Abstract
Mammals have evolved a dramatic diversity of aging rates. Within the single order of Rodentia maximum lifespans differ from four years in mice to 32 years in naked mole rats. Cancer rates also differ significantly, from cancer-prone mice to virtually cancer-proof naked and blind mole rats. Recent progress in rodent comparative biology, in combination with the emergence of whole genome sequence information, has opened opportunities for the discovery of genetic factors controlling longevity and cancer susceptibility.
Introduction
Mammalian species display dramatically diverse maximum lifespans, ranging from the 2 years of a short-tailed shrew to 211 years of a bowhead whale1. This diversity can be exploited to understand the genetic mechanisms of aging and the diseases associated with aging, most notably cancer. Historically, comparative studies of species have provided spectacular advances in biology, the best example being Darwin’s discovery of natural selection as the driving force underlying the species diversity. Subsequently, however, the focus in biological research shifted to the study of a few model species, which yielded significant discoveries in cell and molecular biology, but lacked the species breadth of previous work. Today, with the new molecular tools and genomic approaches available to study the cellular and physiological mechanisms that control aging, a comparative approach can be utilized at a vastly more sophisticated level. Indeed, in recent years there has been a renewed interest in using interspecies comparisons and unconventional, long-lived animal models for aging studies2–4. A dramatic step toward understanding the molecular basis of human aging and its interaction with disease would be achieved if we could discover the mechanisms responsible for the more than 100-fold differences in lifespan between species of mammals.
Here we focus on the genetic factors that underlie the greatly diverse aging rates and cancer risks among rodents. As we discuss, rodents with their relatively close genetic makeup yet profoundly different lifespans proved especially suitable for identifying key longevity assurance systems, including tumor suppressor mechanisms, such as telomere maintenance and global genome maintenance mechanisms. We review recent progress in understanding how the longest-lived rodents, the blind mole rat and the naked mole rats, achieve cancer resistance and a long life. Finally, we discuss recent advances in whole-genome sequencing of very long-lived mammals and how this can increase our understanding of the genetics of human longevity and the resistance to cancer and other age-related diseases.
Rodents as models for comparative studies
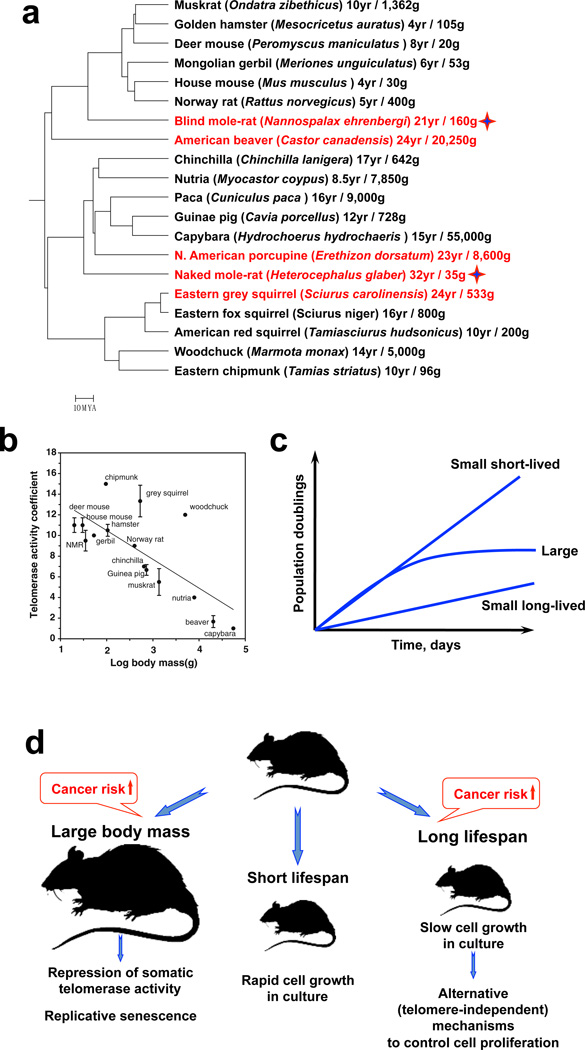
Rodents are the most prevalent mammals on Earth, with ~ 40% of mammalian species being rodents. Rodents are an ideal group of species for pursuing comparative aging studies. These animals are phylogenetically related, yet their lifespans are extremely diverse, ranging from 3–4 years in mice and rats to over 20 years in blind mole rats, beavers, porcupines, and squirrels and over 30 years in naked mole rats (Figure 1a). This nearly ten-fold variance in lifespan among rodent species is greater than the variance observed in other mammalian orders, and is hypothesized to be connected to the variability among rodent species in extrinsic mortality caused by predation (Box 1). The longest-lived rodents: naked mole rat, blind mole rat, beaver, porcupine, and squirrel, all belong to different phylogenetic groups, indicating that slow aging has independently evolved at least four times in rodents2 (Figure 1a). Moreover, a ten-fold range in lifespan far exceeds differences in lifespan observed within a species and is certainly much greater than anything that has been achieved in extending lifespan using genetic, pharmacological, or dietary interventions in mice or rats. Indeed, dietary restriction, the best-documented intervention to extend lifespan, only results in a maximum increase of ~ 40%5.
Figure 1. Evolution of tumor suppressor mechanisms.
a, Slow aging and resistance to cancer have evolved multiple times in rodents. Numbers following the species name indicate maximum lifespan in years/body mass in grams1, 81. Red font highlights slow aging species with maximum lifespan greater than 20 years. Asterisks indicate species for which cancer resistance had been documented. b, Strong negative correlation between telomerase activity in somatic tissues and body mass. Telomerase is repressed in somatic tissues of large rodents. Adapted from82. c, Cell proliferation patterns of primary fibroblasts isolated from species depending on their body mass and maximum lifespan. Small body mass and short lifespan correlate with rapid cell proliferation in vitro and the absence of replicative senescence. Large body mass (>10 kg) correlates with rapid cell proliferation in vitro followed by replicative senescence due to telomere shortening. Finally, cells of small but long-lived (maximum lifespan > 10 years) animals tend to proliferate very slowly, but do not enter replicative senescence. d, The model summarizing evolution of tumor-suppressor strategies depending on lifespan and body mass. When species evolve large body mass the cancer risk is increased due to increased number of cells. To mitigate this risk large body mass coevolves with repression of telomerase activity and replicative senescence. Small and short lived-species require fewer tumor suppressors. Finally, evolution of longer lifespan in small-bodies species is associated with telomere-independent tumor-suppressor mechanisms that stringently control cell proliferation and are characterized by very slow proliferation rate in vitro. Adapted from15.
Box 1 | How do lifespans evolve?
