Abstract
Drug development groups are close to discovering another pot of gold-a therapeutic target-similar to the success of imatinib (Gleevec) in the field of cancer biology. Modern molecular biology has improved cancer therapy through the identification of more pharmaceutically viable targets, and yet major problems and risks associated with late-phase cancer therapy remain. Presently, a growing number of reports have initiated a discussion about the benefits of metabolic regulation in cancers. The Warburg effect, a great discovery approximately 70 years ago, addresses the “universality” of cancer characteristics. For instance, most cancer cells prefer aerobic glycolysis instead of mitochondrial respiration. Recently, cancer metabolism has been explained not only by metabolites but also through modern molecular and chemical biological techniques. Scientists are seeking context-dependent universality among cancer types according to metabolic and enzymatic pathway signatures. This review presents current cancer metabolism studies and discusses future directions in cancer therapy targeting bio-energetics, bio-anabolism, and autophagy, emphasizing the important contribution of cancer metabolism in cancer therapy.
Keywords: Cancer, Metabolism, Cancer therapy
INTRODUCTION
Our scientific logic resides in an empiricism influenced by the 17th century philosophy of Francis Bacon. An inductive method, accompanied by recent technological developments, has led us to dissect cancers using oncogenomics, which includes somatic mutation analysis and whole genome sequencing, in an attempt to determine the underlying cause of cancer cell dysregulation. Somatic mutation represents a “specific problem in certain cancer type due to heterogeneity.” This implies that each tumor exhibits a unique altered signaling pattern due to specific somatic mutations. In other words, there is no universality among tumors; however, scientists continue to seek common mutations and potential drug targets such as BCR-ABL, which was identified in chronic myeloid leukemia. In a recent study, TCGA reported that over 30,000 mutations were identified in breast cancer tissue (Cancer Genome Atlas Network, 2012). The evidence seems to suggest that science based on an inductive method, such as cancer genomics, cannot continue.
In the foggy philosophical forest of oncology, there may be three beacons of hope that will light our path. The first was the discovery that targeting lactate-fueled respiration abrogated tumors in mouse models (Sonveaux et al., 2008). This encouraged us to frame the “universality” of cancer cells in terms of the therapeutic approach. Starvation, along with monocarboxylate transporter 1 (MCT1) inhibition terminated tumor growth in a cervical squamous carcinoma tumor model. The second beacon was accidently sparked by a meta-analysis investigating diabetes mellitus and the risk of breast cancer (Jiralerspong et al., 2009). Women with diabetes had a statistically significantly increased risk of breast cancer. A series of epidemiological studies have shown a decrease in cancer incidence among metformin-treated patients. Metformin activates the AMP-activated protein kinase (AMPK) pathway, a major sensor of the cellular energy status that has been proposed as a promising cancer therapeutic target. This finding represents another successful case in which cancer “universality” has been targeted. Presently, many clinical trials of metformin in cancer are ongoing worldwide. The third beacon was the discovery of the mechanism underlying the failure of mammalian target of rapamycin (mTOR) inhibition in pro-survival autophagy induction (Keystone Symposia, Tumor Metabolism, 2012). For many years after the failed clinical trial of HSP90 inhibition, drug development researchers have focused their energy on rapamycin, which reduces tumor growth via mTOR inhibition. In cell metabolism, mTOR is an important inducer of biosynthesis, a process that is potentiated in cancer cells for excessive growth. However, clinical trials of mTOR inhibition failed because this intervention allowed cancer cells to trigger autophagy as an alternate survival pathway. As most anti-cancer kinase inhibitors induce autophagy in cancer, the suggestion that autophagy inhibition may be required for anti-cancer chemotherapy provides new hope. Again, this might be another successful case of “universality” in the very near future. Therefore, the definition of “universality” in cancer metabolism appears to be now “under a specific context, a common phenomenon of cancer cells for survival regardless of mutations”.
