Abstract
EYS mutations demonstrate great genotypic and phenotypic varieties, and are one of the major causes for patients with autosomal recessive retinitis pigmentosa (ARRP). Here, we aim to determine the genetic lesions with phenotypic correlations in two Chinese families with ARRP. Medical histories and ophthalmic documentations were obtained from all participants from the two pedigrees. Targeted next-generation sequencing (NGS) on 189 genes was performed to screen for RP causative mutations in the two families. Two biallelic mutations in EYS, p.[R164*];[C2139Y] and p.[W2640*];[F2954S], were identified in the two families, respectively. EYS p.R164* and p.F2954S are novel alleles associated with RP, while p.C2139Y and p.W2640* are known mutations. Crystal structure modeling on the protein eyes shut homolog encoded by the EYS gene revealed abnormal hydrogen bonds generated by p.C2139Y and p.F2954S, which would likely affect the solubility and cause significant structural changes of the two mutated proteins. In conclusion, our study expands the genotypic spectrums for EYS mutations, and may provide novel insights into the relevant pathogenesis for RP. We also demonstrate targeted NGS approach as a valuable tool for genetic diagnosis.
Retinitis pigmentosa (RP), the most common form of inherited retinal dystrophies (IRDs), presents a global prevalence of 1 in 3500 to 5000 individuals1, while data from Beijing Eye Research Center shows that the prevalence of RP in China is even higher, which is approximately 1 in 10002. Patients with RP are clinically presented with initial night blindness, followed by constriction of peripheral visual fields (VF), and eventual impaired central vision, or even complete blindness. Typical RP fundus presentations include attenuation of retinal vessels, bone-spicule pigment deposits in the mid-peripheral or far peripheral retina, waxy optic disk, and atrophy of the retina. The biological interplay underlying the clinical manifestations of RP includes the sequential degeneration of rod photoreceptors, cone photoreceptors, and retinal pigment epithelium (RPE).
RP can be transmitted via all three Mendelian inheritance patterns, including autosomal dominant, autosomal recessive, and X-linked. Digenic mutations in the peripherin/RDS and ROM1 genes have also been reported in causing RP3,4. Hitherto, 71 mapped loci involving 64 genes have been found in the etiology of RP (www.retnet.org). To be more specific, mutations in 24, 45, and 3 genes would cause autosomal dominant RP (ADRP), autosomal recessive RP (ARRP), and X-linked RP (XLRP), respectively. Though the number of RP causative genes and loci keeps increasing over time, the disease causative genes remain unclear in 40% to 50% of patients with RP, indicating that lots of unknown disease causative genes still remain to be found. Traditional techniques have their limitations in the detection of specific mutations among a large number of candidate genes. Therefore, a powerful and efficient strategy or platform for RP mutation screening should be established to discover the putative novel disease causative genes, and to increase the possibility of identifying genetic causes for RP patients.
Next-generation sequencing (NGS) technique has recently been developed and has enabled the rapid and systematic identification of variants on a large scale, which will also accelerate the pace of gene discovery and disease diagnosis on a molecular level5. NGS usually includes exome sequencing, targeted gene capture array sequencing, and whole sequencing for mapped chromosomal region. It has enabled investigators to obtain variant information down to single-base resolution in a rapid, high-throughput fashion on the scale of the whole human genome. Enrichment by targeted capture array can rapidly isolate candidate regions of interest ranging from hundreds of kilobases in size or capture the entire protein-coding sequence of an individual for sequencing. NGS with array based target enrichment is efficient to discover disease causative gene, and can screen for mutations in hundreds of loci in genetically heterogeneous diseases. Therefore, NGS has unmatched advantages compared to Sanger sequencing or other routine screening techniques for identifying pathogenic mutations in terms of both accuracy and efficiency. Thus, it can be used for both investigative and diagnostic purposes as this technique can identify the disease causing mutation from hundreds alleles related to RP within a reasonable time frame.
In the present study, by means of a targeted NGS approach, we have revealed four heterozygous mutations in the EYS gene (MIM 612424) as RP causative mutations for the two Chinese families with typical ARRP. The genotype-phenotype correlations were also annotated.