Evolutionary theory of aging was developed by Haldane74, Williams75 and Medawar76, who concluded that the force of natural selection declines with age. In the natural world animals die from predation and accidents, thus genes that confer fitness and longevity beyond the expected lifespan of a given species based on extrinsic risks, would be largely ignored by natural selection. In other words, there is little benefit for a species to invest in maintaining physical fitness and fecundity beyond the time that the individuals of this species are expected to survive based on extrinsic pressures. Hence, one can predict that the differences in aging rates between species are driven by differences in extrinsic mortality due to predation, starvation or accidents. Species that experience high extrinsic mortality gain no advantage from investing resources into somatic maintenance and longevity, and instead they benefit from reproducing prolifically while they are still alive. By contrast, in species that are shielded from predators by specific adaptations, molecular mechanisms evolve that allow them to live longer. Experimental evidence for this theory was provided by classical experiments in Drosophila melanogaster where long-lived flies were artificially selected by only allowing older flies to reproduce77–79 such that the force of natural selection no longer decreased with age. Additionally, studies of opossums showed that the animals living on a predator-free island reproduced later and aged slower than animals of the same species on the more hazardous mainland80. The correlation between lifespan and environmental vulnerability holds true even for a large collection of over 500 mammalian species8, see also19. The evolutionary theory helps to understand the diversity of lifespans in the rodent clade (Figure 1a). For example, the quills of a porcupine make it inaccessible to predators, coinciding with a long maximum lifespan of 24 years. The sturdy lodges and imposing body size of the beaver, which lives for over 24 years, protect the animal from predators. And the subterranean lifestyle of blind mole rats and naked mole rats shields them from the majority of predators. In all these cases a low level of extrinsic mortality is likely to have promoted the evolutionary emergence of pro-longevity and tumor suppressive mechanisms that could be uncovered by an in-depth, comparative genetic analysis of such species.
Rodents also differ dramatically in body mass. The smallest rodents, such as mice, have an average body mass of 20–30 g, while the largest rodent, capybara, weighs 55 kg. This diversity is very useful for comparative aging studies since many aging-related traits show a dependence on body mass6–8. Importantly, rodents include species with all possible combinations of lifespan and body mass i.e. species with large body mass and average lifespan (capybara); large body mass and long lifespan (beaver and porcupine); small body mass and short lifespan (mouse); and small body mass and long lifespan (naked mole rat).
Like in humans, the aging process in rodents is associated with an increased incidence of disease. This is especially relevant for cancer, which is a typical disease of the aged. Short-lived rodents such as mice and rats are prone to cancer with cancer incidence reaching 95% in some strains9, 10. Yet the two longest-lived rodent species, the naked mole rat and the blind mole rat, are remarkably cancer-resistant, with no cases of cancer reported in either species after multiyear observations of large animal colonies11–13. This disparity in cancer rates between phylogenetically so closely related species offers an inimitable opportunity to understand how cancer resistance can be achieved in these related species and possibly in mammals in general.
The independent evolution of long-lived, cancer-resistant rodent species provides a unique opportunity to study what are possibly multiple mechanisms that lead to a long life without cancer. Long-lived rodents are especially interesting due to their relationship to the best-studied aging models – mice and rats. However, in contrast to many long-lived laboratory mouse mutants that cannot compete in the wild11–13, natural, long-lived species are well adapted to their respective environments and offer an opportunity to identify longevity mechanisms that can be modulated without undesirable effects on fitness.
Cross-species biological comparisons
Prior to the emergence of genomic data for the longest-lived rodents important biological insights related to longevity and tumor suppressive mechanisms were obtained through cross-species comparisons.
Telomere maintenance and replicative senescence
Permanent arrest of cell proliferation induced by progressive telomere shortening, named replicative senescence, is one of the key human anticancer mechanisms14. Telomerase activity is repressed in the somatic tissues of humans, while it is constitutively active in mice. The prevailing view had been that long-lived species evolve repression of telomerase activity to enable replicative senescence. This hypothesis was tested using a comparative analysis of 15 rodent species with diverse lifespans. Surprisingly, it was found that telomerase activity does not coevolve with lifespan but instead coevolves with body mass7 (Figure 1b). During evolution, larger animals (body mass > 10 kg) evolved repressed telomerase activity in somatic cells to mitigate increased cancer risk conferred by the large number of cells. Consistent with repressed telomerase activity, fibroblasts from large rodents undergo replicative senescence when cultured in vitro15 (Figure 1c). These findings were confirmed using a wider range of mammalian species16. These studies experimentally demonstrated the importance of body mass as a risk factor for cancer. If cancer results from a random mutation within a cell, cancer risk must be proportional to the number of cells, yet larger animals do not have higher cancer incidence, a concept known as “Peto’s paradox”17. The answer to Peto’s paradox is that larger animals must evolve additional anticancer mechanisms, one of which is replicative senescence. If the increase in size from 30 g in a mouse to 55 kg in a capybara commands additional tumor suppressor mechanisms, one can only speculate how many novel tumor suppressor mechanisms are awaiting to be discovered in animals with even greater body mass, such as elephants and whales.
Mechanisms controlling cell proliferation
Analogous to the evolution of replicative senescence as a tumour suppressor mechanism to counteract the heightened cancer risk from increased body mass, tumour suppressor mechanisms would also be expected to evolve to counteract the increased cancer risk in organisms with longer lifespans. Interestingly, despite the absence of replicative senescence, small rodents with short and long lifespans show a striking difference: cells from small shorter-lived species show a rapid rate of proliferation in vitro, whereas cells from small long-lived species (maximum lifespan > 10 years) proliferate much more slowly in vitro by comparison15 (Figure 1c). Fibroblasts divide infrequently in vivo, whereas in vitro cells are induced to over-proliferate by growth factors that are present in the serum in the culture medium. Therefore in vitro growth may serve as a surrogate test for susceptibility to malignant transformation. The slow growth of fibroblasts from long-lived species in vitro is likely to reflect more stringent cell cycle control mechanisms that restrict over-proliferation of cells. This suggests that cells of small, long-lived rodents, lacking replicative senescence, have evolved alternative tumor-suppressor mechanisms that slow cell growth in vitro and that might prevent proliferation of pre-malignant cells in vivo.
Body mass and lifespan shape tumor suppressor mechanisms
The analysis of diverse rodent species suggests a set of rules for how increased cancer risk conferred by large body mass or long lifespan drives the evolution of tumor suppressor mechanisms (Figure 1d). Both body mass and lifespan contribute to the evolution of tumor suppressor mechanisms, but in different ways. Body mass larger than ~10 kg co-evolves with replicative senescence, while lifespans longer than ~10 years, are associated with the evolution of more stringent cell cycle control mechanisms that increase the sensitivity of cells to growth conditions and manifests slow cell proliferation in culture (Figure 1d). Replicative senescence, while being a potent anticancer mechanism, also contributes to aging of tissues through accumulation of senescent cells18. Thus, small long-lived species are free of the pro-aging effects of replicative senescence while being protected from cancer via alternative mechanisms. Remarkably, members of the Squiridae family, such as the grey squirrel, woodchuck and chipmunk, have particularly high telomerase activity that may be beneficial for wound healing and a robust immune response, two processes requiring rapid cell proliferation7, 15. Studies of such small and long-lived species open new avenues of research into novel tumor suppressor mechanisms.
