Abstract
The study of human T lymphocyte biology often involves examination of responses to activating ligands. T cells recognize and respond to processed peptide antigens presented by MHC (human ortholog HLA) molecules through the T cell receptor (TCR) in a highly sensitive and specific manner. While the primary function of T cells is to mediate protective immune responses to foreign antigens presented by self-MHC, T cells respond robustly to antigenic differences in allogeneic tissues. T cell responses to alloantigens can be described as either direct or indirect alloreactivity. In alloreactivity, the T cell responds through highly specific recognition of both the presented peptide and the MHC molecule. The robust oligoclonal response of T cells to allogeneic stimulation reflects the large number of potentially stimulatory alloantigens present in allogeneic tissues. While the breadth of alloreactive T cell responses is an important factor in initiating and mediating the pathology associated with biologically-relevant alloreactive responses such as graft versus host disease and allograft rejection, it can preclude analysis of T cell responses to allogeneic ligands. To this end, this protocol describes a method for generating alloreactive T cells from naive human peripheral blood leukocytes (PBL) that respond to known peptide-MHC (pMHC) alloantigens. The protocol applies pMHC multimer labeling, magnetic bead enrichment and flow cytometry to single cell in vitro culture methods for the generation of alloantigen-specific T cell clones. This enables studies of the biochemistry and function of T cells responding to allogeneic stimulation.
Keywords: Immunology, Issue 93, T cell, immunology, human cell culture, transplantation, flow cytometry, alloreactivity
Introduction
T lymphocytes are critical components of the adaptive immune system. T cells are responsible for not only directly mediating protective immune responses to pathogens through a variety of effector mechanisms, but also actively maintaining immunological self-tolerance and directing the responses of other cells in the immune system. These functions are directed through a number of integrated signals, including T cell receptor (TCR) ligation, cytokines and chemokines, and metabolites1. Of these signals, the TCR is of particular importance, as it provides the characteristic specificity that defines the T cell’s role in adaptive immunity. A TCR interacts with linear peptide antigens presented by MHC (human ortholog HLA) molecules (pMHC complexes) in a highly specific and sensitive manner to provide the signals that initiate T cell effector function. The biochemical parameters of TCR interactions with pMHC ligands provide not only the specificity for T cell activation, but also have a qualitative impact on subsequent T cell function2. Thus, studying T cell function often requires examining the responses of clonal T cells with defined antigenic specificity.
The human T cell compartment, containing approximately 1012αβ T cells, contains an estimated 107– 108 distinct αβTCRs3-4. This diverse repertoire provides opportunity for recognition of the vast array of peptides from potential pathogens that would necessitate a T cell response for protective immunity. It is estimated that the frequency of T cells responding to a given foreign antigen presented by self-MHC is on the order of 10-4– 10-7 in the absence of prior immune response to that antigen5. The naive T cell repertoire is shaped by thymic selection to ensure the ability to recognize self-MHC presenting peptide antigens and limit reactivity against self-peptide antigens, maximizing the potential utility for mediating protective immunity2. However, in violation of this designed reactivity, a relatively large frequency, 10-3– 10-4, of T cells from immunologically naive individuals respond to stimulation with allogeneic cells, recognizing both the foreign MHC molecules as well as the endogenous peptides they present6. The recognition of allogeneic pMHC ligands is structurally similar to the recognition of foreign antigens presented by self-MHC; the TCR makes critical biochemical interactions with both the allogeneic MHC molecule as well as the presented peptide7. The robust nature of the response of T cells to allogeneic stimulation results from the diversity of pMHC complexes present on the surface of allogeneic cells8. It is estimated that each MHC presents approximately 2 x 104 different endogenous peptide antigens9. This breadth of response to allogeneic stimulation is a significant aspect of the clinically-relevant pathology, such as allograft rejection or graft versus host disease (GVHD), resulting from T cell alloreactivity.
