Abstract
Asini Corii Collas (ACC; donkey glue) is a crude drug used to promote hematopoiesis and arrest bleeding. Because adulteration of the drug with substances from other animals such as horses, cattle, and pigs has been found, we examined PCR methods based on the sequence of the cytochrome b gene for source species identification. Two strategies for extracting DNA from ACC were compared, and the ion-exchange resin procedure was revealed to be more suitable than the silica-based one. Using DNA extracted from ACC by the ion-exchange resin procedure, PCR methods for species-specific detection of donkey, horse, cattle, and pig substances were established. When these species-specific PCR methods were applied to ACC, amplicons were obtained only by the donkey-specific PCR. Cattle-specific PCR detected as little as 0.1 % admixture of cattle glue in the ACC. These results suggest that the species-specific PCR methods established in this study would be useful for simple and easy detection of adulteration of ACC.
Electronic supplementary material
The online version of this article (doi:10.1007/s11418-013-0790-z) contains supplementary material, which is available to authorized users.
Keywords: Asini Corii Collas, Donkey glue, Species identification, Cytochrome b
Introduction
Asini Corii Collas (ACC; donkey glue) is a crude drug made of dry skin, bone, tendons, and ligaments of donkeys (Equus asinus) by decoction, degreasing, and concentration according to the Japanese standards for non-Pharmacopoeial crude drugs (non-JP crude drug standards) 2012 [1]. The main component is collagen. ACC has mainly been used in Kampo formulas such as Choreito and Unkeito in Japan, used for promoting hematopoiesis and arresting bleeding.
ACC is mainly produced in Shangton, China. In the Chinese Pharmacopoeia as well as in the non-JP crude drug standards, it is specified as being made of dry skin derived from donkeys [2]. However, adulteration with skins or bones from other animals such as horses, cattle, and pigs has been found [3, 4], because ACC is relatively expensive. Under these circumstances, the establishment of a method of discriminating the animal origin of ACC was needed. Several spectroscopy-based methods have been reported [5–7]; however, these methods have not reached the stage of practical application due to sample-processing variability.
Along with advances in DNA sequence technology, various methods for species identification of feed or processed food have been developed [8]. DNA-based authentication is one of the methods for identifying the animal source of ACC, but DNA extraction from ACC is considered to be difficult because its DNA is severely degraded during processing. In recent years, however, Lv et al. [3] succeeded in the extraction and detection of severely degraded DNA from ACC. Furthermore, they reported a PCR method for authentication of ACC using SINEs (short interspersed nuclear elements) [4]. A SINE is a kind of retroposon, which is a short repetitive sequence with a length of about 300 bp, and has more than 106 total copies per haploid genome [9]. The method of donkey DNA detection using SINE consists of two PCR steps: the first PCR is based on the equine-specific ERE-1 region [10], and the second is based on the horse-specific satellite DNA region [11]. From donkey DNA, a PCR product should be generated by the first PCR, but not by the second. However, this method allows only indirect detection of donkey DNA, and cannot distinguish whether animal glues are derived from a mixture of donkey and horse or from 100 % horse. Although mitochondrial DNA has fewer copy numbers (several thousands) compared with those of SINE, it is widely used in species determination of animal meat and processed food; it has also been useful for assessing the evolutionary relationship between different species [12–14]. Furthermore, it can distinguish donkey from other animals including its close relatives, the horse [15]. In this study, we developed a more effective method of extracting DNA from ACC with reference to the method reported by Lv et al., and established a PCR method for source species identification of ACC based on the sequence differences of the cytochrome b gene in the mitochondrial DNA of several common mammals.
Materials and methods
Materials
ACC and cattle glue were purchased from Shandong Donge E-jiao Co. of Shandong Province.
A donkey hair sample was kindly provided by Hamura Zoo, Japan, with the aid of Dr. Sugita-Konishi. Meat samples from a horse, cow, and pig were purchased from local supermarkets or internet-based store. Their origins were identified by DNA analyses. One sample of each of these animal species was used in this study.
DNA extraction
Before DNA extraction, each side of solid glue samples was irradiated with UV light for 2 h in order to avoid a false-positive result due to the external DNA on the sample surfaces.