It is important to emphasize that the rules discussed above relate to interspecies comparisons, where body mass is an average body mass for the species. The correlations between longevity and cancer resistance between and within species show opposing trends. Larger species tend to be longer-lived than smaller species, a trait shaped by millions of years of evolution8, 19. For example, the longest-lived mammals, besides human, are elephants and whales. However, larger individuals within a given species are often shorter-lived. This is particularly striking when comparing large and small dog breeds, with larger breeds being shorter-lived and more susceptible to cancer20–22. Similarly, dwarf mice with mutations in genes involved in insulin-like growth factor 1 (IGF1)–growth hormone (GH) axis are longer-lived and resistant to cancer23 and human patients with Laron syndrome characterized by GH deficiency are protected from cancer24.
Lifespan and genome stability
Genome maintenance capacity has long been implicated in the evolution of species-specific maximum lifespan25. For example, cells from short-lived species, such as mice and rats, as compared to cells from long-lived humans are less capable of repairing DNA damage induced by ultraviolet light26 or repairing DNA double-strand breaks27, possibly due to the observed lower expression levels of the DNA repair factors PARP1 and DNA-PK28, 29. In spite of these deficiencies, mouse or rat cells have survival characteristics after treatment with DNA damaging agents that are very similar to those of human cells30. This raises the possibility that cells from short-lived species have a reduced stringency of DNA damage control at the cost of increased mutations31. Finally, a mouse cell in culture has a much higher probability of undergoing karyotypic changes and of becoming neoplastically transformed than does a human cell32, which also points toward a superior genome maintenance system in longer-lived species. Spontaneous immortalization never occurs in primary populations of human cells33.
Naked mole rats and blind mole rats
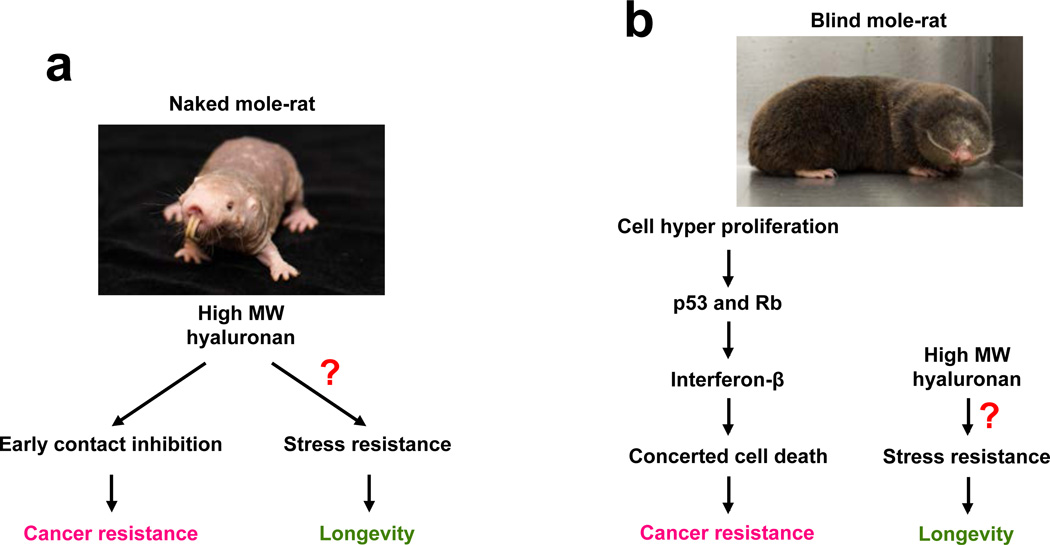
The naked mole rat (Heterocephalus glaber) and the blind mole rat (Spalax ehrenbergi) represent two striking examples of animals that live in protected environments and display extreme longevity and resistance to cancer12, 34–36. The naked mole rat is a eusocial rodent that lives in large colonies in Africa37, while the blind mole rat is a solitary animal found in the Middle East38. The two species are phylogenetically distant from each other, with the naked mole rat being closer to guinea pig and the blind mole rat closer to mice and rats (Figure 1a). Strikingly, however, the protective environments of these two mole rat species allowed them to evolve independent mechanisms of exceptional longevity and cancer resistance11–13, 39, 40 (Figure 2).
Figure 2. Longevity and anticancer adaptations in two mole rat species that independently evolved longevity and resistance to cancer.
a, The naked mole rat is the longest lived rodent that is virtually cancer-proof11, 41. Cancer resistance in the naked mole rat is mediated by high molecular weight hyaluronan resulting in “early contact inhibition” — that is, hypersensitivity of naked mole rat cells to contact inhibition. High molecular weight hyaluronan may also contribute to naked mole rat longevity by increasing stress resistance due to hyaluronan’s antioxidant and cytoprotective properties. b, The blind mole rat is one of the longest-lived rodents that is also resistant to cancer. Cancer resistance in the blind mole rat is mediated by an interferon-mediated necrotic cell death mechanism. Blind mole rat cells produce high molecular weight hyaluronan, but in contrast to the naked mole rat, do not display early contact inhibition. Antioxidant properties of high molecular weight hyaluronan in the blind mole rat can increase stress resistance and contribute to longevity in this species.
Hyaluronan mediates naked mole rat tumor resistance
Cancer resistance in the naked mole rat is mediated by the extreme sensitivity of their cells to contact inhibition41. Normal animal cells arrest their proliferation when they come into contact with each other, which serves as a powerful anticancer mechanism. This growth arrest is associated with the activation of the naked mole rat INK4 locus41.
The trigger for the contact inhibition of naked mole rat cells is a polysaccharide, hyaluronan42. Hyaluronan is an abundant molecule in the body and a major non-protein component of the extracellular matrix termed “extracellular goo”43, 44. Hyaluronan is produced by hyaluronan synthases HAS1, 2, and 3, that differ in the size of HA produced45. The widely distributed form of HA in normal tissue is a high molecular mass hyaluronan, which forms a highly viscous network. At the sites of inflammation, injury, and in tumors hyaluronan may be present in a low molecular mass form. Both high and low molecular mass forms bind CD44, but trigger distinct biological outcomes46. High molecular mass hyaluronan binding to CD44 arrests cell cycle by repressing mitogenic signaling47. In contrast, low molecular mass hyaluronan binding to CD44, promotes cell cycle progression48. In addition, high molecular mass hyaluronan has anti-inflammatory properties, while low molecular mass hyaluronan promotes proliferation and inflammation46. Thus, high molecular mass hyaluronan has antitumor activity while the low molecular mass form may promote tumorigenesis. Naked mole rat tissues contain hyaluronan of extremely high molecular mass, which is five times as long as the hyaluronan of mice or humans. Naked mole rat cells cannot be malignantly transformed by a combination of oncoproteins that cause mouse cells to form tumors49. However, when the HAS2 gene responsible for hyaluronan synthesis was knocked down, or the hyaluronoglucosaminidase 2 (HYAL2) gene responsible for breaking down hyaluronan was overexpressed, naked mole rat cells readily formed tumors indicating that high molecular mass hyaluronan is a key to this species’ cancer resistance. Two mechanisms contribute to high hyaluronan levels in the naked mole rat. The hyaluronan synthase gene in the naked mole rat has a unique sequence, which may confer an enhanced activity to the enzyme, and the rate of hyaluronan turnover is very slow in naked mole rat tissues. It is anticipated that finding ways to manipulate hyaluronan degradation or alter the hyaluronan synthase activity may lead to new therapies to treat or prevent cancer in humans.