ELUSIVE OUTCOMES IN THE WAR ON CANCER
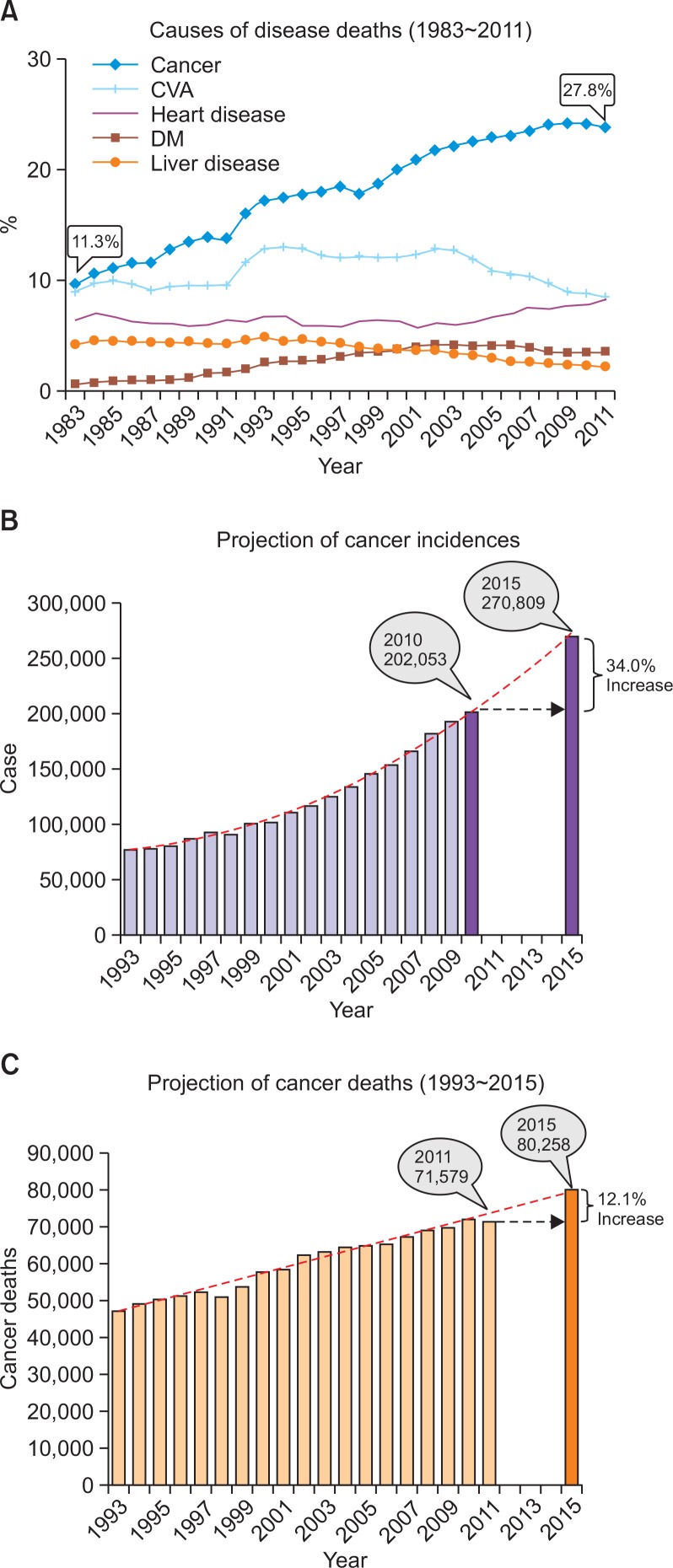
Are we winning the war on cancer? We have been informed that modern anti-cancer treatments are beneficial in terms of increasing overall survival. However, the overall survival among patients with late-stage cancer has remained steady for more than 40 years. We agree that the best chance for cancer survival depends on an early diagnosis followed by early surgical removal. Imatinib therapy confers a 95% survival rate upon patients with chronic myelogenous leukemia (CML) who harbor the BCR-ABL mutation. However, in the absence of a specific therapeutic regimen, most late-stage patients may benefit from radiation therapy or general chemotherapy in terms of extending survival, although these therapies are not curative. Why are such universal targets, such as that in CML, not found in all cancers? Among CML cases, 95% harbor the same chromosomal anomaly as the Philadelphia chromosome, which was discovered in 1960 by Peter Nowell at the University of Pennsylvania School of Medicine and David Hungerford at the Fox Chase Cancer Center. In this case, CML possesses a universal phenotype, such as a genetic disease characteristic, that is favorable for targeted therapy. Unfortunately, most cancers exhibit heterogeneous disease characteristics that often demonstrate the occurrence of Darwinian selection. In such cases, we have observed the failure of monotherapyin clinical trials. Despite all of our efforts, which have included combinational therapy, overall survival among late-stage cancer patients has not changed for many years. Cancer has been the leading cause of death in Korea since 1983, when it accounted for 11.3% of all deaths (Fig. 1). The frequency of death from cancer has increased steadily to account for 27.8% of all deaths in 2011. In comparison, the frequencies of deaths related to other diseases, including heart and liver diseases, have remained stable or decreased steadily (Fig. 1). Why have we upheld the same scientific philosophy while continuing to lose the war on cancer?
Fig. 1.
Cancer incidence and mortality in Republic of Korea. (A) Causes of death. Cancer has been the leading cause of death in Republic of Korea, rising steadily since 1983 and accounting for 11.3% of the total number of deaths in 1983 and 27.8% in 2011 (STATISTICS KOREA, 2012). (B) Projection of cancer incidence. The total number of cancer cases is expected to increase from 202,053 in 2010 to 270,809 in 2015, a 34.0% increase over the five-year period (National Cancer Center, Korea, 2010). (C) Projection of cancer death. The total number of cancer deaths is expected to grow from 71,579 in 2011 to 80,258 in 2015, a 12.1% increase in the next four-years (National Cancer Center, Korea, 2010).