Study of human T cell alloreactive responses has traditionally relied upon examining polyclonal responses of naive T cells following stimulation with allogeneic cells. Repeated stimulation with the same allogeneic cell line combined with limiting dilution analyses is capable of generating clonal T cells with defined recognition of allogeneic HLA10. However, this approach is problematic for examining responses to individual allogeneic pMHC ligands, as the large and diverse repertoire of endogenous pMHC complexes present for a given allogeneic HLA stimulates a broad repertoire of T cells. This bulk population stimulation and limiting dilution approach would require screening of large numbers of clones to isolate T cells with the desired reactivity against a single pMHC ligand. Additionally, the frequency of T cells responding to an individual allogeneic pMHC ligand is relatively low among naive T cell populations, which presents a barrier to efficient generation of human T cell clones responsive to a given antigen.
Identification and isolation of antigen-specific T cells from polyclonal populations have been enabled by the development of fluorophore-labeled pMHC multimers11. This approach utilizes specific peptide antigens loaded into recombinant soluble biotinylated MHC molecules, which are labeled by binding to a streptavidin-labeled fluorophore. Multimerization of pMHC increases the avidity, compensating for the intrinsically low (µM) affinity of TCR for soluble pMHC ligands. Labeled cells can be identified and isolated by flow cytometry. However, this approach is still limited by the low frequency of antigen-specific T cells among naive T cell populations, which are typically orders of magnitude less than the limit of accurate identification and quantification on most flow cytometers. To address this limitation, a method of pMHC tetramer labeling and subsequent magnetic bead enrichment for tetramer-labeled cells has been developed12. This method has demonstrated reliable detection, enumeration, and isolation of low-frequency antigen-specific T cells.
This protocol describes an effective protocol for the generation of human T cell clones that specifically respond to individual allogeneic pMHC ligands. The protocol applies pMHC (HLA) multimer labeling and enrichment for the isolation of alloantigen-specific human T cells with flow cytometry cell sorting and a robust method for in vitro culture of human T cells to enable production of T cell clones from single sorted cells (overview in Figure 1).
Protocol
NOTE: This protocol requires use of peripheral blood samples from human volunteers. All research with human subjects should be reviewed and approved by a Human Studies Institutional Review Board to ensure compliance with the Declaration of Helsinki (2013) and the Health Insurance Portability and Accountability Act of 1996.
1. Isolation of T cells from Whole Blood
Prior to starting, warm the density gradient medium to room temperature. Aliquot 4 ml of density gradient medium into 2-4 sterile 15 ml conical centrifuge tubes (1 tube will be used for each 10 ml total volume of diluted blood).
Obtain 10-20 ml of blood in 1-2 sodium heparin spray coated (green-top) venous blood collection tubes. Collect human specimens under the supervision of trained individuals and according to an Institutional Review Board approved protocol.
Wipe the outside of the tubes with 70% ethanol. Carefully remove the tops of the filled collection tubes and decant the blood into a sterile 50 ml centrifuge tube.
Add 10 ml sterile phosphate buffered saline (PBS) to each blood collection tube. Collect the PBS, add to the decanted whole blood, and mix gently.
Add 25 µl Human T Cell Enrichment Cocktail / 2 ml total volume. Incubate at room temperature for 20 min.
Using a 10 ml pipette, layer up to 10 ml of the 1:1 diluted blood gently on top of the density gradient medium. Be careful to not disrupt the surface of the density gradient medium.
Centrifuge layered blood and density gradient medium at 1,200 x g for 20 min at 20 oC.
Remove the tubes from the centrifuge, taking care to not disrupt the interface between the density gradient medium and the layer of leukocytes at the interface between the density gradient medium and the diluted plasma. Using a 5 ml pipette, carefully collect the leukocyte layer and transfer to a new sterile 50 ml conical tube.
Add PBS to bring the volume of the collected PBL to 50 ml and mix gently.
Centrifuge at 600 x g for 5 min at 20 oC. Decant.