Accurate protein synthesis in the naked mole rat
Another bizarre feature of the naked mole rat molecular biology is that its 28S ribosomal RNA is cleaved into two fragments40. Such 28S ribosomal RNA cleavage has been described for only one other species of vertebrates50. This unusual structural feature of the naked mole rat ribosome is associated with high fidelity of translation compared to a mouse. Thus, naked mole-rat cells produce fewer aberrant proteins, supporting the hypothesis that a more stable proteome in the naked mole rat contributes to its longevity40.
Interferon mediates blind mole rat cancer resistance
Anticancer mechanisms in the blind mole rat evolved independently and have taken a different path than in the naked mole rat. Remarkably, as an adaptation to hypoxic conditions underground, blind mole rats have evolved an amino acid change in the p53 gene that corresponds to a mutation frequently found in human tumors51. Blind mole rat p53 lost the ability to induce Apaf1 transcription, and increased the induction of Mdm2 transcription52. Thus the blind mole rat p53 cannot induce apoptosis, making the tumor resistance of these animals even more surprising. Unlike naked mole rats, blind mole rat cells do not display early contact inhibition, but instead use a “scorched earth” strategy to kill pre-cancerous cells and their neighbors. Premalignant blind mole rat cells secrete interferon β1 that mediates massive necrosis of the surrounding cells13. An independent study confirmed that blind mole rat cells secrete a soluble factor that inhibits proliferation and triggers death of cancerous cells39. Activation of a similar interferon-mediated cell death mechanism has been observed in mice with defective p53, where it is driven by the activation of transcription of repetitive elements and noncoding RNAs53.
Recent analysis of the whole genome sequence of the blind mole rat showed that Ifnb1, the gene encoding interferon β1, underwent a duplication event when compared with mouse, rat and naked mole rat54. The Mx1 genes from the interferon signaling pathway, and multiple genes involved in regulation of cell death and inflammation (Nfkb, Tnfrsf1a, Birc3, Fem1b and Aifm1), also underwent expansion in the blind mole rat. Furthermore, three genes involved in necrosis and inflammation (Tnfrsf1a, Tnfsf15 and Nfkb1) show evidence of positive Darwinian selection54. Collectively, these studies suggest that blind mole rat evolved a “weak” p53, possibly as an adaptation to hypoxia. To compensate for the insufficient function of p53, blind mole rats then evolved a very efficient cancer-resistance mechanism relying on heightened immunoinflammatory response via gene amplification within the interferon β1 pathway.
In addition, blind mole rat was shown to express alternatively spliced form of heparanase that acts as a dominant negative repressing heparan sulphate degradation in the extracellular matrix55. This heparanase splice variant has antitumor activity55. The mechanism leading to more stable extracellular matrix in the blind mole rat resembles the high molecular mass hyaluronan combined with slow hyaluronan turnover mediating cancer resistance of the naked mole rat.
Hyaluronan evolved in long-lived subterranean rodents
Interestingly, blind mole rat cells, similar to cells from naked mole rats, produce high molecular mass hyaluronan, but do not show hypersensitivity to contact inhibition. Besides regulating cell proliferation, hyaluronan is also a potent antioxidant. It is possible that hyaluronan contributes to longevity of the mole rats by ameliorating oxidative stress. Naked mole rat heart, for example, contains very high levels of hyaluronan42, which could delay heart disease. Why did these two species evolve high molecular weight hyaluronan? We hypothesize that hyaluronan production was initially upregulated as an adaptation for subterranean life to provide elastic skin needed for the life in underground tunnels. Later this trait might have been co-opted to provide cancer resistance and longevity. In conclusion, the studies of naked and blind mole rats represent a successful example of how moving research efforts to non-canonical long-lived rodent models can lead to discoveries that may benefit human health.
Comparative genomics of aging and cancer
Genomic and genetic approaches provide opportunities for unbiased discovery of genes and pathways as well as molecular mechanisms behind the exceptional longevity and cancer resistance observed in some rodents. Particularly appealing is the fact that these traits evolved independently, e.g., in naked mole rats, blind mole rats, and possibly some other species. Comparative genomics of rodents also benefits from the fact that these animals are among the best represented by completely sequenced genomes. Owing to their small body size (compared to most other mammalian orders), great diversity and abundance, and availability of tissues, rodents have also been subject to many omics approaches, most notably transcriptome analyses. Finally, with mice and rats being rodents and having short lifespans and high susceptibilities to cancer, rodents offer a direct experimental system to examine the predictions arising from comparative genomics studies.
Strategies for comparative genomics
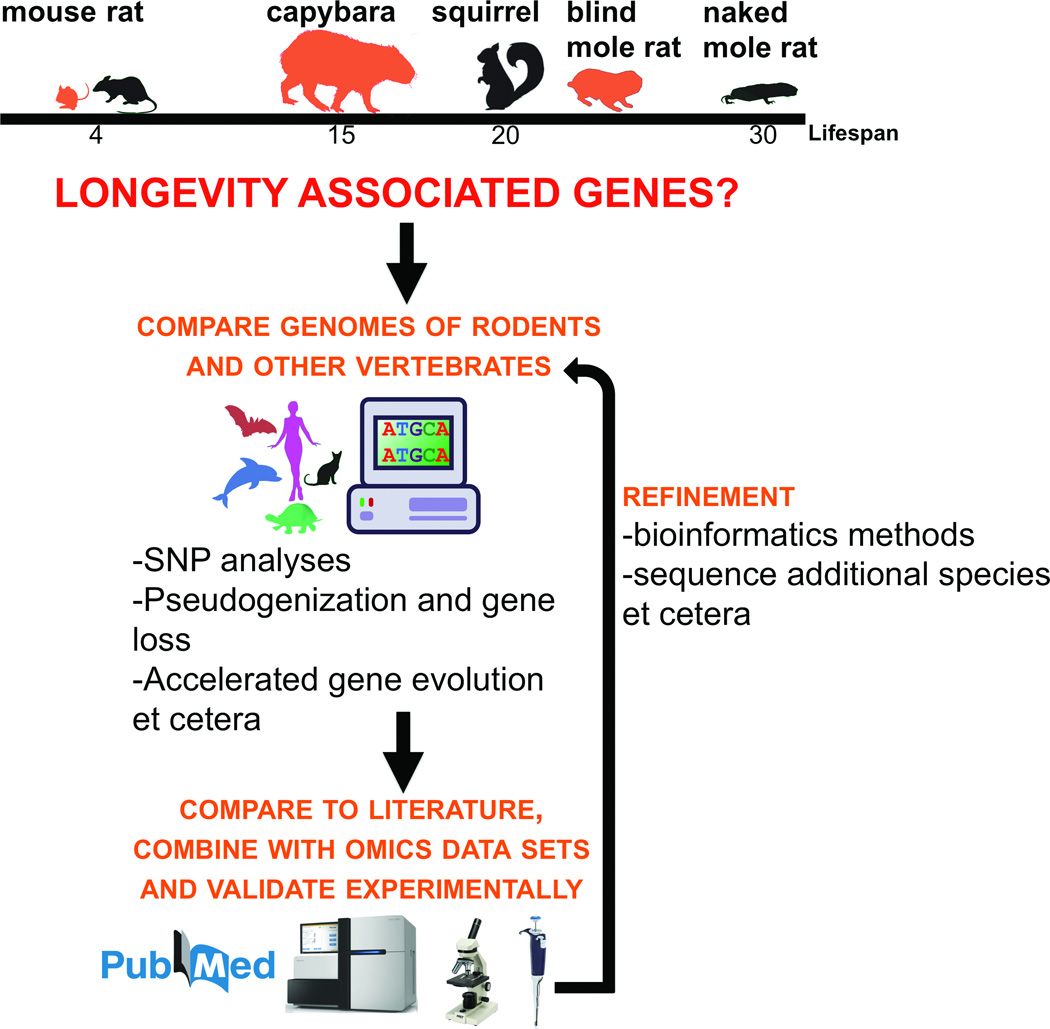
Analyses of rodent genomes and the associated biology, such as unique and common traits of the examined species, may proceed at several levels and utilize diverse approaches (Figure 3)56, 57. Genome alignments, such as the UCSC genome browser, that include both closely related and distant species are helpful for comparative studies as they provide both genome-wide and gene-centric views. One approach in comparative genomics is to search for patterns of lost and pseudogenized genes, which may provide information on the processes and systems that are no longer needed by the species of interest. For example, occupying the subterranean niche, naked mole rats lost many genes that are involved in visual functions. It is also useful to analyze the lost genes and pseudogenes with regard to the pathways and systems in which they have biological roles. Analyses of synteny often accompany such searches to ensure that the identified pseudogenes correspond to functional orthologs in other species.