Results
Clinical Assessments
Two patients from family ARRP05 and three from family ARRP06 were included in the present study with their detailed clinical information summarized in Table 1. In family ARRP05, only two patients agreed to participate in our study, both of whom were presented with typical RP fundus presentations. Although patients ARRP05-II:2 and ARRP05-II:5 declined to donate their blood samples or to have further ophthalmic examinations, they both declared that their disease courses were quite similar to their siblings. All four patients from family ARRP05 reported to have night blindness in their early 20 s, and the disease progressed fast since then. Their VF decreased rapidly and their central visions were significantly affected shortly after the occurrence of night blindness. All patients were presented with very poor central visions at their last visit to our hospital, indicating a form of RP with late onset age and rapid progression.
Table 1. Clinical features of attainable patients.
| Patient ID | Mutation | Onset Age (year) | Age (year)/Sex | BCVA (logMAR) | Fundus Appearance | ERG | VF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O.D. | O.S. | |||||||||||||||
| O.D. | O.S. | MD | OD | AA | PD | MD | OD | AA | PD | O.D. | O.S. | |||||
| ARRP05-II:2 | NA | 23 | 44/F | LP | LP | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| ARRP05-II:3 | c.[490C>T];[6416G>A] | 22 | 40/M | FC | FC | Severe | Waxy | Yes | Yes | Severe | Waxy | Yes | Yes | D | NA | NA |
| ARRP05-II:5 | NA | 20 | 37/M | HM | HM | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| ARRP05-II:7 | c.[490C>T];[6416G>A] | 25 | 32/M | 0.1 | 0.1 | Mild | Waxy | Yes | Yes | Mild | Waxy | Yes | Yes | D | NA | NA |
| ARRP06-II:2 | c.[7919G>A];[8861T>C] | 18 | 54/M | LP | LP | Severe | Waxy | Yes | Yes | Severe | Waxy | Yes | Yes | D | NA | NA |
| ARRP06-II:4 | c.[7919G>A];[8861T>C] | 35 | 52/M | LP | LP | Severe | Waxy | Yes | Yes | Severe | Waxy | Yes | Yes | D | NA | NA |
| ARRP06-II:8 | c.[7919G>A];[8861T>C] | 32 | 49/F | 0.8 | 0.8 | Normal | Waxy | Yes | Yes | Normal | Waxy | Yes | Yes | R | <10° | <10° |
Abbreviations: M: male; F: female; BCVA: best corrected visual acuity; logMAR: logarithm of the minimum angle of resolution; O.D.: right eye; O.S.: left eye; LP: light perception; FC: finger count; HM: hand movement; MD: macular degeneration; OD: optic disk; AA: artery attenuation; PD: pigment deposits; ERG: electroretinography; NA: not available; D: diminished; R: reduced; VF: visual field.
Unlike patients in family ARRP05, clinical features of the three patients in family ARRP06 varied greatly from each other. The proband, patient ARRP06-II:2, suffered from night blindness since age 18, while the other two patients didn't have any visual problem until their early 30 s. Noteworthy, patient ARRP06-II:4 was presented with the eldest RP onset age but the most rapid RP progression. Typical RP fundus was presented by all three included patients, while macular degeneration was only found in two of them. Patient ARRP06-II:8 showed relatively normal macular presentations with her central visions preserved. However, VF test revealed severe constriction for her both eyes.
Genetic Findings
We selectively performed targeted NGS approach on patients ARRP05-II:3, ARRP05-II:7, ARRP06-II:2, and ARRP06-II:8. The detailed NGS results were provided in Table S1. Our targeted NGS strategy reached an average mean depth of 113.18-fold for the four tested samples. In addition, each sample reached an equal to or over 99.88% coverage of targeted region and an equal to or below 0.18% nucleotide mismatch rate (Table S1). A total of 3359 variants, including 2966 SNVs and 393 Indels were initially disclosed in family ARRP05, and 3390 variants, including 2989 SNVs and 401 Indels, were identified in family ARRP06 (Table 2). All identified variants were then submitted to bioinformatics analyses. Variants were annotated to the 5 SNPs databases, and those with MAF over 0.01 or found homozygous in any of the 5 databases were discarded. A total of four putative pathogenic heterogeneous variants in the EYS gene were identified in the two investigated families, including two novel variants and two recurrent mutations. All the four variations were further confirmed absent in 100 controls.