- Resuspend pelleted cells in 10 ml sterile flow cytometry sorting buffer (sterile-filtered PBS containing 1% BSA). Sample 10 µl of cell suspension, dilute 1:10 (add to 90 μl) trypan blue to count cells using a hemocytometer (expected yield 1 – 4 x 106 cells / tube of whole blood).
- Remove 1 ml aliquot, transfer to 5 ml tube, and keep on ice for analysis of T cells not labeled by tetramer. Keep cell suspension on ice.
2. Magnetic Enrichment of Alloantigen-specific T cells
Dilute alloantigen pMHC tetramer to 1:100 in sterile sort buffer.
Centrifuge cell suspension (9 ml from step 1.11) at 600 x g for 5 min at 20 oC. Decant.
Add 50 µl of diluted alloantigen pMHC tetramer to pelleted cells (up to 107 cells). Mix by gentle pipetting. Transfer to a sterile 5 ml tube. Incubate for 30 min at room temperature.
Add 2 ml sort buffer. Centrifuge at 600 x g for 5 min at 20 oC. Decant.
Resuspend cells in 100 µl sort buffer. Add 10 µl Biotin Selection Cocktail and incubate for 15 min at room temperature.
Add 5 µl Magnetic Nanoparticles and incubate for 10 min at room temperature.
Add 2.5 ml sort buffer and mix gently by pipetting. Remove 100 µl aliquot, transfer to 5 ml tube, and keep on ice for analysis of pre-enrichment cells.
Place the 5 ml tube containing the cell suspension in the cell separation magnet. The cap should be loosely atop the tube. Incubate for 5 min at room temperature.
Gently remove the cap from the tube. Holding the tube and magnet together, decant the tube contents into a fresh 5 ml tube. Do not tap or shake the tube contents to remove the last drops of liquid.
Remove the tube from the magnet. Add 2 ml cold sort buffer and mix gently by pipetting. Sample 10 µl of cell suspension, dilute 1:2 (add to 10 µl) trypan blue to count cells using a hemocytometer.
3. Preparation of T cells for Single-cell Flow Cytometry Cell Sorting
Add cold sort buffer to all tubes of cells (tetramer unlabeled, tetramer-labeled un-enriched, and tetramer-labeled enriched) to bring volume to 3 ml.
Centrifuge at 600 x g for 5 min at 4 oC and decant buffer to recover cells.
Resuspend pelleted cells in 25 µl cold sort buffer. Add 5 µl human Fc block. Incubate on ice for 20 min.
Prepare antibodies for flow cytometry sorting. Mix 15 µl sort buffer, 15 µl anti-CD5, 15 µl anti-CD14, and 15 µl anti-CD19.
Add 20 µl of antibody mixture to each tube of cells. Incubate on ice for 20 min.
Add 2 ml of cold sort buffer to each tube.
Centrifuge at 600 x g for 5 min at 4 oC and decant buffer to recover cells.
Resuspend cells at a concentration of 1 -2 x 106 cells/ml in sort buffer.
Aliquot 100 µl of human T cell culture medium (Iscove’s DMEM supplemented with 2 mM Glutamax, 10 mM HEPES, 50 µg/ml gentamycin, 50 µM 2-mercaptoethanol, 10% heat-inactivated human AB serum, and 2.5 ng/ml recombinant human IL-2) to each well of a 96-well round bottom cell culture plate. Keep on ice.
4. Isolation of Tetramer-labeled T cells by Single-cell Flow Cytometry Sorting
Establish flow cytometry gating parameters to identify alloantigen-pMHC tetramer labeled T cells (Figure 2.A). Use the pre-enrichment tetramer labeled fraction to establish gating strategies to eliminate doublets, gate on lymphocytes, exclude CD19 expressing B cells and CD14 expressing monocytes, and identify T cells by CD5 expression. Use the sample not labeled with tetramer as a fluorescence minus one control to establish the gating for identification of tetramer-binding T cells (with 0% of CD5+CD14-CD19- cells from the sample not labeled by tetramer should fall within this gate). NOTE: Gating strategies that are not adequately stringent will result in isolation of non-T cells or T cells that are not antigen specific, while overly restrictive gating strategies will reduce the number of T cells isolated.