Figure 3. Comparative genomics of aging.
The figure illustrates strategies for comparative genomic analyses of rodents, starting with the genomes of organisms with widely different lifespans and focusing on genetic adaptations of long-lived species, such as the naked mole rat and the blind mole rat. Approaches are shown that may lead to the identification of functionally relevant genes that contribute to the examined traits. For example, these approaches may uncover lineage-specific genetic changes associated with longevity and cancer resistance. In addition, the use of omics approaches may support analyses across rodents, thereby characterizing common strategies employed by these organisms to regulate species lifespan and cancer susceptibility.
Another approach is to carry out analyses of positive selection, unique amino acid replacements or accelerated evolution58. Applied across whole mammalian genomes, these searches identify a multitude of genes (and sites within genes), and hence require robust filtering methods to remove false positives, such as genes with repeats. Further prioritization of the list of candidate genes and regions may be guided by pathway enrichment, underlying biology or other considerations.
A third approach is to integrate these analyses with omics datasets (e.g., gene expression, proteomics or metabolite profiling) and contrasting the species or lineages with broader groups of organisms or examining the consequences of system perturbations or interventions59. In this case, prioritization of molecular targets for further studies may be improved when several methods point to a particular gene, pathway or mode of regulation. The omics studies may also be appropriately designed to improve their impact. For example, studies on aging and longevity may examine the omics data across lifespan or under conditions that affect lifespan.
Finally, another approach is to focus the searches on genes with known connections to the biological processes of interest60. For example, known aging-related genes and cancer driver genes could be examined in the genomes of interest when one focuses on longevity and cancer. In such focused studies, it is easier to analyze the associated regulatory regions, such as promoters, enhancers, allosteric sites and untranslated regions, as well as gene variants. There is currently no universally accepted strategy to uncover genomic features that underlie the biology, but methods are rapidly advancing, and the already available methods and approaches are numerous.
Genomics of the naked mole rat
The completed genome of the naked mole rat56 represents the first case of an animal genome sequenced with the explicit purpose of explaining the exceptional longevity and cancer resistance. Comparative genome analyses, either within vertebrates or mammals including several rodents, revealed several genes and processes that may contribute to the unique traits of the naked mole rat. For example, these animals evolved a shortened version of the tumor suppressor p16INK4A, a protein, which was found to contribute to early contact inhibition of naked mole rat fibroblasts41. The modified p16INK4A structure could make the cell cycle machinery more sensitive to hyaluronan-mediated contact inhibition, which is controlled by p16INK4A. Another important protein that uniquely changed in the naked mole rats is the thermogenesis regulator UCP1, which is altered at a conserved site that is regulated by fatty acids and nucleotides. Being poikilothermic, naked mole rats are unique among mammals in the inability to maintain stable body temperature, and the detection of UCP1 as an altered protein is consistent with this phenotype56. However, additional studies are needed to determine how this change affects UCP1 activity and/or regulation. Pseudogenes of the naked mole rat are highly enriched for visual perception function, which is consistent with the strictly subterranean life of these animals56. Naked mole rats also pseudogenized both melatonin receptors and altered two proteins involved in telomere maintenance56. Although a causal role of these changes in longevity is unknown, the findings offer direct molecular targets for subsequent experimental analyses. The recently completed genome of the blind mole rat54 allows for comparison of the two species that independently evolved adaptations to subterranean life, long lifespans and resistance to cancer. The analysis revealed that Clock genes responsible for adaptation to darkness showed convergent evolution between these species54. The proton-gated nociceptor sodium channel Nav1.7 gene also shows convergent evolution, which may represent an adaptation to high CO2 environment by blocking pain induced by tissue acidosis61, 62. However, no obvious common patterns of evolution of genes involved in cancer and aging were identified. This suggests that either these species evolved different mechanisms to achieve longevity and resistance to cancer or a more sophisticated data analysis involving greater number of species is needed to obtain this insight.
Comparative genomics of rodents and other mammals
High-quality genome sequences are available for numerous short-lived mice and rats, and several additional rodent genomes have been sequenced for the species with short and intermediate lifespans56, 63. There is no doubt that the availability of rodent genomes will increase in the coming years. For example, it would be very useful to obtain genome sequences of the Damaraland mole rat, a species closely related to the naked mole rat. With these genomic resources, the longevity trait should be amenable to the analyses of convergent evolution of mammalian traits as was recently demonstrated by the studies of echolocation in bats and dolphins64, 65, adaptation to aquatic life in whales66, horse evolution67, and the comparative genomics across 29 mammals63.
One hypothesis that can be more rigorously tested by genomic studies is whether species-specific enhanced longevity is related to superior genome maintenance efficacy, as was suggested by the studies discussed above. Here, rodent species with their broad range of lifespans offer a unique opportunity. Indeed, with complete genome sequences available for many rodent species it is possible to comparatively analyze genome maintenance genes in relation to lifespan. Beyond rodents, a recent analysis of two bat genomes showed that DNA repair and DNA damage signaling genes ATMh, TP53 (which encodes p53), RAD50 and KU70 are under selection in bats, suggesting that genome maintenance systems are under selective pressure in longer-lived species68. It would be interesting to determine whether such repair genes have unique signatures in long-lived species. Both germline and somatic mutation frequency can now be analyzed directly using whole-genome sequencing of parent–offspring trios69 and single somatic cells70. Identifying genes and pathways that are responsible for more efficient genome maintenance in long-lived species may help to develop strategies to increase genome stability in humans.