Table 2. Variations identified in each family.
| Items | ARRP05 | ARRP06 | ||
|---|---|---|---|---|
| SNVs/Indels | SNVs | Indels | SNVs | Indels |
| Initial variants | 2966 | 393 | 2989 | 401 |
| Excluded variants found in SNP databases | 2880 | 377 | 2863 | 378 |
| Remaining variants | 86 | 16 | 126 | 23 |
| Excluded non-coding variants | 75 | 15 | 92 | 21 |
| Remaining variants | 11 | 1 | 34 | 2 |
| Excluded variants not shared by the two sequenced samples | 8 | 1 | 29 | 2 |
| Remaining variants for Sanger sequencing | 3 | 0 | 5 | 0 |
| Excluded false positive variants | 0 | - | 0 | - |
| Remaining variants | 3 | - | 5 | - |
| Excluded variants failed intrafamilial cosegregation analysis | 1 | - | 3 | - |
| Remaining variants | 2 | - | 2 | - |
| Found in 100 unrelated controls | 0 | - | 0 | - |
| Identified RP causative mutations | 2 | - | 2 | - |
Abbreviations: SNVs: single nucleotide variations; Indels: insertions and deletions.
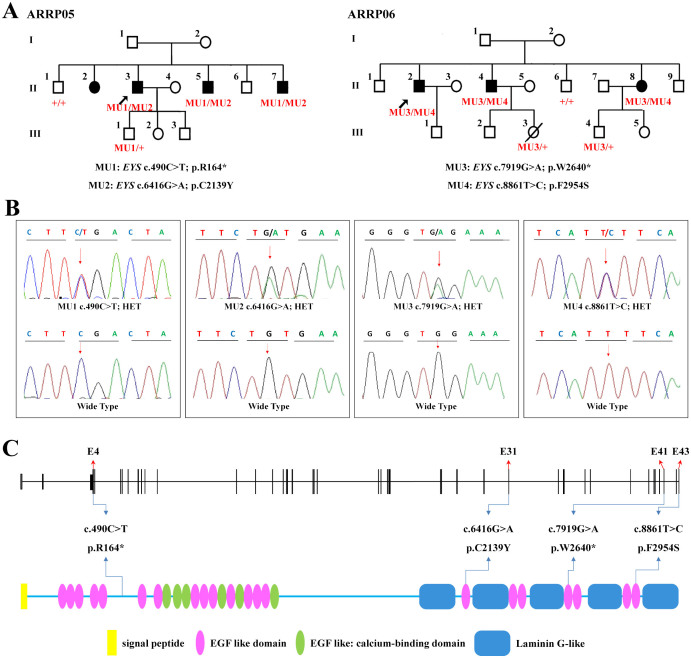
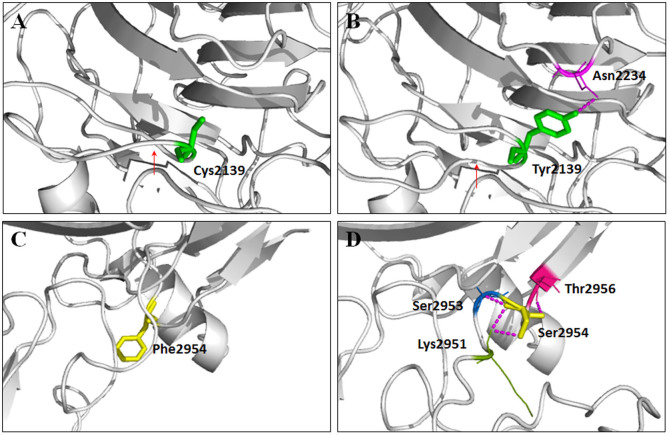
Two nonsense and two missense mutations were revealed by targeted NGS approach, and both families carried biallelic heterozygous EYS mutations (Figures 1A–1C, and Table 3). Biallelic mutations, EYS c.[490C>T];[6416G>A], were found as RP causative in the two included patients from family ARRP05. The former substitution from a C to a T at exon 4 of the EYS gene would lead to the generation of a premature termination codon (PTC) at residue 164 of the protein eyes shut homolog, protein encoded by the EYS gene (Figure 1C). The latter one was a missense change from cysteine to tyrosine at residue 2139 of the protein. This variant was predicted to be deleterious by three types of online predictive software (Table 3). Structural modeling revealed the generation of a novel hydrogen bond between the mutated tyrosine at residue 2139 and asparagine at residue 2234, and the consequent vanishing of the original twist (Figures 2A–2B).