TWO SCIENTIFIC METHODS: INDUCTIVE AND DEDUCTIVE METHODS
Why have we focused on targeted therapy for decades? The answer dates to the 17th century, when Francis Bacon introduced the inductive scientific method and a birth of reasoning during which the acceptance of scientific “Truth”, as imagined, began to decline. Bacon’s philosophy, which is simple and straightforward, became the basis of modern science. According to his philosophy, scientific truth can be accepted only when the three major rules of reductive matierialism, inductive method and empiricism are satisfied. Regarding empiricism, Bacon insisted that all theories regarding scientific truth must be proven through experimentation and result in the same phenomena; he defined the inductive method as the explanation of all scientific phenomena by causative factors and reductive materialism as the reduction of all causative factors to the smallest fragments. This scientific philosophy influenced many great discoveries in science such as Lavoisier’s law of conservation in the 18th century, Mendel’s law of inheritance in the 19th century, and Darwin’s law of natural selection in the 20th century. Progression of the inductive method has led development in all scientific fields, including oncology, for centuries.
TRENDS IN CANCER RESEARCH
Oncogenomics
The discovery of oncogenes several decades ago initiated a race to identify cancer genes and targets. The oncogene theory is based on an inductive method through which the theory must be proven by an etiologic disease factor; however, cancers contain many somatic mutations. This theory was influenced by the viral infection theory of tumorigenesis, which was discovered by Dr. Peyton Rous in 1911. A series of oncogenes and cancer driver genes were introduced up to the completion of human genome sequencing. On February 15, 2001, the Human Genome Project (HGP) consortium published the first human genome sequence in the journal Nature, followed one day later by a similar publication by the Celera Corporation in Science (Lander et al., 2001; Venter et al., 2001). The HGP identified and mapped the approximately 20,000–25,000 genes contained in the human genome. Human genome sequencing was completed in 2003. This technology led to the field of oncogenomics and attempts to identify the “smoking gun” of cancer. Oncogenomics is a relatively new sub-field of genomics that applies high-throughput technologies to characterize cancer-associated genes. Oncogenomics is synonymous with “cancer genomics.” Oncogenomists have proposed that cancer is a genetic disease caused by the accumulation of DNA mutations that lead to uncontrolled cell proliferation. The goal of oncogenomics is to identify new oncogenes or tumor suppressor genes that might provide new insights into cancer diagnosis, clinical cancer prognosis, and new cancer therapeutic targets.
Cancer develops towing to an accumulation of DNA mutations. These mutations accumulate randomly, and thus different DNA mutations and combinations of mutations are present in different individuals with the same type of cancer. Accordingly, the identification and targeting of the specific mutations that have occurred in an individual patient might lead to more effective cancer therapy. However, a recent study subjected 510 tumors from 507 breast cancer patients to whole-exome sequencing and identified 30,626 somatic mutations in breast cancer (Cancer Genome Atlas Network, 2012). If we were to select the 20 most important cancer targets from which to identify disease-causing combinations, we would need to crossbreed more than one million combinations in 20 genetically altered mouse models. According to Bacon’s philosophy, identical cancer development must be demonstrated experimentally in genetically mutated mice. Otherwise, mutation cannot be accepted as a scientific truth of cancer cause.
The pharmaceutical sector was doomed by the uncertainty of genomic drug development. We are not able to determine the “smoking gun” of cancer through genetics. This reality influenced a drive toward personalized and combination therapies in the pharmaceutical anti-cancer drug development sector. The article “Drug Makers’ New Reality: 'Innovate or die', published in the New York Times in 2002, reported that more than 80% of drugs in the pipelines of global pharmaceutical companies had failed. Unexpectedly, this agonizing dilemma was rescued by the success of imatinib (Gleevec).
Targeted cancer therapy
Gleevec rescued the pharmaceutical sector. Imatinib, which is marketed by Novartis as Gleevec, is a competitive tyrosine-kinase inhibitor used to treat multiple cancers, most notably Philadelphia chromosome-positive (Ph+) CML (Goldman and Melo, 2003). Like all tyrosine-kinase inhibitors, imatinib inhibits a central tyrosine kinase enzyme, in this case BCR-ABL, thus preventing the growth of cancer cells (Goldman and Melo, 2003). Because BCR-ABL exists only in cancer cells, imatinib works as a form of targeted therapy such that only cancer cells are killed through the drug's action (Fausel, 2007). In this regard, imatinib was among first cancer therapies to demonstrate the potential of targeted action and is often cited as a new paradigm in “targeted cancer therapy” research (Stegmeier et al., 2010).