Program the plate parameters for the single cell sort. Direct the sorter to place 1 CD5+CD14-CD19-tetramer+ cell in each well. Direct the plate set up to contain 1 negative control well (no cells sorted into the well) and 1 positive control well (100 CD5+CD14-CD19-tetramer- cells) in each row (Figure 2.B).
Using the established flow cytometry gating strategy and plate setup, isolate individual tetramer-binding T cells directly into the 96-well plate using a flow cytometric cell sorter.
5. Culture and Expansion of Alloantigen-specific T cell Clones
After completion of cell sorting, centrifuge plate at 600 x g for 5 min at 20 oC. Place culture plate at 37 oC in a 6% CO2 incubator.
Vortex stimulatory anti-CD3/anti-CD28 microbeads for 30 sec to resuspend beads.
Calculate volume of activator beads required for stimulation of collected T cells (0.5 µl of activator microbeads / well). Transfer calculated volume of stimulator beads to a sterile 5 ml tube. Add 1 ml human T cell culture medium and vortex.
Place tube with microbead suspension in magnet. The cap should be loosely atop the tube. Incubate for 2 min at room temperature.
Gently remove the cap from the tube. Holding the tube and magnet together, decant tube contents into a fresh 5 ml tube. Do not tap or shake the tube contents.
Remove the tube from the magnet. Add human T cell culture medium (100 µl medium / 0.5 µl of microbeads initially added).
Aliquot 100 µl of activator bead suspension to each well of the 96-well plate. Incubate culture plate at 37 oC in a 6% CO2 incubator.
Monitor cell growth daily by microscopic examination. Small clusters of proliferating cells may be observed microscopically after 5 – 7 days of culture (Figure 3.A). NOTE: Visualization of T cell cultures at this point can be difficult, and the absence of observable cell clusters at this stage may not be indicative of a lack of growing T cells.
At 14 days after cell isolation, feed cultures by carefully removing 100 µl of media off of the top of the culture and replace with 100 µl fresh human T cell culture medium. Continue incubating culture plate at 37 oC in a 6% CO2 incubator.
Monitor cell growth daily by microscopic examination and evaluation of the color of the culture medium. Large cell clusters should be visible microscopically 2 – 3 days after the medium change. Macroscopically, cell pellets should become visible during the 14 days following the medium change.
At 28 days following cell isolation, identify growing clones by microscopic examination. Transfer the 200 µl volume of the growth-positive 96-well plate cultures to individual wells of a 48-well tissue culture plate containing 200 µl human T cell culture medium.
Add 100 µl containing 12.5 µl of stimulatory anti-CD3/anti-CD28 microbeads (prepared as described in sections 5.2 – 5.6). Incubate culture plate at 37 oC in a 6% CO2 incubator.
Monitor culture growth daily by microscopic examination and evaluation of the color of the culture medium. When T cell culture reaches >2x106/ml (typically 3-5 days after re-stimulation, can be estimated by the day that the media begins turning straw-yellow), transfer culture to a well of a 24-well tissue culture plate and add 500 µl human T cell medium.
At 10-14 days follow re-stimulation, collect 200 µl of T cell culture to assess alloantigen specificity by flow cytometry analysis of pMHC tetramer binding (using the labeling and gating strategy described in sections 3 and 4.1).
6. Long-term Re-stimulation and Culture of T cell Clones
Continually re-stimulate and expand clonal T cell cultures with the desired specificity can be every 14 days. Collect T cell cultures into a sterile 5 ml tube at day 14 following the last anti-CD3/CD28 microbead stimulation. Combine multiple wells of the same T cell clone in a single tube, up to a volume of 2.5 ml.
Add medium to bring the final volume to 2.5 ml and mix gently by pipetting.