Figure 1. Pedigrees and Identified Mutations.
(A) Pedigrees of families ARRP05 and ARRP06 are demonstrated with EYS genotypes annotated for each included family members of the two families. Probands are indicated by arrows. Circles represent females, and squares, males. Filled symbols are for affected patients, and empty symbols, normal controls. (B) DNA sequencing profiles of the identified mutations (upper) and their wild type form (below). (C) Schematic representation of the linear location of the four identified EYS mutations in context of genome (upper) and protein (below).
Table 3. Mutations identified in the present study.
| Family ID | Variation | Exon | Bioinformatics Analysis | HGMD/Novel | Frequency in Controls | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino Acid | Type | Status | SIFT | PolyPhen | CONDEL | Proven | ||||
| ARRP05 | c.490C>T | p.R164* | Nonsense | Het | 4 | NA | NA | NA | NA | Novel | 0/100 |
| c.6416G>A | p.C2139Y | Missense | Het | 31 | D | PD | NE | DE | CM102730 | 0/100 | |
| ARRP06 | c.7919G>A | p.W2640* | Nonsense | Het | 41 | NA | NA | NA | NA | CM085393 | 0/100 |
| c.8861T>C | p.F2954S | Missense | Het | 43 | T | B | DE | NE | Novel | 0/100 | |
Abbreviations: Het: heterozygous; NA: not available; D: damaging; PD: probably damaging; NE: neutral; DE: deleterious; T: tolerated; B: benign; HGMD: the Human Gene Mutation Database.
Figure 2. Predicted crystal structures.
Predicted crystal structures of the wild type (A and C) and mutant protein eyes shut homolog (B and D).
Another two heterozygous mutations, EYS c.7919G>A and c.8861T>C, were identified as RP causative for the three patients in family ARRP06. EYS c.7919G>A was a recurrent nonsense mutation creating a PTC at residue 2640. The other mutation c.8861T>C was a novel missense substitution from the hydrophobic phenylalanine to the hydrophilic serine at residue 2954. Similar to the missense mutation found in family ARRP05, this mutation would also induce three additional hydrogen bonds between serine and lysine at residue 2951, serine at residue 2953, and threonine at residue 2956, respectively (Figures 2C–2D). However, unlike EYS c.7919G>A, the predicted crystal structure of the mutated protein indicated that this substitution would significantly interfere with the interaction among distinct amino acids and thus transform its three-dimensional structure. Structural modeling was constructed on the basis of the crystal structure of template 3poy.1A6, which demonstrated the sequence identity of 19.56% with the protein eyes shut homolog.
Discussion
Mutations in the EYS gene are recognized as major causes for ARRP in multiple ethnic groups7,8,9,10,11. Reportedly, EYS mutations account for nearly 5% of ARRP patients with western European ancestry12, while 15.9% of ARRP patients in the Spanish population9. The estimated prevalence of EYS involvement in the pathogenesis of ARRP in the Japanese population is even higher, which has reached 18%8. Additionally, studies have also revealed the relationship between EYS mutations and autosomal recessive cone-rod dystrophy (CRD)13,14. In our study, we report two biallelic EYS mutations, including two novel variants and two recurrent mutations, as disease causative for two Chinese families with ARRP.