The success of Imatinib (Gleevec) influenced the development of Trastuzumab (Herceptin), which targets the epidermal growth factor receptor (EGFR) family member Her2, and encouraged the field of oncogenomics to elucidate new cancer treatment targets (Strausberg et al., 2004). A series of targeted therapeutics have been developed by global pharmaceutical companies, including gefitinib (Iressa), which targets EGFR for lung cancer and was developed by AstraZeneca (United States Food and Drug Administration [US FDA] approved in 2003) (Armour and Watkins, 2010); erlotinib (Tarceva), which targets EGFR in lung and pancreatic cancers and was developed by Genentech/Roche/OSI (US FDA approved in 2004) (Buck et al., 2006); and sorafenib (Nexavar), which targets vascular endothelial growth factor receptor (VEGFR) in kidney cancer and was developed by Bayer (U.S. FDA approved in 2005) (Wilhelm et al., 2008) (Table 1). However, the cost-effectiveness of these therapies is not logical for late-stage and terminal cancer patients.
Table 1.
United States Food and Drug Administration-approved targeted cancer drugs
| Year | Drug | Trade Name | Key targets for therapeutic activity | US FDA-approved indication | Company |
|---|---|---|---|---|---|
| 2001 | Imatinib | Gleevec | BCR-ABL, PDGFR, KIT | CML and GIST | Novartis |
| 2003 | Gefitinib | Iressa | EGFR | Lung cancer | AstraZeneca |
| 2004 | Erlotinib | Tarceva | EGFR | Lung and pancreatic cancer | OSI/Genen-tech/Roche |
| 2005 | Sorafenib | Nexavar | VEGFR2, b-RAF, PDGFR | Kidney and liver cancers | Onyx/Bayer |
| 2006 | Dasatinib | Sprycel | BCR-ABL | CML | BMS |
| 2006 | Sunitinib | Sutent | VEGFR2, PDGF, KIT | Kidney cancer and GIST | Sugen/Pfizer |
| 2007 | Lapatinib | Tykerb | EGFR, ERBB2 | Breast cancer | GSK |
| 2007 | Temsirolimus | Torisel | mTOR | Kidney cancer | Wyeth/Pfizer |
| 2008 | Nilotinib | Tasigna | BCR-ABL | CML | Novartis |
| 2009 | Pazopanib | Votrient | VEGFR2, PDGFR, KIT | Kidney cancer | GSK |
| 2009 | Everolimus | Afinitor | mTOR | Kidney cancer | Novartis |
| 2011 | Crizotinib | Xalkori | EML-ALK, Met | NSCLC | Pfizer |
| 2011 | Vemurafenib | Zelboraf | b-RAF | Melanoma | Roche/Plexxicon |
| 2011 | Ruxolitinib | Jakafi | JAK1/2 | Myelofibrosis | Incyte |
Personalized and combination therapies
Imatinib (Gleevec) certainly demonstrated that a focus on critical cancer targets might confer a broad beneficial effect on the survival of patients who usually share general mutations. This traditional scientific approach, which incorporated the inductive method, demonstrated therapeutic benefits in the small proportion of cancer patients harboring homogeneous mutations. Often, cancer heterogeneity underlies the occurrence of drug resistance after standard therapeutic approaches. To overcome drug resistance in chemotherapy, two alternative strategies have been considered in the clinical sector. A new trend in cancer treatment takes an approach in which the tissue of origin and histology direct the choice of therapy toward a strategy in which knowledge regarding oncogenic mutations is used to select patients eligible for treatment involving highly selective drugs. This shift was enabled by two major developments over the past decades: “personalized therapy” and “combination therapy.”
In the first, the emergence of next-generation DNA sequencing technologies has enabled the identification of recurrent mutations in a variety of cancers. Clinicians can find algorithms for biomarkers (e.g., genomic mutations, epigenetics, and microarrays) that can be used to separate patients into groups according to good responses or non-responses to specific therapeutic regimens. Despite the lack of a scientifically logical link between biomarkers and therapeutic regimens, researchers can identify methods for improving patient survival rates. This does not exactly fit the definition of “personalized therapy” but does allow the broad use of clinical trials. In the second, the development of highly selective inhibitors of the products of genes activated by these frequent genomic alterations has provided a larger number of targeted cancer therapeutics. Therefore, researchers expect that the inhibition of additional oncogenic signaling pathways might increase the survival rate. This is known as “combination therapy.” However, the determination of therapeutic combinations is a fine art. For example, AKT inhibition leads to the activation of multiple receptor tyrosine kinases (RTKs), thus attenuating the effects of AKT inhibition. Therefore, AKT inhibition may have a better cancer therapeutic effect when combined with RTK inhibition. However, despite the avalanche of recent publications regarding single and combination targeted cancer therapies, the vast majority of cancer patients are still treated with conventional chemotherapies, the clinical benefits of which are virtually impossible to predict in the individual patient.