DNA extraction was done using a silica-membrane column (QIAquick spin column; Qiagen) according to the report of Lv et al. [3], and using an ion-exchange resin column (Qiagen Genomic tip 20/G or 100/G; Qiagen) with reference to the method for examining genetically modified foods (Ministry of Health, Labour and Welfare, Japan) [16].
For scale-up DNA preparation using the Genomic tip 100/G, 5 g solid glue was ground into powder and transferred into a 50-mL centrifuge tube with 16 mL of G2 buffer (Qiagen), 10 μL of RNase (100 mg/mL), and 200 μL of Proteinase K (20 mg/mL). After incubating the tube for 2 h at 50 °C with occasional shaking, the tube was centrifuged for 15 min at 6,000g at 4 °C.The supernatant was transferred into a 15-mL tube, and centrifuged again for 5 min at 10,000g at 4 °C.After equilibration of the Genomic tip 100/G with 4 mL of QBT buffer (Qiagen), the supernatant was applied to the tip and allowed to pass through the resin by gravity flow. The tip was then washed with 22 mL of QC buffer (Qiagen), and DNA was eluted with 5 mL of QF buffer prewarmed to 50 °C.The eluted DNA was precipitated by adding 3.5 mL of isopropanol, and centrifuged for 15 min at 10,000g at 4 °C. The pellet was washed with 5 mL of 70 % ethanol, and centrifuged. After drying the pellet at 65 °C, the pelleted DNA was suspended in 100 μL of distilled water, and stored at −20 °C until use.
DNA extraction from hair and meat samples was performed using Genomic tip 20/G according to the manufacturer’s protocol.
For the comparison of DNA extraction methods, PCR was performed as reported by Lv et al. [3], using Ass-up/down primers.
Species-specific PCR
Multiple sequences of cytochrome b genes of donkeys, horses, cattle, and pigs were obtained from DDBJ/EMBL/Genbank, and the PCR primers listed in Table 1 were designed on the basis of the sequences specific to each animal species. A schematic of each primer is shown in Fig. S1. In designing the EaCytB-f primer, adenine (A), the fifth nucleotide from the 3′ end, was substituted for thymine (T) to enhance the specificity for donkey against the other three species [17].
Table 1.
Sequence information of primers for species-specific PCR
| Target animal | Primer name | Primer sequence (5′–3′) | Product length |
|---|---|---|---|
| Donkey (Equus asinus) |
EaCytB-f EaCytB-r |
GCCCTTATCCTTTCCATCTTA GCTTCGTTGTTTTGTCATG |
66 bp |
| Horse (Equus caballus) |
EcCytB-f EcCytB-r |
CCCATTCCACCCATATTAT GAGTTAGTAGGAGCAAGATC |
71 bp |
| Cattle (Bos taurus) |
BtCytB-f BtCytB-r |
CCATCGGACAACTAGCATCTG TGTGCCGGCCGTTGGTATTAGC |
68 bp |
| Pig (Sus scrofa) |
SsCytB-f SsCytB-r |
CTAGTAGCAGACCTCATTA GTTGGCCGATGATGATGAAC |
76 bp |
The underlined nucleotide in the donkey sequence was the one that was replaced by another nucleotide to enhance the specificity
PCR amplification was performed in a 25-μL reaction mixture containing 12.5 μL of Ampdirect Plus (Shimadzu), 0.75 U of Ex Taq Hot Start Version (Takara), 0.4 μM each of forward and reverse primer, and 5 ng (hair or meat samples) or 50 ng (glue sample) of template DNA. Amplification was conducted in a DNA Engine thermal cycler, PTC-200 (Bio-Rad) with the following conditions: 94 °C for 4 min; 35 cycles of 94 °C for 30 s, 60 °C (for donkey, horse, and cattle detection) or 64 °C (for pig detection) for 30 s, and 72 °C for 10 s; 72 °C for 2 min. PCR products were electrophoresed on a 4.5 % NuSieve GTG Agarose gel, stained with GelRed Nucleic Acid Gel Stain (Biotium). The amplicons were purified using MinElute PCR purification kit (Qiagen), subcloned into a 2.1-TOPO vector (Invitrogen/Life Technologies), and sequenced. Cycle sequencing was performed using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems/Life Technologies), and the results were analyzed using an ABI Prism 3130 genetic analyzer (Applied Biosystems/Life Technologies).