An example of the application of comparative genomics to reveal the molecular basis for mammalian traits is the evolution of echolocation in mammals64, 65. Similar comparative genomics approaches can be applied to longevity and cancer resistance. In addition, these approaches can be extended to the analyses of gene expression71, proteomics, ribosome profiling72 and metabolite profiling across the group of mammals. Here, in contrast to species- or lineage-specific adaptations highlighted above using the examples of the naked mole rat, the blind mole rat and the Brandt’s bat, longevity or cancer-resistance traits can be characterized that operate across multiple clades. These studies may be especially informative about the coordinate changes in the levels and activity of cellular components, such as transcripts, proteins and metabolites. These components can be further assessed at the level of pathways and networks, and integrated into cellular models of extended longevity. Overall, future studies may reveal both common adaptations that contribute to the longevity trait in many species and lineage-specific adaptations, as exemplified by UCP1 in naked mole rats, interferon in blind mole rats, and the insulin-like growth factor 1 (IGF1)–growth hormone (GH) axis in microbats (Box 2).
Box 2 | Comparative genomics reveals longevity adaptations of the Brandt’s bat.
The genome of an exceptionally long-lived mammal, the Brandt’s bat, has recently been completed64. This 4–8 gram mammal can live for more than 40 years. Comparative genomics analyses revealed unique changes in the transmembrane domains of growth hormone (GH) receptor and insulin-like growth factor 1 (IGF1) receptor, which might have contributed to the evolution of long lifespan in the Brandt’s bat and other microbats, and perhaps to the evolution of their small body mass. Interestingly, these genes are well known for their role in regulation of longevity. The IGF1–GH axis has been implicated in the extended lifespan of various dwarf mice as well as in the decreased incidence of age-related diseases in humans with Laron syndrome, which is characterized by GH receptor dysfunction. Of note is that this adaptation to longevity is distinct from those found in naked mole rats and blind mole rats.
Beyond studies on evolution of lifespan, microbats have recently been the focus of comparative genomics approaches targeting other unique traits, such as echolocation, and these studies have identified the genes involved64. Another study used this approach to uncover genes involved in DNA repair and genome maintenance in microbats68, whereas a study that examined placental mammals identified altered telomere maintenance genes in microbats44.
Conclusions
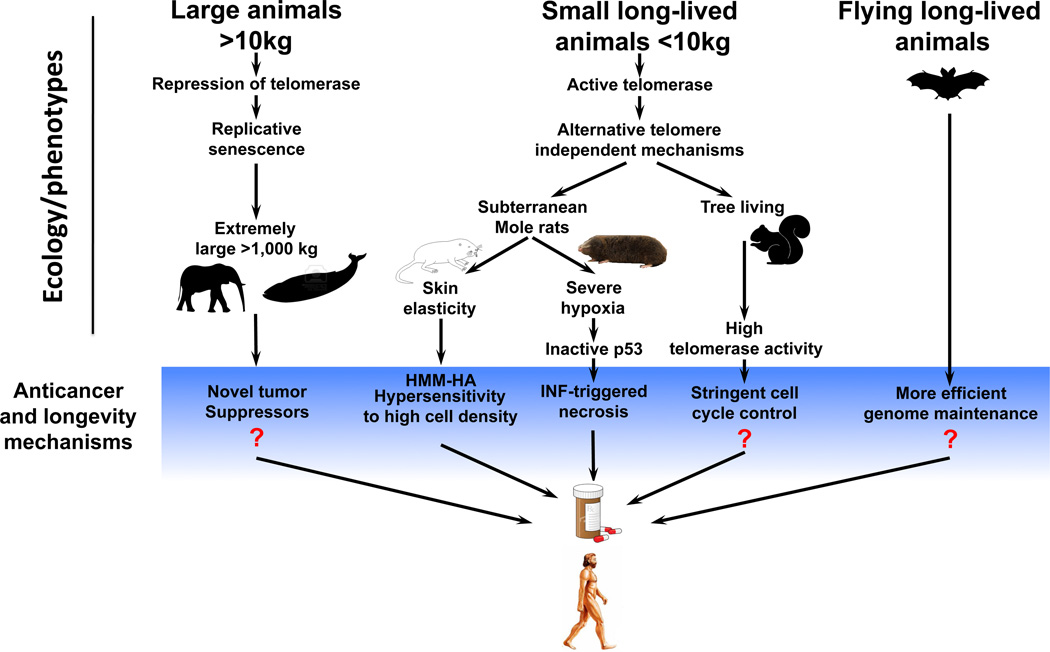
Increased insight into the molecular mechanisms at the interface of aging and age-related disease, such as cancer, is critical for making further progress in improving human health. In this review we showed that comparative studies of rodents are highly informative in this respect and have already led to a new understanding of how cancer can be suppressed in an animal. The paradigm that emerges from these initial studies suggests that species ecology contributed to the evolution of lifespan and body mass, which together have driven the evolution of distinct tumor suppressor strategies in different species such as the high molecular weight hyaluronan-mediated contact inhibition in the naked mole rat and the interferon-mediated cell death mechanism in the blind mole rat (Figure 4).
Figure 4. Lineage-specific mechanisms of longevity and cancer resistance that evolved in species with diverse ecology could be adapted to benefit human health.
The upper part of the figure depicts three groups of species whose ecology or phenotype is associated with evolution of longevity and anticancer adaptations. The blue banner below highlights such adaptations. On the left, body size >10 kg is associated with evolution of replicative senescence. The giant mammals such as elephants and whales are hypothesized to evolve novel tumor suppressor mechanisms that are absent in smaller species including human. Shown in the middle are small long-lived species. This group is characterized by diverse anticancer adaptations such as high molecular mass hyaluronan (HMM-HA), interferon (IFN)-triggered necrosis, or stringent cell cycle control. On the right, are long-lived bats that possibly evolved more efficient DNA repair and DNA damage systems and alterations in IGF1–GH axis. Question marks indicate adaptations for which exact molecular mechanisms are still unknown.
Interestingly, other species within the rodent clade likely harbor additional, novel tumor suppression mechanisms. For example, the grey squirrel is a long-lived, diurnal rodent that lives above ground. In tissue culture, grey squirrel cells do not secrete high molecular weight hyaluronan, but instead display extremely high telomerase activity combined with slow proliferation15, indicating the presence of a yet-to-be discovered cell cycle control mechanism. Such a mechanism, which prevents telomerase-positive cells from undergoing malignant transformation, can potentially be exploited to prevent cancer from arising in cells with naturally active telomerase, such as human stem cells and germline. Another example of species with unique adaptations are animals with extremely high body mass. As the studies of rodents have shown, body masses above ~10 kg are associated with telomerase repression in somatic cells and replicative senescence. It is plausible that species with even greater body mass such as elephants and large whales possess unique anticancer adaptations yet to be discovered. Since unique species ecology determines the evolutionary path for longevity and anticancer adaptations, humans would not be expected to possess all of these mechanisms, and could benefit from them. Thus studying such species-specific adaptations and then “importing” these strategies to humans opens new avenues for cancer prevention and lifespan extension. One very successful example of this strategy is the invention of antibiotics where an antibacterial strategy that evolved in fungi is used to benefit humans. This may be a powerful alternative to the studies focusing on conserved mechanisms of longevity that operate across rodents and beyond. For example, high molecular mass hyaluronan can be directly administered to humans, or pharmacological inhibitors of HYAL enzymes responsible for hyaluronan degradation could be developed to increase the levels and molecular mass of endogenous hyaluronan.