However, although more than a decade has passed since the adoption of this new therapeutic approach to personalized medicine, the death toll from cancer has not improved (Fig. 1).
Cancer heterogeneity and the deductive method
Pioneering chemical cancer treatments have been introduced via the deductive method. Dr. Paul Ehrlich, a German physician who received the Nobel Prize in 1908, initiated and also named the concept of chemotherapy before 1900 and advocated the use of animal models to study the effects of drugs on diseases. Sidney Farber’s study of the effects of folic acid on pediatric leukemia led to the development of antifolates as anticancer drugs. His findings generated an effective anti-leukemic drug, although the functional mechanism remained unknown (Sausville and Johnson, 2000; Chabner and Roberts, 2005). Successes with nitrosourea drugs and antifols led to the development of new drugs based on analogs of these compounds (Jackson, 1987).
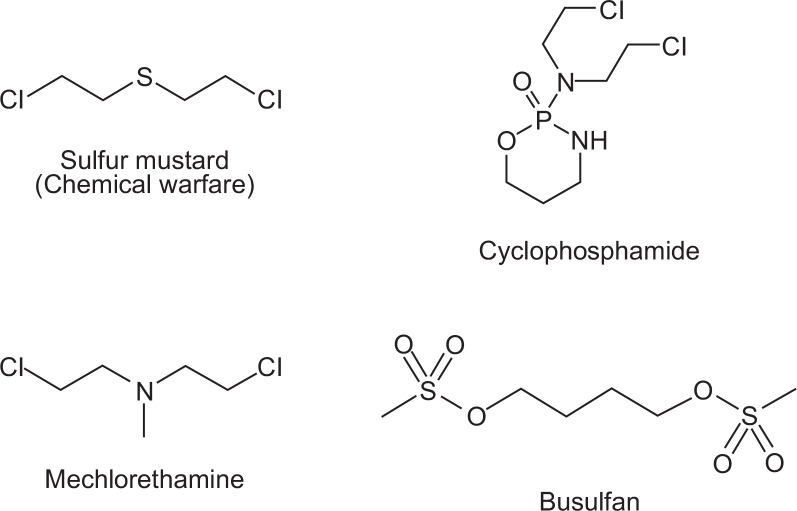
Prior to the 1950s, most cancers were treated with surgery and radiation. Louis Goodman and Alfred Gilman were pioneers in clinical trials of chemotherapy. Goodman and Gilman used nitrogen mustard to treat a patient with non-Hodgkin’s lymphoma; this work was based on findings of lymphoid cell destruction during the autopsies of soldiers who succumbed to sulfur mustard gas exposure during the First World War (Gilman, 1963) (Fig. 2). By defining the molecular action of this mustard compound, formation of an alkylating intermediate was determined to be a key mechanism. This finding led to the development of safe and reactive alkylating agents, including mechlorethamine, which was used mainly to treat Hodgkin’s disease and other lymphomas (Jacobs et al., 1968). Cyclophosphamide is the most commonly used alkylating agent (Foley and Kennedy, 1964); this agent is used in the treatment of Hodgkin’s and other lymphomas, acute lymphocytic leukemia, and a variety of solid tumors. During the 1950s, available cancer therapeutic drugs such as mechlorethamine (Edmonson et al., 1976), mercaptopurine (Regelson et al., 1964), methotrexate (Fisher and Elliott, 1965), melphalan (Brook et al., 1964), and busulfan (Arduino and Mellinger, 1967) (Table 2) exhibited a cytotoxic mechanism of action. Although the role of mechlorethamine in cancer cells was unknown, effective killing of leukemia cells was observed. In fact, this early stage of anti-cancer chemotherapy was developed using a deductive scientific method. Arsenic trioxide, which exhibits universal cancer cytotoxicity, was approved by the FDA in 2000. Interestingly, just over 50 years of anti-cancer drug development based on universal cytotoxic targets had passed since the FDA approved mechlorethamine in 1949. Imatinib (Gleevec) was introduced one year after the approval of arsenic trioxide (Table 2).
Fig. 2.
The first anti-cancer chemotherapeutics approved by the U.S. FDA. The destruction of lymphoid cells was discovered following autopsies of soldiers that died from sulfur mustard gas exposure during World War I. The mustard compound formed an alkylating intermediate, representing the key mechanism of action. This finding led to safe and reactive alkylating agents, including mechlorethamine, cyclophosphamide, and busulfan.
Table 2.