The EYS gene, located on 6q12, contains 43 exons and covers a genomic fragment of over 2 Mb. EYS encodes the protein eyes shut homolog, which comprises 3165 amino acids and is expressed specifically in the outer segment of the photoreceptor cell layer7. Biologically, this protein interacts selectively and non-covalently with calcium ions (Ca2+), and is involved in visual perception in which a light stimulus is received and converted into a molecular signal7. Its homological protein spam in Drosophila melanogaster interacts with another protein prom and functions in the morphogenesis of retina. Defects in spam and prom can cause a failure of inter-rhabdomeral-space separation15. The protein eyes shut homolog starts with a signal peptide containing 21 amino acids, and contains five Laminin G-like domains and 27 EGF like domains. Of all 27 EGF like domains, six are calcium-binding domains, and three of all four RP causative mutations identified in this study locate in the EGF like domains. The four identified mutations included two missense, p.C2139Y and p.F2954S, and two nonsense variations, p.R164* and p.W2640*. Nonsense variants would probably lead to the generation of a truncated protein; however, such truncated proteins may degrade in vivo via nonsense mediated mRNA decay (NMD). Crystal structural modeling suggests that both missense variants would result into the generation of hydrogen bonds and further alter the spatial conformation of the protein. In addition, those hydrogen bonds would also potentially affect the solubility of the protein.
Mutations in EYS correlate with a wide panel of phenotypes. The disease onset ages varied greatly from 6- to 62-year-old12,15. Two biallelic EYS mutations are revealed in the present study. Biallelic EYS mutations have been reported in multiple ethnicities13,14,16,17,18. Two mutations have been previously reported. EYS p.W2640* has been found in a Spanish family while the clinical details were not reported7. The other mutation, EYS p.C2139Y, was found in both Caucasians and Chinese14,16,17, while the clinical details were only discussed in a Chinese family carrying compound EYS p.[C2139Y];[G2186E]17. Patients in this family showed remarkable intrafamilial phenotypic diversity with their disease onset ages varied from 20- to 43-year-old, which were distinct from family ARRP05 in this report carrying EYS p.[R164*];[C2139Y]. All patients in family ARRP05 developed RP at their early 20 s and the disease progressed fast since they got their first symptom, indicating the interfamilial phenotypic diversity for EYS mutations. Similar to the previously reported Chinese family, the RP onset ages and disease progressions varied greatly among patients in family ARRP06, further supporting the significant intrafamilial phenotypic varieties for EYS mutations. Based on the observed interfamilial and intrafamilial phenotypic diversities, further investigations on the biological interplays of EYS are needed for a better understanding of the pathogenesis of EYS defects and a better look into the genotype-phenotype correlations.
RP shows significant clinical and genetic heterogeneities. Identifying genetic causes and developing advanced and applicable molecular diagnostic tools for RP are essential to lower the prevalence of RP and to find the therapeutic method for RP. However, the large number of RP causative genes and the limitations of routine techniques hinder investigators from further studying the genetic causes of RP. Our study indicates that targeted genes capture with NGS yield high sensitivity and speed for mutation detection in RP patients. When compared with traditional techniques, targeted NGS approach presents tremendous advantages. In addition, the development of a powerful molecular diagnostic platform for RP aims to improve the detection rate of causative genes/mutations in RP patients, to further investigate the genetic causes for RP, to better understand the pathological basis of RP, and to promote the fast development of molecular diagnosis globally. Meanwhile, it will have significance for the clinical and prenatal diagnosis of RP, and thus providing rationale for gene therapy on RP.
In summary, by means of targeted NGS approach, we have identified biallelic heterozygous EYS mutations in two Chinese RP families with their genotype-phenotype correlations discussed. Our study extends the mutational spectrums for EYS, and supports the application of targeted NGS approach in the molecular diagnosis for RP patients.
Methods
Participants and Clinical Evaluations
Ten participants from two unrelated families, including five patients and five unaffected family members, were recruited from the First Affiliated Hospital of Nanjing Medical University (Figure 1A). Peripheral blood samples were collected from each participant using 5 mL tubes with ethylene diamine tetraacetic acid (EDTA). Genomic DNA was extracted from leukocytes with a QIAmp DNA blood kit (Qiagen, Valencia, CA) according to the manufacturer's protocols. Detailed ophthalmic examinations, including best-corrected visual acuities (BCVAs) measurements, slit-lamp bio-microscopy, fundus photos, VF evaluations, and electroretinography (ERG) tests, were performed on each included members. Additionally, another 100 controls free of retinal dystrophies or other major ocular diseases were included with their blood samples collected19. This study adhered to the tenets of the Declaration of Helsinki, and was approved and prospectively reviewed by the Ethics Committee on Human Research of Nanjing Medical University. Written informed consents were signed by all participants or their statutory guardian before their participation.