Recent advances in DNA sequencing technologies have enabled comparative genomics using whole-genome sequencing, and these studies indicate that multiple DNA damage and repair genes are under selection in long-lived species73. Understanding how more efficient genome maintenance is achieved may open avenues for improving human DNA repair and genome stability. Comparative genomics also offers an unbiased view on other adaptations of long-lived animals and their unique genetic strategies. The first two genomes of exceptionally long-lived mammals, the naked mole rat and the Brandt’s bat (Box 2), revealed different lineage-specific mechanisms employed by these mammals and implicated the insulin-like growth factor 1 (IGF1)–growth hormone (GH) axis, genome maintenance and thermogenesis in lifespan control. The availability of additional genomes of long-lived animals will undoubtedly offer additional insights into the genomics of longevity and cancer resistance.
In conclusion, it is important to diversify the "bestiary" of standard model organisms and include various long-lived and cancer-resistant species. This is crucial because the short-lived and cancer-prone mice and rats may be missing the mechanisms for longevity and cancer resistance. Future studies, focused on comparative genomics and in-depth molecular studies of long-lived rodent species, promise to drive the aging and cancer fields forward.
Acknowledgements
We thank Daniel Promislow and Michael van Meter for comments on the manuscript and Inge Seim for help with preparing the figure. The work in the authors’ laboratories is supported by the US National Institutes of Health (all authors); the Life Extension Foundation (V.G. and A.S.); and the Glenn Foundation for Medical Research (J.V.). The authors thank past and present members of their laboratories for their insights.
Glossary terms
- Maximum lifespan
the maximum documented lifespan achieved by a representative of a species; typically the maximum lifespan is documented in a captive environment protected from predators.
- Telomerase
ribonucleoprotein enzyme that elongates telomeres by synthesizing the telomeric repeat sequence using an RNA templete.
- Pseudogenized
a gene becomes presudogenized when it loses its functional gene product, e.g., via accumulation of frameshifts or stop codons. Pseudogenes also arise when a gene is processed by a retrotransposon such that a portion of the mRNA transcript of a gene is reverse transcribed back into DNA and inserted into chromosomal DNA.
- Synteny
Shared genomic organization between related species. It is usually seen as a shared relative order of genes or other functional elements on a portion of a chromosome.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
References
- 1. Tacutu R, et al. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–D1033. doi: 10.1093/nar/gks1155. This is an excellent well curated database integrating information on species' longevity and life histories.
- 2.Austad SN. Diverse aging rates in metazoans: targets for functional genomics. Mech Ageing Dev. 2005;126:43–49. doi: 10.1016/j.mad.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Miller RA. Biomedicine. The anti-aging sweepstakes: catalase runs for the ROSes. Science. 2005;308:1875–1876. doi: 10.1126/science.1114393. [DOI] [PubMed] [Google Scholar]
- 4.Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5:525–532. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- 5.Swindell WR. Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev. 2012;11:254–270. doi: 10.1016/j.arr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenzini A, Tresini M, Austad SN, Cristofalo VJ. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech Ageing Dev. 2005;126:1130–1133. doi: 10.1016/j.mad.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 7. Seluanov A, et al. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. The first analysis of telomerase activity across species in relation to lifespan and body mass.
- 8.Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- 9.Lipman R, Galecki A, Burke DT, Miller RA. Genetic loci that influence cause of death in a heterogeneous mouse stock. J Gerontol A Biol Sci Med Sci. 2004;59:977–983. doi: 10.1093/gerona/59.10.B977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burek JD, Hollander CF. Incidence patterns of spontaneous tumors in BN/Bi rats. J Natl Cancer Inst. 1977;58:99–105. doi: 10.1093/jnci/58.1.99. [DOI] [PubMed] [Google Scholar]
- 11.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol [B] 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 12.Delaney MA, Nagy L, Kinsel MJ, Treuting PM. Spontaneous Histologic Lesions of the Adult Naked Mole Rat (Heterocephalus glaber): A Retrospective Survey of Lesions in a Zoo Population. Vet Pathol. 2013 doi: 10.1177/0300985812471543. [DOI] [PubMed] [Google Scholar]
- 13.Gorbunova V, et al. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc Natl Acad Sci U S A. 2012;109:19392–19396. doi: 10.1073/pnas.1217211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:S27–S31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 15. Seluanov A, et al. Distinct tumor suppressor mechanisms evolve in rodent species that differ in size and lifespan. Aging Cell. 2008;7:813–823. doi: 10.1111/j.1474-9726.2008.00431.x. This paper identified rules that control evolution of tumor suppressors depending on lifespan and body mass.
- 16.Gomes NM, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peto R, Roe FJ, Lee PN, Levy L, Clack J. Cancer and ageing in mice and men. Br J Cancer. 1975;32:411–426. doi: 10.1038/bjc.1975.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Healy K, et al. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc Biol Sci. 2014;281:20140298. doi: 10.1098/rspb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus C, Pavard S, Promislow DE. The size-life span trade-off decomposed: why large dogs die young. Am Nat. 2013;181:492–505. doi: 10.1086/669665. [DOI] [PubMed] [Google Scholar]
- 21.Selman C, Nussey DH, Monaghan P. Ageing: it's a dog's life. Curr Biol. 2013;23:R451–R453. doi: 10.1016/j.cub.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Fleming JM, Creevy KE, Promislow DE. Mortality in north american dogs from 1984 to 2004: an investigation into age-, size-, and breed-related causes of death. J Vet Intern Med. 2011;25:187–198. doi: 10.1111/j.1939-1676.2011.0695.x. [DOI] [PubMed] [Google Scholar]
- 23.Bartke A, Sun LY, Longo V. Somatotropic signaling: trade-offs between growth, reproductive development, and longevity. Physiol Rev. 2013;93:571–598. doi: 10.1152/physrev.00006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guevara-Aguirre J, et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart RW, Sacher GA, Hoskins TL. DNA repair in a short- and a long-lived rodent species. J Gerontol. 1979;34:808–817. doi: 10.1093/geronj/34.6.808. [DOI] [PubMed] [Google Scholar]
- 26.Hanawalt PC. Revisiting the rodent repairadox. Environ Mol Mutagen. 2001;38:89–96. doi: 10.1002/em.1057. [DOI] [PubMed] [Google Scholar]
- 27.Lorenzini A, et al. Significant correlation of species longevity with DNA double strand break recognition but not with telomere length. Mech Ageing Dev. 2009;130:784–792. doi: 10.1016/j.mad.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 30.Ganesan AK, Spivak G, Hanawalt PC. In: Manipulation and expression of genes in Eukaryotes. Nagley P, Linnane AW, Peacock WJ, Pateman JA, editors. Australia: Academic Press; 1983. pp. 45–54. [Google Scholar]
- 31.Vijg J. Aging of the Genome. Oxford, New York: 2007. [Google Scholar]
- 32.Woo RA, Poon RY. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 2004;18:1317–1330. doi: 10.1101/gad.1165204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 34.Lewis KN, Andziak B, Yang T, Buffenstein R. The Naked Mole-Rat Response to Oxidative Stress: Just Deal with It. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buffenstein R. The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 36.Azpurua J, Seluanov A. Long-lived cancer-resistant rodents as new model species for cancer research. Front Genet. 2012;3:319. doi: 10.3389/fgene.2012.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarvis JU. Eusociality in a mammal: cooperative breeding in naked mole-rat colonies. Science. 1981;212:571–573. doi: 10.1126/science.7209555. [DOI] [PubMed] [Google Scholar]
- 38.Nevo E, Ivanitskaya I, Beiles A. Adaptive Radiation of Blind Subterranian Mole Rats. The Netherlands: Backhuys, Leiden; 2001. [Google Scholar]
- 39.Manov I, et al. Pronounced cancer resistance in a subterranean rodent, the blind mole-rat, Spalax: in vivo and in vitro evidence. BMC Biol. 2013;11:91. doi: 10.1186/1741-7007-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azpurua J, et al. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seluanov A, et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. USA. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tian X, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–349. doi: 10.1038/nature12234. This study deciphered a molecular mechanism of naked mole rat's resistance to cancer.