United States Food and Drug Administration-approved anti-cancer drugs with assistance from the National Cancer Institute Developmental Therapeutics Program between 1949 and 2004, most anti-cancer drugs were developed on the basis of universal cytotoxicity (http://dtp.nci.nih.gov/timeline/flash/index.htm)
| 1949 | 1970 | 1991 |
| Mechlorethamine (NSC 762) | FUDR (NSC 27640) | Fludarabine Phosphate (NSC 312887) |
| Ethinyl Estradiol (NSC 71423) | Mithramycin (NSC 24559) | Pentostatin (NSC 218321) |
| 1953 | o-p′-DDD (NSC 38721) | 1992 |
| TEM (NSC 9706) | 1973 | Chorodeoxyadenosine (NSC 105014) |
| Mercaptopurine (NSC 755) | Bleomycin (NSC 125066) | Taxol (NSC 125973) |
| Methotrexate (NSC 740) | 1974 | Teniposide (NSC 122819) |
| 1954 | Adriamycin (NSC 123127) | 1994 |
| Busulfan (NSC 750) | Mitomycin C (NSC 26980) | Navelbine (NSC 608210) |
| 1957 | 1975 | 1995 |
| Chlorambucil (NSC 3088) | Dacarbazine (NSC 45388) | All-t-retinoic acid (NSC 122758) |
| 1959 | 1976 | Porfimer Na (NSC 603062) |
| Cyclophosphamide (NSC 26271) | CCNU (NSC 9037) | 1996 |
| Thiotepa (NSC 6396) | 1977 | Gemcitabine (NSC 613327) |
| 1961 | BCNU (NSC 409962) | Gliadel (NSC 714372) |
| Vinblastine (NSC 49842) | 1978 | Irinotecan (NSC 616348) |
| 1962 | cis-Platinum (NSC 119875) | Taxotere (NSC 628503) |
| Uracil Mustard (NSC 34462) | 1979 | Topotecan (NSC 609699) |
| Fluorouracil (NSC 19893) | Daunomycin (NSC 82151) | 1998 |
| 1963 | Tamoxifen (NSC 180973) | Herceptin (NSC 688097) |
| Vincristine (NSC 67574) | 1982 | Ontak (NSC 697979) |
| 1964 | Streptozotocin (NSC 85998) | 2000 |
| Melphalan (NSC 8806) | 1983 | Arsenic Trioxide (NSC 706363) |
| Actinomycin D (NSC 3053) | Etoposide (NSC 141540) | Celebrex (NSC 719627) |
| 1966 | 1987 | 2001 |
| Pipobroman (NSC 25154) | Mitoxantrone (NSC 301739) | Gleevec (NSC 716051) |
| Thioguanine (NSC 752) | 1988 | 2003 |
| 1967 | Ifosfamide (NSC 109724) | Velcade (NSC 681239) |
| Hydroxyurea (NSC 32065) | 1989 | 2004 |
| 1969 | Carboplatin (NSC 241240) | Clolar (NSC 606869) |
| Ara-C (NSC 63878) | 1990 | Erbitux (NSC 632307) |
| Procarbazine (NSC 77213) | Hexamethylmelamine(NSC 13875) | |
| Idarubicin (NSC 256439) | ||
| Levamisole (NSC 177023) |
EMERGENCE OF CANCER METABOLISM
The deductive method in cancer biology
Current cancer treatments are highly inefficient and deliver benefits to an average of only 25% of patients (Spear et al., 2001). In addition, in economic terms, the conventional approach is unsustainable. Globally, annual spending on cancer drugs was approximately $49 billion in 2011, of which by inference, approximately $37 billion was spent to induce adverse effects without delivering patient benefits. This inefficient outcome has led to the hope of better outcomes with combination targeted therapies (Bernards, 2012). However, good outcomes cannot be fully expected.
Regardless of the many other scientific issues with combination therapy, drugs cause so many adverse effects that patients are unable to overcome drug toxicity and survive. This is a real and serious issue in clinical trials of multi-combination cancer therapies, thereby limiting the number of two- and three-way drug combinations that are practically feasible in the clinic while maintaining proper suppression of the intended drug targets. Furthermore, in combination therapies, drug dosages must be lowered to levels lower than those used for single therapies, which can result in poorer outcomes than those achieved with maximum dosages of single therapies. Whereas cancer research and therapy are restricted by the limits of the philosophic and scientific methods, three important discoveries have redirected cancer biology to “cancer metabolism.” Cancer metabolism appears to be a beacon of hope for cancer research because it demonstrates the possibility of overcoming all of the missing links in cancer therapy. Indeed, cancer metabolism is no longer considered a passive change in metabolic status but has instead achieved the status of a core hallmark of cancer (Ward and Thompson, 2012).