Detection sensitivity analysis of cattle DNA
We mixed 0.1 % of cattle glue with ACC and used 5 g of the mixture for DNA extraction as described above. Using the DNA as a template, PCR with cattle-specific primers was performed to investigate the detection sensitivity of cattle DNA.
Results
Examination of DNA extraction method from ACC
Lv et al. [3] compared three strategies, the “SDS/proteinase K” (phenol/chloroform) method, the “Wizard magnetic DNA purification system for food” method, and the modified silica-based method, for extracting DNA from ACC. They concluded that the modified silica-based method was the best because it achieved higher DNA yield and purity than the others. In this study, we compared the silica-based method by Lv et al. with the ion-exchange resin-based method (Qiagen Genomic tip 20/G), which was recommended for DNA extraction from genetically modified foods [16].
The DNA extracted by each method from 1 g of donkey glue differed little in purity as estimated from the A260/280 value. Ten ng of each DNA was amplified by PCR with primers based on ERE-1 (equine SINE) in the same way as reported by Lv et al., which resulted in the generation of each PCR amplicon with the predicted size. However, more amplicons were obtained from DNA extracted by the ion-exchange resin method than by the silica-based method, though the same amount of DNA was used in PCR (Fig. S2).
Lv et al. had reported that the SINE region was amplified, but mitochondrial regions such as cytochrome b were not amplified using DNA extracted by the silica-based method [4]. As the ion-exchange resin method was shown to be effective in obtaining more amplicons of the SINE region, we tried to amplify the cytochrome b region, and were able to obtain amplicons using 50 ng of DNA extracted by the ion-exchange resin method. These results suggest that the ion-exchange resin method is superior to the silica-based one for extraction from ACC.
As mentioned above, mitochondrial DNA sequences can distinguish donkey from horse sources, unlike those of SINE. Because it was confirmed that the cytochrome b region can be amplified from DNA of ACC, we selected that region as the target in species-specific PCR.
Establishment of species-specific PCR
ACC adulterated with substances from other animals such as horses, cattle, and pigs has been found in Chinese markets [3, 4]. We tried to establish PCR methods for species-specific detection of substances derived from donkey as well as horse, cattle, and pig-.
As DNA extracted from donkey glue is highly degraded, amplicons more than 100 bp long are difficult to obtain [3]. The primers were designed based on the species-specific sequences in the cytochrome b gene so that the size of amplicons was less than 100 bp (Table 1; Fig. S1). PCR was performed using 5 ng of DNA extracted from each animal sample with the designed species-specific primers. As a result, amplicons specific to each animal were obtained (donkey, 66 bp; horse, 71 bp; cattle, 68 bp; pig, 76 bp) (Fig. 1a–d). Sequencing analysis after subcloning of the PCR products confirmed that all of the sequences showed good identity with cytochrome b genes of the corresponding target animals.
Fig. 1.
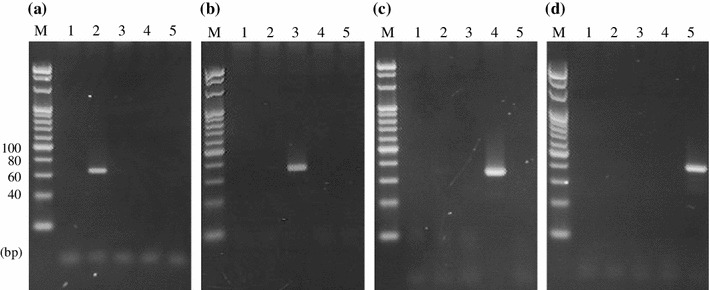
Specificity of PCR for donkey, horse, cattle, and pig DNA with each of their specific primers. a Donkey-specific PCR. b Horse-specific PCR. c Cattle-specific PCR. d Pig-specific PCR. Lane 1 is a no-template control, and lanes 2–5 show the DNA from donkey, horse, cattle, and pig samples, respectively. M, 20-bp ladder marker
These species-specific PCR methods were applied to DNA extracted from ACC known to be derived from donkeys only. Amplicons could be obtained only from the donkey-specific PCR (Fig. 2a–d). These results suggest that the species-specific PCR methods established in this study would be useful for simple and easy identification of the animal origin of ACC.
Fig. 2.