Place the 5 ml tube containing the T cell culture into the cell separation magnet to remove old microbeads from the culture. The cap should be loosely atop the tube. Incubate for 5 min at room temperature.
Gently remove the cap from the tube. Holding the tube and magnet together, decant the cell suspension into a fresh 5 ml tube. Do not tap or shake the tube contents to remove the last drops of liquid.
Add medium to bring the final volume to 4 ml. Sample 10 µl to count cells using a hemocytometer.
Recover T cells by centrifugation at 600 x g for 5 min at room temperature.
Decant supernatant. Resuspend T cells at 106 cells/ml human T cell culture medium. Transfer 1 ml aliquots to each well of a 24-well tissue culture plate.
Re-stimulate T cells by adding 100 µl media containing 12.5 µl of stimulatory anti-CD3/anti-CD28 microbeads (as described in sections 5.2 – 5.6). Incubate at 37 oC in a 6% CO2 incubator.
Representative Results
This protocol describes the generation of clonal human T cell cultures with defined alloantigen specificity via a magnetic bead enrichment and single-cell flow cytometry sorting strategy. Figure 1 provides an outline of the process.
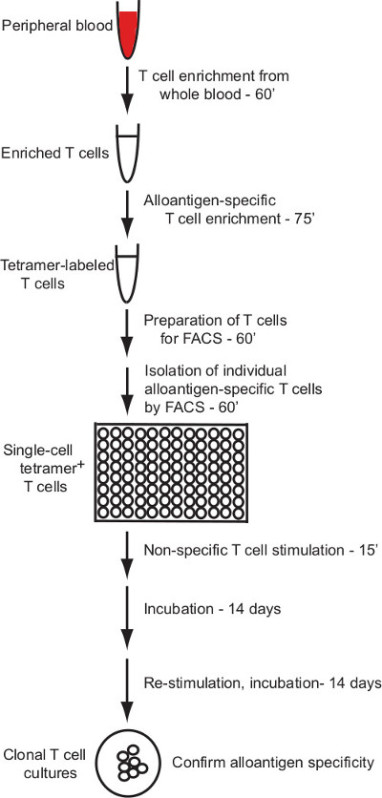 Figure 1: Protocol overview. The protocol described here provides a reliable method for generation of alloantigen-specific human T cell clones from peripheral blood. The process of single T cell isolation and setting up the cell culture is expected to take approximately 4.5 hr. Expansion of the T cell clones requires multiple rounds of non-specific T cell stimulation and culture, each taking 14 days. Clonal cultures can be tested for alloantigen specificity 28 days after isolation, and cultures can be further expanded for additional testing by repeated rounds of stimulation.
Figure 1: Protocol overview. The protocol described here provides a reliable method for generation of alloantigen-specific human T cell clones from peripheral blood. The process of single T cell isolation and setting up the cell culture is expected to take approximately 4.5 hr. Expansion of the T cell clones requires multiple rounds of non-specific T cell stimulation and culture, each taking 14 days. Clonal cultures can be tested for alloantigen specificity 28 days after isolation, and cultures can be further expanded for additional testing by repeated rounds of stimulation.
The expected yield of alloantigen-specific T cells will depend on the donor, the alloantigen used, and the corresponding frequency of reactive T cells in the donor. An average healthy donor will have 4.5 - 10.0 x 106 leukocytes/ml of blood, with approximately 20% T lymphocytes. Figure 2 illustrates the results of measuring the binding of T cells from a HLA-DR4 (HLA-DRB1*04:01)-negative donor to a specific alloantigen, amino acid residues 30-48 of the HLA-A2 protein (DTQFVRFDSDAASQRMEPR, A230-48) presented by the class II molecule HLA-DR413.