The first hope: targeting metabolism in a tumor model
In 2008, Dr. Pierre Sonveaux identified MCT1 as a component of the prominent lactate uptake pathway in a human cervical squamous carcinoma cell line that preferentially utilized lactate for oxidative metabolism. Inhibiting MCT1 in these cells using α-cyano-4-hydroxycinnamate (CHC) or siRNA induced a switch from lactate-fueled respiration to glycolysis (Sonveaux et al., 2008). A similar switch from lactate-fueled respiration to glycolysis was observed in oxygenated tumor cells in both a mouse model of lung carcinoma and xenotransplanted human colorectal adenocarcinoma cells after the administration of CHC. CHC retarded tumor growth because the hypoxic/glycolytic tumor cells died from glucose starvation and rendered the remaining cells sensitive to irradiation. Since then, MCT1 inhibitors (Sonveaux et al., 2012) or toxic molecules specifically transported by MCT1 (Birsoy et al., 2013) have been successful in many pre-clinical animal models. This is an outstanding success in cancer treatment with a focus on the “universality” concept rather than targeting of cancer driver genes.
The second hope: discovery of reduced risk of breast cancer with metformin
In 2009, Dr. Sao Jiralerspong reported that diabetic patients with breast cancer who were treated with metformin (anti-diabetic drug) and neoadjuvant chemotherapy had a higher pathologic complete response rate than did similar patients not treated with metformin (Jiralerspong et al., 2009). This result was consistent with epidemiologic data demonstrating that among diabetics, metformin use decreases both cancer incidence and mortality (Evans et al., 2005). This result was also consistent with the known inhibitory effect of metformin on growth in both cancer cell lines (Zakikhani et al., 2006) and animal tumor models (Ben Sahra et al., 2008). Although the effects of diabetes on breast cancer are complex and have been the subject of recent investigation, diabetes was found to be a risk factor for breast cancer. Therefore, metformin treatment is another successful case in which the “universality” of cancer was targeted. Additional studies to evaluate the potential of metformin as an antitumor agent are warranted.
The third hope: the major reason for mTOR inhibition failure, autophagy
mTOR was a much-researched cancer therapy target for more than a decade because mTOR is a serine/threonine protein kinase that regulates cell growth, proliferation, motility, and survival as well as protein synthesis and gene transcription (Hay and Sonenberg, 2004). mTOR belongs to the phosphatidylinositol 3-kinase (PI3K)-related kinase protein family. The antiproliferative effects of rapamycin may play a role in cancer treatment. A combination therapy involving doxorubicin and rapamycin was shown to drive AKT-positive lymphomas into remission in a mouse model (Chan, 2004). The mode of action of rapamycin, which is similar to that of tacrolimus, is binding of the cytosolic protein FK-binding protein 12 (FKBP12). However, unlike the tacrolimus-FKBP12 complex, which inhibits calcineurin, the sirolimus-FKBP12 complex inhibits the mTOR pathway by directly binding the mTOR complex 1 (mTORC1).
However, in 2012 Novartis announced unexpected results from a clinical trial of rapamycin. In particular, rapamycin failed in clinical trials. The major reason for this failure was autophagy induction induced by mTOR inhibition (Takeuchi et al., 2005). This finding directed attention to autophagy within the field of cancer research.
Researchers considered one major role of mTOR, specifically the induction of protein biosynthesis, including HIF-1α and PPAR-γ, which induce angiogenesis and lipogenesis respectively. However, another major role of mTOR, strong suppression of the autophagy process that drives biosynthetic pathways, was overlooked. Therefore, mTOR inhibition provoked the uncontrolled induction of autophagy, thus allowing cancer cells a survival opportunity. According to this finding, combining rapamycin treatment with autophagy inhibition may provide better clinical trial results.
FUTURE DIRECTIONS OF CANCER THERAPY
Multiple promising targets have been reported for cancer metabolism regulation. However, metabolic flux in cancer is not well understood. The greatest challenge to our understanding is that the metabolic switches in cancer cells differ from the set metabolic profiles of normal cells. Many leading scientists have attempted to identify the most efficient combinations of metabolic regulation in cancers. Current studies of cancer metabolism regulation have revealed general study directions, including combination therapy involving targeted drugs and drugs against bioenergetics, bio-anabolism, and autophagy. Other useful efforts may include the adoption of combination therapies directed at two bioenergetics targets such as hexokinase and mitochondrial complex I via treatment with 2-deoxyglucose and metformin. Additional evidence and clever combinations will be available in the very near future from powerful arsenals based on metabolic disease treatments.