- 43.Laurent TC, Fraser JR. Hyaluronan. Faseb J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 44.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 45.Jiang D, Liang J, Noble PW. Hyaluronan in tissue injury and repair. Annu Rev Cell Dev Biol. 2007;23:435–461. doi: 10.1146/annurev.cellbio.23.090506.123337. [DOI] [PubMed] [Google Scholar]
- 46.Pure E, Assoian RK. Rheostatic signaling by CD44 and hyaluronan. Cell Signal. 2009;21:651–655. doi: 10.1016/j.cellsig.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kothapalli D, et al. Hyaluronan and CD44 antagonize mitogen-dependent cyclin D1 expression in mesenchymal cells. J Cell Biol. 2007;176:535–544. doi: 10.1083/jcb.200611058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kothapalli D, Flowers J, Xu T, Pure E, Assoian RK. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. J Biol Chem. 2008;283:31823–31829. doi: 10.1074/jbc.M802934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melen GJ, Pesce CG, Rossi MS, Kornblihtt AR. Novel processing in a mammalian nuclear 28S pre-rRNA: tissue-specific elimination of an 'intron' bearing a hidden break site. EMBO J. 1999;18:3107–3118. doi: 10.1093/emboj/18.11.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashur-Fabian O, et al. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci U S A. 2004;101:12236–12241. doi: 10.1073/pnas.0404998101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avivi A, et al. P53 in blind subterranean mole rats--loss-of-function versus gain-of-function activities on newly cloned Spalax target genes. Oncogene. 2007;26:2507–2512. doi: 10.1038/sj.onc.1210045. [DOI] [PubMed] [Google Scholar]
- 53.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang X, et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nature Communications. 2014 doi: 10.1038/ncomms4966. In Press. [DOI] [PubMed] [Google Scholar]
- 55.Nasser NJ, et al. Alternatively spliced Spalax heparanase inhibits extracellular matrix degradation, tumor growth, and metastasis. Proc Natl Acad Sci U S A. 2009;106:2253–2258. doi: 10.1073/pnas.0812846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. This paper reports the naked mole rat genome and a plethora of findings on its biology and longevity.
- 57.Gladyshev VN, Zhang G, Wang J. The naked mole rat genome: understanding aging through genome analysis. Aging (Albany NY) 2011;3:1124. doi: 10.18632/aging.100417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, de Magalhaes JP. Accelerated protein evolution analysis reveals genes and pathways associated with the evolution of mammalian longevity. Age (Dordr) 2013;35:301–314. doi: 10.1007/s11357-011-9361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu C, et al. RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat when compared to mice. PLoS One. 2011;6:e26729. doi: 10.1371/journal.pone.0026729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasaikina MV, et al. Reduced utilization of selenium by naked mole rats due to a specific defect in GPx1 expression. J Biol Chem. 2011;286:17005–17014. doi: 10.1074/jbc.M110.216267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park TJ, et al. Selective inflammatory pain insensitivity in the African naked mole-rat (Heterocephalus glaber) PLoS Biol. 2008;6:e13. doi: 10.1371/journal.pbio.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith ES, et al. The molecular basis of acid insensitivity in the African naked mole-rat. Science. 2011;334:1557–1560. doi: 10.1126/science.1213760. [DOI] [PubMed] [Google Scholar]
- 63. Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. This is the first comparative genomics study of mammals based on 29 high and low resolution genome sequences.
- 64.Seim I, et al. Genome analysis reveals insights into physiology and longevity of the Brandt's bat Myotis brandtii. Nat Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parker J, et al. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013 doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yim HS, et al. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 2014;46:88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 68.Zhang G, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gundry M, Li W, Maqbool SB, Vijg J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic Acids Res. 2012;40:2032–2040. doi: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. This is the first analysis of gene expression across mammals.
- 72.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Magalhaes JP, Kean M. Endless paces of degeneration--applying comparative genomics to study evolution's moulding of longevity. EMBO Rep. 2013;14:661–662. doi: 10.1038/embor.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haldane JBS. New Paths in Genetics. London, UK: Allen and Unwin; 1941. [Google Scholar]
- 75.Williams GG. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 76. Medawar PB. An Unsolved Problem of Biology. London, UK: 1952. Medawar was the first to formally work out a complete model of aging as a process that naturally emerges from the decline in efficacy of natural selection during the life course.
- 77.Luckinbill LS, Clare MJ. Selection for life span in Drosophila melanogaster. Heredity (Edinb) 1985;55(1):9–18. doi: 10.1038/hdy.1985.66. [DOI] [PubMed] [Google Scholar]
- 78.Rose MR. Genetics of increased lifespan in Drosophila. BioEssays. 1989;11:132–135. doi: 10.1002/bies.950110505. [DOI] [PubMed] [Google Scholar]
- 79.Rose MR. Evolutionary Biology of Aging. New York: Oxford University Press; 1991. [Google Scholar]
- 80.Austad S. Retarded senescence in an insular population of Virginia opossums (Didelphis virginiana) Journal of Zoology. 1993;229:695–708. [Google Scholar]
- 81.de Magalhaes JP, et al. The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell. 2009;8:65–72. doi: 10.1111/j.1474-9726.2008.00442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gorbunova V, Bozzella MJ, Seluanov A. Rodents for comparative aging studies: from mice to beavers. Age (Dordr) 2008;30:111–119. doi: 10.1007/s11357-008-9053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]