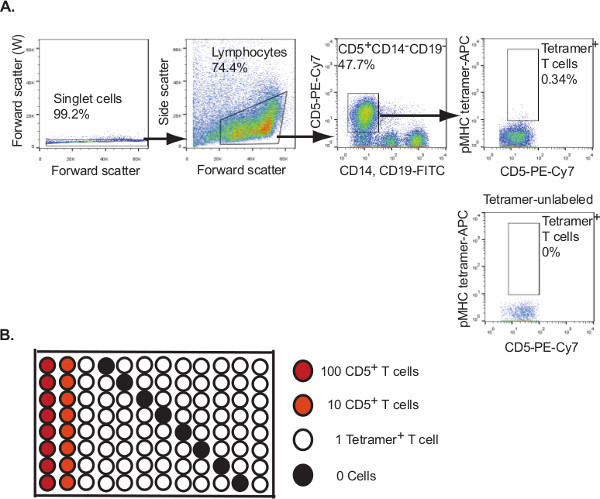 Figure 2: Flow cytometry sorting of alloantigen tetramer-labeled cells for generating single cell cultures. A. Flow cytometric identification and isolation of A230-48/HLA-DR4 tetramer-labeled T cells from a HLA-DR4-negative donor. T cells enriched for tetramer-labeled cells by magnetic bead selection were sorted by flow cytometry, gating on singlet CD5+CD14-CD19-tetramer+ lymphocytes as illustrated. Tetramer-unlabeled cells were used as a fluorescence-minus one control to establish gating for tetramer+ cells. B. Tetramer+ T cells were sorted into a 96-well round-bottom tissue culture plate containing 100 µl human T cell culture medium. The plate set-up included positive and negative control wells as indicated.
Figure 2: Flow cytometry sorting of alloantigen tetramer-labeled cells for generating single cell cultures. A. Flow cytometric identification and isolation of A230-48/HLA-DR4 tetramer-labeled T cells from a HLA-DR4-negative donor. T cells enriched for tetramer-labeled cells by magnetic bead selection were sorted by flow cytometry, gating on singlet CD5+CD14-CD19-tetramer+ lymphocytes as illustrated. Tetramer-unlabeled cells were used as a fluorescence-minus one control to establish gating for tetramer+ cells. B. Tetramer+ T cells were sorted into a 96-well round-bottom tissue culture plate containing 100 µl human T cell culture medium. The plate set-up included positive and negative control wells as indicated.
From a starting number of 2.0 x 107 leukocytes we found a frequency of alloantigen-specific cells of 1/4776 T cells:
1.1x106 tetramer-enriched cells x 0.992 (singlets) x 0.744 (lymphocytes) x 0.477 (CD5+CD14-CD19-) x 0.0034 (tetramer+ T cells) / 2.0 x 107 peripheral blood leukocytes x 0.314 (pre-enrichment CD3+)
From the starting 2.0x107 leukocytes we were able to isolate 88 individual tetramer-labeled T cells for culture. After 2 rounds of anti-CD3/CD28 microbead stimulation and culture as described we identified 2 growth-positive cultures by microscopic examination (representative example of growth 10 days after single cell isolation and culture shown in Figure 3.A). Clonal cultures were expanded to 1 ml as described, and alloantigen specificity was assessed by examining binding of the A230-48/HLA-DR4 tetramer (Figure 3.B).
 Figure 3. Evaluation of T cell cultures. A. Microscopic examination of 96-well plate T cell cultures. Representative example of T cell culture 10 days after isolation of individual T cells by flow cytometry sorting. Cells were stimulated with 0.5 µl anti-CD3/CD28 microbeads/well (examples indicated by arrowhead) in a volume of 200 µl human T cell culture medium. B. T cell alloantigen specificity of clonal T cell cultures evaluated by flow cytometry analysis of pMHC tetramer labeling. Clonal T cell cultures of flow cytometry isolated CD5+CD14-CD19- A230-48/HLA-DR4 tetramer+ lymphocytes were examined for A230-48/HLA-DR4 tetramer binding after 4 weeks of in vitro culture. Tetramer-unlabeled cells were used as a fluorescence-minus one control to establish gating for tetramer+ cells.