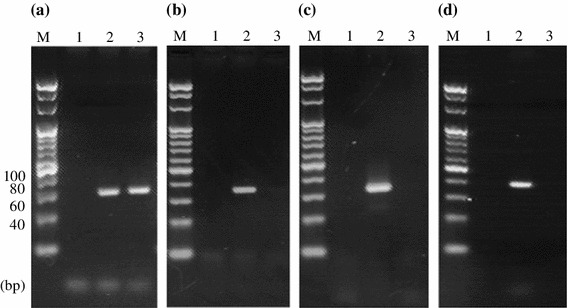
Specificity of PCR for DNA from ACC with each of four animal-specific primers. a Donkey-specific PCR. Lane 2, positive control (PC; donkey DNA), b Horse-specific PCR. Lane 2, PC (horse DNA). c Cattle-specific PCR. Lane 2, PC (cattle DNA). d Pig-specific PCR. Lane 2, PC (pig DNA). Lanes 1 and 3 indicate no-template control and ACC, respectively, in all panels. M, 20-bp ladder marker
Detection sensitivity of cattle DNA
In order to estimate the detection sensitivity of the PCR methods in this study, the detection limit was analyzed using cattle glue as a contaminant. DNA extracted from ACC blended with 0.1 % of cattle glue was applied to the cattle-specific PCR, and the cattle-specific amplicon was detected (Fig. 3). Thus, the PCR method was confirmed to be able to detect as little as 0.1 % admixture of cattle glue in ACC.
Fig. 3.
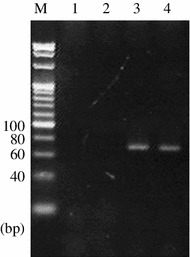
Evaluation of cattle-specific PCR assay sensitivity. Lane 1, no-template control; lane 2, ACC; lane 3, 0.1 % cattle glue in ACC; lane 4, cattle glue; M, 20-bp ladder marker
Discussion
In this study, new PCR methods were established to identify the animal origins of ACC. First, the silica-based and the ion-exchange resin methods were compared as methods for extracting DNA from ACC, and the ion-exchange resin method was revealed to be more suitable. This may be because the ion-exchange resin procedure was so gentle that DNA fragments were not degraded further during preparation, or was superior for removing polysaccharides and phenols that inhibit the activity of the PCR enzyme.
PCR methods based on the cytochrome b gene were established to detect DNA specific to donkeys, horses, cattle, and pigs. The methods were shown to be effective in identifying the animal origins of ACC. Unlike the SINE-based PCR, the cytochrome b-based method could directly distinguish donkey- from horse-derived substances, and should be considered as a new method for simple and easy identification of the animal origins of ACC.
The detection sensitivity of the PCR methods against ACC blended with DNA from other animal species was also investigated. Because horse glue and pig glue could not be obtained, only cattle glue was used for detection limit analysis. As a result, a 0.1 % mixture of cattle was detected in the cattle-specific PCR, which was as sensitive as in the SINE-based method and sensitive enough to examine impurity contamination. On the other hand, the cattle-specific PCR did not have quantitative capability, given the fact that there was little difference in the amount of amplicons from 0.1 and 100 % cattle glue. One of the reasons may be that we detected the amplicons at the endpoint of amplification in the PCR method. For quantitative analysis, further investigations into quantitative PCR would be needed.
Electronic supplementary material
Supplementary material 1 Fig. S1 Schematic of each species-specific PCR primer in the cytochrome b region (600–1,140 bp). DDBJ/EMBL/Genbank accession numbers of the sequences of Equus asinus (donkey), Equus caballus (horse), Bos taurus (cattle), and Sus scrofa (pig) were JF718884, D32190, EF693798, and GU211924, respectively. (EPS 3648 kb)
Supplementary material 2 Fig. S2 PCR amplification using the primers based on ERE-1. a Gel image b Electropherograms. N, no-template control; 1 and 2 indicate the amplicons from DNA by the silica-based and ion-exchange resin method, respectively. LM and UM mean lower and upper markers, respectively. Arrows represent amplicons of ERE-1. Electrophoresis was done by MCE-202 (Shimadzu). (EPS 2296 kb)
Acknowledgments
We thank Dr. Sugita-Konishi and the members of Hamura Zoo (Tokyo, Japan) for kindly providing us with a hair sample from a donkey. This work was supported by Health and Labour Sciences Research Grants in Japan.