Targeted NGS Approach, Bioinformatics Analyses, and Sanger Sequencing
Two patients from each included family were selected for the targeted NGS approach. A previously described microarray targeting 180 IRDs causative and 9 candidate genes1,19,20,21,22,23 were applied for mutation screening in patients ARRP05-II:3, ARRP05-II:7, ARRP06-II:2, and ARRP06-II:8. Library preparation, qualification, and NGS were further conducted on the Illumina Hiseq2000 platform (Illumina, Inc., San Diego, CA, USA) in collaboration with BGI-Shenzhen (Shenzhen, Guangdong, China) as detailed previously20. Bioinformatics analyses, including reads alignment and calculations of coverage and depth, were also conducted using a previously described protocol1,19,20,22,23. The following 5 databases were then used for annotation of all identified variants, including dbSNP137 (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/snp137.txt.gz.), HapMap Project (ftp://ftp.ncbi.nlm.nih.gov/hapmap), 1000 Genome Project (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp), YH database (http://yh.genomics.org.cn/), and Exome Variant Server (http://evs.gs.washington.edu/EVS/). For variants passed the initial filtration, we used Sanger sequencing for verification of variants within all attainable family members and for prevalence test in the above-mentioned 100 unrelated controls. Sanger sequencing was conducted as indicated previously24 with primer information listed in Table S2.
In Silico Analyses
Four types of online predictive software were applied to predict impacts caused by the mutations, including Sorting Intolerant From Tolerant (SIFT; http://sift.jcvi.org/)25, Polymorphism Phenotyping v2 (PolyPhen-2, v.2.2.2; http://genetics.bwh.harvard.edu/pph2/)26, Consensus Deleteriousness score of missense SNVs (CONDEL; http://bg.upf.edu/condel/home)27, and Protein Variation Effect Analyzer (PROVEAN, v.1.1.3; http://provean.jcvi.org/index.php)28. The crystal structures of the wild type and mutant proteins were predicted using SWISS-MODEL online server29,30, and displayed by PyMol software (version 1.5).
Author Contributions
Study design: X.C., K.Z. and C.Z. Collected the samples and performed the experiments: X.C., X.L., X.S., X.G., X.Z., Z.L., H.L., Y.L. and W.R. Data interpretation and analysis: X.C. and X.L. Wrote the manuscript: X.C. and C.Z. All authors have read and approved the final manuscript.
Supplementary Material
Supplementary information file
Acknowledgments
The authors thank all patients and their family members for their participation. The authors also appreciate all clinicians for collecting of samples and clinical data, and are grateful to Liping Guan, Jingjing Xiao, and Jingjing Jiang from BGI-Shenzhen for technical support. This work was supported by National Key Basic Research Program of China (973 Program, 2013CB967500); National Natural Science Foundation of China (81222009, 81170856, 81160124, 81260154, and 81170867); Thousand Youth Talents Program of China (to C. Z.); Jiangsu Outstanding Young Investigator Program (BK2012046); Jiangsu Province's Key Provincial Talents Program (RC201149); the Fundamental Research Funds of the State Key Laboratory of Ophthalmology (to C. Z.); Jiangsu Province's Scientific Research Innovation Program for Postgraduates (CXZZ13_0590); and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Chen X. et al. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet 23, 2926–2939 (2014). [DOI] [PubMed] [Google Scholar]
- Xu L., Hu L., Ma K., Li J. & Jonas J. B. Prevalence of retinitis pigmentosa in urban and rural adult Chinese: The Beijing Eye Study. Eur J Ophthalmol 16, 865–866 (2006). [DOI] [PubMed] [Google Scholar]
- Dryja T. P., Hahn L. B., Kajiwara K. & Berson E. L. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest Ophthalmol Vis Sci 38, 1972–1982 (1997). [PubMed] [Google Scholar]
- Kajiwara K., Berson E. L. & Dryja T. P. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 264, 1604–1608 (1994). [DOI] [PubMed] [Google Scholar]
