Benfey et al., 1993; Scheres et al., 1995). In these mutants there is no abnormal cell expansion equivalent to that seen in the CORE mutants.
Because plant growth regulators can have a dramatic effect on plant cell expansion we grew the CORE mutants on different concentrations of auxin, cytokinin, gibberellic acid, abscissic acid, the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), silver ions (which inhibit ethylene action), the auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA), and the gibberellic acid biosynthesis inhibitor, ancymidol. The root expansion phenotype was not noticeably affected by any of these treatments (data not shown).
Other mutations have been identified that affect root cell expansion. In the sabre mutant there is a shift in orientation of expansion primarily in root cortex cells (Benfey et al., 1993; Aeschbacher et al., 1995). It was found that reduction in effective ethylene levels resulted in partial rescue of the phenotype. This suggested that the SABRE gene and ethylene regulated two counteracting processes that determined the extent of radial expansion of certain cells (Aeschbacher et al., 1995). A temperature sensitive screen identified three mutants with abnormal root expansion (Baskin et al., 1992). At the restrictive temperature these mutants have abnormal aerial growth as well. Complementation analysis revealed that these mutants are not allelic to the CORE mutants (R.Williamson, M.-T.Hauser and P.Benfey unpublished results).
Double mutant analysis suggests CORE genes may have similar functions
The mutant phenotype of all of the CORE mutants is conditional on the root growing at a maximum rate. Unlike a simple temperature-sensitive mutation in which a protein folding defect would be implicated, the CORE mutant conditional phenotype suggests that there may be conditional redundancy of function. In this hypothesis, under submaximal growth conditions, if any one of the CORE gene activities is removed another gene activity can substitute for it. However, under maximum growth conditions all of the CORE genes would be necessary for regulated root cell expansion. A prediction of this hypothesis is that combinations of the mutations at redundant loci should result in a loss of the conditional phenotype. This is what we found for all tested double mutant combinations of the CORE mutants. This is evidence for the CORE mutants providing partially redundant functions which might be accomplished if they acted in a multimeric complex or in a similar developmental process.
In addition to loss of the conditional phenotype, novel phenotypes appeared in some double mutant combinations. These included complete cessation of root growth and severely affected aerial organs. Cessation of root growth was particularly evident in double mutant combinations in which stele tissue was affected in both single mutants. Thus, this may be a secondary effect of impairment of vascular tissue function. The aerial phenotypes may also be attributable to impaired root function. Alternatively, they may indicate that the CORE genes play important functions in shoot cell expansion but that under our growth conditions a single gene mutation is always masked by functional redundancy.
The double mutant combinations of the CORE mutants also revealed possible epistatic relationships. For the character of root expansion, lit was epistatic to all the other mutants tested, cud was epistatic to all mutants except lit while the double mutant phenotypes of the other four mutants cob, qui, pom1 and pom2, were essentially additive. Interpretation of these results should take into account whether the allele used in the double mutant analysis represented the null phenotype. In the absence of molecular data or deletion tester lines it is impossible to make such designations. Thus, extreme caution must be exercised in interpreting these results. We note, nonetheless, that the apparent epistasis of lit to the other six mutants would be plausible, in that lit is impaired in its ability to increase cell volume and therefore to elongate properly and expand. It seems reasonable that this phenotype would be epistatic to either that of cob, in which the orientation of expansion is primarily affected, or the other four mutants, in which there is too much expansion. It is important to note that this apparent epistasis is only for the root expansion character. These lit double mutants also lose their conditional phenotype and have abnormal aerial organs. An alternative possibility is that the double mutant phenotypes are the result of combinations of hypomorphic mutations in the same pathway.
In summary we have identified six loci that regulate either the orientation, the extent or both aspects of expansion in root cells. The loss of the conditional phenotype when the mutations in these loci are combined indicates that they may act in a similar pathway. Molecular analysis of the CORE gene products should shed additional light upon the genetic control of cell expansion.