Figure 3. Evaluation of T cell cultures. A. Microscopic examination of 96-well plate T cell cultures. Representative example of T cell culture 10 days after isolation of individual T cells by flow cytometry sorting. Cells were stimulated with 0.5 µl anti-CD3/CD28 microbeads/well (examples indicated by arrowhead) in a volume of 200 µl human T cell culture medium. B. T cell alloantigen specificity of clonal T cell cultures evaluated by flow cytometry analysis of pMHC tetramer labeling. Clonal T cell cultures of flow cytometry isolated CD5+CD14-CD19- A230-48/HLA-DR4 tetramer+ lymphocytes were examined for A230-48/HLA-DR4 tetramer binding after 4 weeks of in vitro culture. Tetramer-unlabeled cells were used as a fluorescence-minus one control to establish gating for tetramer+ cells.
Discussion
T cell alloreactivity is a long-studied and clinically-relevant phenomenon. The robust proliferative and effector responses of T cells to allogeneic stimulation has enabled extensive analyses of human T cell responses in vitro through relatively straightforward mixed lymphocyte reactions of peripheral blood T cells against inactivated allogeneic cells. However, these primary alloreactive T cell responses are oligoclonal, comprised of a large number of individual T cells responding to specific alloantigens. This diversity in the T cell response impairs the ability to examine the biochemistry of pMHC ligand recognition driving alloreactive T cell responses. The standard solution to this challenge has been to generate clonal T cell lines through repeated stimulation with a given allogeneic cell line and limiting dilution analysis. While this approach is capable of generating clonal T cell lines with a single specificity, it may not reflect the original composition of the oligoclonal response of naive T cells. Additionally, this approach does not directly identify the specific pMHC alloantigen recognized (even narrowing the response to a single HLA molecule still presents the challenge of finding the stimulatory peptide from among the estimated 2x104 endogenous peptides presented under steady-state conditions), complicating biochemical characterization of T cell ligand recognition in alloreactivity.
The protocol presented here utilizes fluorescently-labeled pMHC multimers, magnetic bead enrichment, and flow cytometry cell sorting to identify and isolate human T cells specific for individual allogeneic pMHC ligands. This technique has become a powerful tool for examining antigen-specific T cell responses12. Our increasing understanding of the highly peptide-specific nature of T cell recognition of allogeneic pMHC complexes7,8 has enabled us to adapt this approach to the study of alloreactive T cells in both mice14 and humans15. This approach uniquely enables examination of the composition of the naive T cell populations that comprise the initial oligoclonal alloreactive response. Furthermore, combination of the pMHC multimer labeling approach with in vitro tissue culture facilitates generation of clonal T cell cultures that can be used to dissect the biochemistry of allogeneic pMHC ligand recognition, a question that cannot adequately be addressed via studying polyclonal T cell responses and is significantly impaired by traditional T cell cloning methods.
Using this approach, it can be expected that T cells specific for a given alloantigen pMHC should be identifiable for isolation and culture. However, the efficacy of this approach is dependent on multiple variables. First, the frequency of T cells from an immunologically naive donor that recognize a single alloantigen pMHC is relatively low (on the order of 1/103– 1/106) and varies between individuals. The ability to isolate pMHC-specific T cells depends on their abundance, as not all isolated cells will successfully expand to generate clonal cultures. The efficiency of the protocol can be influenced by several factors including the nature of the peptide used and the viability of the cells during preparation and isolation. The success of this technique also requires identification of allogeneic pMHC ligands capable of being re-folded into fluorescently-labeled multimers from recombinantly expressed HLA molecules and synthesized peptides. While there are multiple descriptions in the literature of identified peptides presented by specific HLA alleles, not all peptide re-fold in solution equally well. The efficacy of pMHC re-folding depends on several factors, including peptide length, solubility, and affinity for the MHC peptide binding pocket16. The ability of pMHC multimers to label antigen-specific T cells is also dependent on the peptide used. The affinity of the peptide for the MHC as well as the affinity of the TCR for the pMHC complex influences the binding of the fluorophore-labeled pMHC multimer. Weak binding can result in low shifts in fluorescence intensity, which can lead to difficulty in identifying antigen-specific cells by flow cytometry. These are significant technical hurdles that can impair the efficient isolation of alloantigen-specific T cells. In addition to these technical limitations, a significant biological caveat to this approach should be noted. T cell binding of pMHC tetramers is limited to TCRs with relatively high ligand affinity. This feature results in excluding T cells with low affinity for a ligand from identification and analysis. This is significant, as these low-affinity T cells are capable of substantial contribution to primary T cell responses17. However, even with these limitations, the approach described here is an efficient tool for the generation of clonal human T cells with defined antigen specificity. While the focus presented here is identification and isolation of human T cells specific for allogeneic pMHC ligands, the T cell identification, isolation, and culture techniques can be easily adapted to other pMHC specificities.