- Buermans H. P. & den Dunnen J. T. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta 1842, 1932–1941 (2014). 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Miller M. T. et al. The crystal structure of the alpha-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure 19, 767–778 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El-Aziz M. M. et al. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat Genet 40, 1285–1287 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono K. et al. Two novel mutations in the EYS gene are possible major causes of autosomal recessive retinitis pigmentosa in the Japanese population. PLoS One 7, e31036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan I. et al. Mutation spectrum of EYS in Spanish patients with autosomal recessive retinitis pigmentosa. Hum Mutat 31, E1772–1800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandah-Rozenfeld D. et al. Novel null mutations in the EYS gene are a frequent cause of autosomal recessive retinitis pigmentosa in the Israeli population. Invest Ophthalmol Vis Sci 51, 4387–4394 (2010). [DOI] [PubMed] [Google Scholar]
- Huang Y. et al. Identification of a novel homozygous nonsense mutation in EYS in a Chinese family with autosomal recessive retinitis pigmentosa. BMC Med Genet 11, 121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littink K. W. et al. Mutations in the EYS gene account for approximately 5% of autosomal recessive retinitis pigmentosa and cause a fairly homogeneous phenotype. Ophthalmology 117, 2026–2033, 2033 e2021-2027 (2010). [DOI] [PubMed] [Google Scholar]
- Katagiri S. et al. Autosomal recessive cone-rod dystrophy associated with compound heterozygous mutations in the EYS gene. Doc Ophthalmol 128, 211–217 (2014). [DOI] [PubMed] [Google Scholar]
- Audo I. et al. EYS is a major gene for rod-cone dystrophies in France. Hum Mutat 31, E1406–1435 (2010). [DOI] [PubMed] [Google Scholar]
- Zelhof A. C., Hardy R. W., Becker A. & Zuker C. S. Transforming the architecture of compound eyes. Nature 443, 696–699 (2006). [DOI] [PubMed] [Google Scholar]
- Huang X. F. et al. Genotype-phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet Med; 10.1038/gim.2014.138 (2014). [DOI] [PubMed] [Google Scholar]
- Abd El-Aziz M. M. et al. Identification of novel mutations in the ortholog of Drosophila eyes shut gene (EYS) causing autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci 51, 4266–4272 (2010). [DOI] [PubMed] [Google Scholar]
- Jinda W. et al. Whole exome sequencing in Thai patients with retinitis pigmentosa reveals novel mutations in six genes. Invest Ophthalmol Vis Sci 55, 2259–2268 (2014). [DOI] [PubMed] [Google Scholar]
- Rong W. et al. Novel and recurrent MYO7A mutations in Usher syndrome type 1 and type 2. PLoS One 9, e97808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Targeted sequencing of 179 genes associated with hereditary retinal dystrophies and 10 candidate genes identifies novel and known mutations in patients with various retinal diseases. Invest Ophthalmol Vis Sci 54, 2186–2197 (2013). [DOI] [PubMed] [Google Scholar]
- Sheng X. et al. A novel homozygous BEST1 mutation correlates with complex ocular phenotypes. Ophthalmology 120, 1511–1512 e1512 (2013). [DOI] [PubMed] [Google Scholar]
- Pan X. et al. Mutation analysis of pre-mRNA splicing genes in Chinese families with retinitis pigmentosa. Mol Vis 20, 770–779 (2014). [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Targeted next-generation sequencing reveals novel USH2A mutations associated with diverse disease phenotypes: implications for clinical and molecular diagnosis. PLoS One 9, e105439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. et al. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum Genet 119, 617–623 (2006). [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S. & Ng P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A. & Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 88, 440–449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G. E., Murphy S., Miller J. R. & Chan A. P. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J. & Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- Kiefer F., Arnold K., Kunzli M., Bordoli L. & Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37, D387–392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information file