Acknowledgments
We thank K. Roberts, P. Linstead and C. Duckett for their kind help and support. We received expert technical assistance from H. Bondar, V. Blanc, D. Tang, M. Yung, K. Schultheiss, K. Gallagher, J. Lim, and M. S. Hsieh. We thank M. Huelskamp and C. Schultz for stimulating discussions. This publication benefitted enormously from the comments and criticisms of L. Di Laurenzio, J. Wysocka-Diller, J. Malamy, R. Aeschbacher, J. Schiefelbein and the reviewers. We thank J. Schiefelbein for providing the qui-3, pom1-9 and pom1-11 alleles, K. Feldmann for providing the pom1-1 allele and C. Schultz and V. Blanc for the pom1-10 allele. We thank J. Ecker for providing X-irradiated seed, C. Leonard for help with confocal microscopy, S. Ruzin for confocal staining protocols, R. White for use of his slide scanner. M-T. H. was supported by a Schroedinger fellowship from “Fonds zur Foerderung der wissenschaftlichen Forschung” (J0676-MOB). The work in P. N. B.’s laboratory was supported by a grant (RO1-GM43788) from the NIH.
REFERENCES
- Aeschbacher RA, Hauser M-T, Feldmann KA, Benfey PN. The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 1995 doi: 10.1101/gad.9.3.330. in press. [DOI] [PubMed] [Google Scholar]
- Baluska F, Brailsford RW, Hauskrecht M, Jackson MB, Barlow PW. Cellular dimorphism in the maize root cortex: involvement of microtubules, ethylene and gibberellin in the differentiation of cellular behaviour in postmitotic growth zones. Acta Bot. 1993;106:394–403. [Google Scholar]
- Baskin TI, Betzner AS, Hoggart R, Cork A, Williamson RE. Root morphology mutants in Arabidopsis thaliana. Aust. J. Plant Physiol. 1992;19:427–438. [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 1994;35:935–942. [PubMed] [Google Scholar]
- Baskin TI, Williamson RE. Ethylene, microtubules and root morphology in wild-type and mutant Arabidopsis seedlings. Curr. Top. Pl. Biochem. Physiol. 1992;11:118–130. [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;18:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser M-T, Aeschbacher RA. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Cho SO, Wick S, Sundaresan V. Localization of the functional p34-cdc2 homolog of maize in root tip and stomatal complex cells: Association with predicted division sites. Pl. Cell. 1993;5:1011–1111. doi: 10.1105/tpc.5.9.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. How do plant cell walls extend? Plant Physiol. 1993;102:1–6. doi: 10.1104/pp.102.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J, Zhang H, Traas J. Indirect Immunofluorescence on Arabidopsis seedlings. In: Flanders D, Dean C, editors. Arabidopsis, The Complete Guide. AFRC Institute of Plant Science Research; Norwich, U.K.: 1993. [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Giddings TH, Jr., Staehelin LA. Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In: Lloyd CW, editor. The Cytoskeletal Basis of Plant Growth and Form. Academic Press; London: 1991. pp. 85–100. [Google Scholar]
- Hauser M-T, Benfey PN. Genetic Regulation of Root Expansion in Arabidopsis thaliana. In: Pugdomenech P, Coruzzi G, editors. NATO-ASI Plant Molecular Biology Series. Springer-Verlag; New York: 1993. pp. 31–40. [Google Scholar]
- Kaplan DR, Hagemann W. The relationship of cell and organism in vascular plants. BioScience. 1991;41:693–703. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- Roberts K. The plant extracellular matrix. Current Opinion in Cell Biology. 1989;1:1020–1027. doi: 10.1016/0955-0674(89)90074-4. [DOI] [PubMed] [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser M-T, Janmaat K, Weisbeek P, Benfey PN. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the radial axis. Development. 1995;121:53–62. [Google Scholar]
- Schiefelbein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. Pl. Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge University Press; Cambridge: 1989. [Google Scholar]
- Valvekens D, Montagu MV, Lusebettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]