Disclosures
The authors declare no competing financial interests.
Acknowledgments
The author would like to thank the NIH Tetramer Core Facility for tetramer production. The author would also like to thank E.D. O’Connor and K.E. Marquez at the UCSD Human Embryonic Stem Cell Core Facility flow cytometry laboratory for assistance in cell sorting. This work was funded by National Institutes of Health grant K08AI085039 (G.P.M.).
References
- Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu. Rev. Immunol. 2009;27(1):591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat. Immunol. 2012;13(2):121–128. doi: 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- Arstilla TP, et al. A direct estimation of the human αβ T cell receptor diversity. Science. 1999;286(5441):958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- Robbins HS, et al. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanio C, Lemaitre F, Law HKW, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115(18):3718–3725. doi: 10.1182/blood-2009-10-251124. [DOI] [PubMed] [Google Scholar]
- Suchin EJ, et al. Quantifying the frequency of alloreactive T cells in vivo: new answers to an old question. J. Immunol. 2001;166(2):973–981. doi: 10.4049/jimmunol.166.2.973. [DOI] [PubMed] [Google Scholar]
- Felix NJ, Allen PM. Specificity of T-cell alloreactivity. Nat. Rev. Immunol. 2007;7(12):942–953. doi: 10.1038/nri2200. [DOI] [PubMed] [Google Scholar]
- Morris GP, Ni PP, Allen PM. Alloreactivity is limited by the endogenous peptide repertoire. Proc. Natl. Acad. Sci. USA. 2011;108(9):3695–3700. doi: 10.1073/pnas.1017015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri A, et al. In APCs, the autologous peptides selected by the diabetogenic I-Ag7 molecule are unique and determined by the amino acid changes in the P9 pocket. J. Immunol. 2002;168(3):1235–1243. doi: 10.4049/jimmunol.168.3.1235. [DOI] [PubMed] [Google Scholar]
- Yssl H, Spits H. Generation and maintenance of cloned human T cell lines. Curr. Protoc. Immunol. 2002;7 doi: 10.1002/0471142735.im0719s47. [DOI] [PubMed] [Google Scholar]
- Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Moon JJ, et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27(2):203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicz RM, et al. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J. Exp. Med. 1993;178(1):27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni PP, Allen PM, Morris GP. The ability to rearrange dual TCRs enhances positive selection, leading to increased allo- and autoreactive T cell repertoires. J. Immunol. 2014;In press doi: 10.4049/jimmunol.1400532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Uy GL, Donermeyer D, DiPersio JF, Allen PM. Dual receptor T cells mediate pathologic alloreactivity in patients with acute graft-versus-host disease. Sci. Transl. Med. 2013;5(188) doi: 10.1126/scitranslmed.3005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman JD, Reay PA, Davis MM. Formation of functional peptide complexes of class II major histocompatibility complex proteins from subunits produced in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1993;90(21):10330–10334. doi: 10.1073/pnas.90.21.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino JJ, Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J. Exp. Med. 2011;208(1):81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]