References
- 1.The Japanese standards for non-Pharmacopoeial crude drugs (non-JP crude drug standards) (2012) Ministry of Health, Labour and Welfare, Japan
- 2.Pharmacopoeia of the People’s Republic of China, English Edition 2010. China Medical Science Press, Beijing, China
- 3.Lv P, Zhou XS, You JH, Ye BC, Zhang YX (2009) Extraction of trace amount of severely degraded DNA. Z Naturforsch 64c: 581–589 [DOI] [PubMed]
- 4.Lv P, Zhao YJ, Qi F, Zhou XS, You JH, Qin YF, Zhang YX. Authentication of equine DNA from highly processed donkey-hide glue (Colla Corii Asini) using SINE element. J Food Drug Anal. 2011;19:123–130. [Google Scholar]
- 5.Xu CH, Zhou Q, Sun SQ, Wang BQ. The identification of Ejiao by two dimensional correlation infrared spectroscopy. Chinese J Anal Chem. 2005;33:221–224. [Google Scholar]
- 6.Qu HB, Yang HL, Chen YY. Fast and nondestructive discrimination of donkeyhide glue by near-infrared spectroscopy. Spectrosc Spect Anal. 2006;26:60–62. [PubMed] [Google Scholar]
- 7.Wang WJ, Guan Y, Zhu YY. Novel identification of donkeyhide glue by X-ray fluorescence analysis. Spectrosc Spect Anal. 2007;27:1866–1868. [PubMed] [Google Scholar]
- 8.Ballin NZ, Vogensen FK, Karlsson AH. Species determination—can we detect and quantify meat adulteration? Meat Sci. 2009;83:165–174. doi: 10.1016/j.meatsci.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Shedlock AM, Takahashi K, Okada N. SINEs of speciation: tracking lineages with retroposons. Trends Ecol Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Sakagami M, Hiromura K, Chemnick LG, Ryder OA. Distribution of the ERE-1 family in Perissodactyla. Mamm Genome. 1999;10:930–933. doi: 10.1007/s003359901117. [DOI] [PubMed] [Google Scholar]
- 11.Pauciullo A, Kubickova S, Cernohorska H, Petrova K, Di Berardino D, Ramunno L, Rubes J. Isolation and physical localization of new chromosome-specific centromeric repeats in farm animals. Vet Med. 2006;51:224–231. [Google Scholar]
- 12.Krcmar P, Rencova E. Identification of species-specific DNA in feedstuffs. J Agric Food Chem. 2003;51:7655–7658. doi: 10.1021/jf034167y. [DOI] [PubMed] [Google Scholar]
- 13.Pascoal A, Prado M, Calo P, Cepeda A, Barros-Velazquez J. Detection of bovine DNA in raw and heat-processed foodstuffs, commercial foods and specific risk materials by a novel specific polymerase chain reaction method. Eur Food Res Technol. 2005;220:444–450. doi: 10.1007/s00217-004-1088-x. [DOI] [Google Scholar]
- 14.Martin I, Garcia T, Fajardo V, Lopez-Calleja I, Hernandez PE, Gonzalez I, Martin R. Species-specific PCR for the identification of meat species in feedstuffs. Meat Sci. 2007;75:120–127. doi: 10.1016/j.meatsci.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Chrisholm J, Conyers C, Booth C, Lawley W, Hird H. The detection of horse and donkey using real-time PCR. Meat Sci. 2005;70:727–732. doi: 10.1016/j.meatsci.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.The method for examining genetically modified foods, final revision (2008) Ministry of Health, Labour and Welfare, Japan
- 17.Latorra D, Campbell K, Wolter A, Hurle JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 Fig. S1 Schematic of each species-specific PCR primer in the cytochrome b region (600–1,140 bp). DDBJ/EMBL/Genbank accession numbers of the sequences of Equus asinus (donkey), Equus caballus (horse), Bos taurus (cattle), and Sus scrofa (pig) were JF718884, D32190, EF693798, and GU211924, respectively. (EPS 3648 kb)
Supplementary material 2 Fig. S2 PCR amplification using the primers based on ERE-1. a Gel image b Electropherograms. N, no-template control; 1 and 2 indicate the amplicons from DNA by the silica-based and ion-exchange resin method, respectively. LM and UM mean lower and upper markers, respectively. Arrows represent amplicons of ERE-1. Electrophoresis was done by MCE-202 (Shimadzu). (EPS 2296 kb)