Targeting bio-energetics
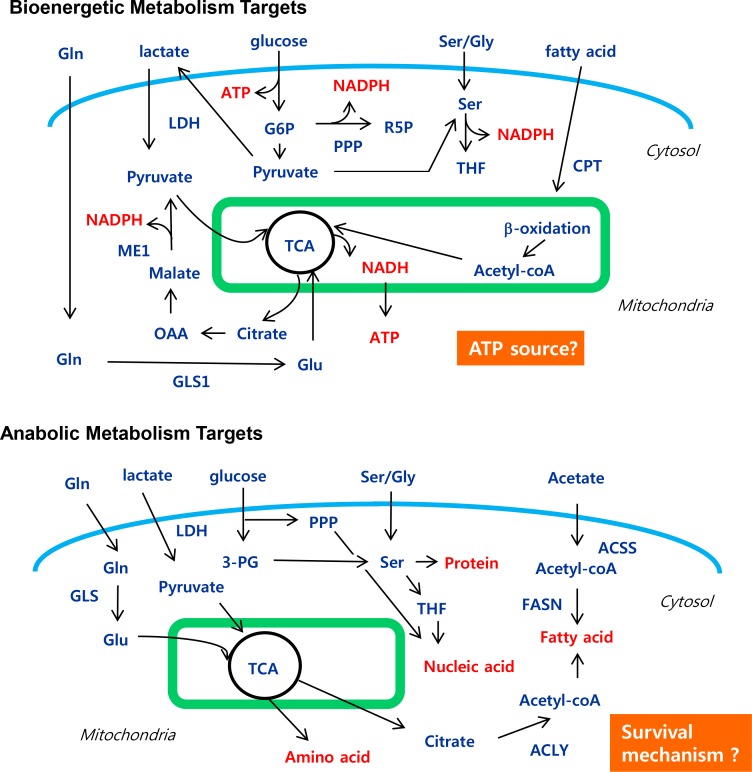
Rapidly growing cancer cells require abundant energy to support biosynthesis and motility (Fig. 3). Glucose metabolism represents a central resource of energy for the cell. Many enzymes contribute to the series of reactions necessary for the glycolytic breakdown of glucose (Table 3). Glycolytic inhibition in the context of the glycolytic pathway has been tested as an anticancer strategy, with an expansion of targets to include glucose transporters (GLUTs), hexokinase (Klionsky et al. 2012), pyruvate kinase M2 (PKM2), and lactate dehydrogenase (LDHA). Recent reviews have summarized clearly the role of each component of glycolysis, available inhibitors, and in vitro and in vivo results (Vander Heiden, 2011). Glucose metabolism is an essential central energetic resource for both normal and cancer cells, although cancer cells reroute anabolic metabolism via the Warburg effect. There are two strategic approaches to cancer growth inhibition. The first approach is the reduction of further energy production via the inhibition of mitochondrial activity using biguanide along with glycolysis inhibitors. This provided better outcomes both in vitro and in vivo in comparison with single chemotherapy. For example, pyruvate dehydrogenase kinase (PDK) inhibition using dichloroacetate (DCA) combined with metformin-based mitochondrial activity inhibition yielded a synergistic effect in breast cancer cells (Haugrud et al., 2014). PDK suppresses pyruvate dehydrogenase (PDH), an enzyme that converts pyruvate into acetyl-CoA. DCA inactivates PDK, leading to the reactivation of PDH and flipping a metabolic switch from glycolysis to mitochondrial respiration (Zhao et al., 2011). Preclinical trials of DCA have demonstrated its effectiveness in tumors via the induction of apoptosis (Michelakis et al., 2008). However, PDK inhibitor monotherapy is limited in ongoing clinical trials. DCA potentiates the anticancer effects of metformin via oxidative damage and attenuation of lactate production (Haugrud et al., 2014).
Fig. 3.
Multiple promising targets for regulating cancer metabolism. Targets for cancer metabolism are divided into two primary groups, including bio-catabolism (bio-energetics) and bio-anabolism. Although increased glycolysis, termed the Warburg effect, contributes to cancer growth through biomass production, the main energy source of cancer remains unknown. The inhibition of anabolism may induce cancer cell death. However, the mechanism of cell death is unclear. Notably, all targets are context-dependent and do not work for all cancers. Gln, glutamine; Glu, glutamate; LDH, lactate dehydrogenase; ME1, NADP-dependent malic enzyme; G6P, glucose-6-phosphate; R5P, ribose-5-phosphate; PPP, pentose phosphate pathway; Ser, serine; OAA, oxaloacetic acid; GLS1, kidney type glutaminase; TCA, tricarboxylic acid cycle, Kreb cycle; THF, tetrahydofolate; CPT, carnitine palmitoyltransferase; 3-PG, 3-phospho-glycerate; Gly, glycine; ACSS, acetyl CoA synthase; FASN, fatty acid synthase; ACLY, ATP-citrate lyase.
Table 3.
A list of therapeutic targets against cancer metabolism
| Targeting Bioenergetic Metabolism | Targeting Anabolic Metabolism | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Targets | Pathway | Agents or approaches (company)* | Targets | Pathway | Agents or approaches (company)* |
| CPT1 | B-oxidation | -Etomoxir | Choline | Lipid | -CK37 |
| -Oxfenicine | Kinase | Biosynthesis | -TCD-717 (TCD Pharma) | ||
| -Perhexiline | HMGCR | Mevalonate | -Statins | ||
| Complex l | Mitochondria | -Metformin | Pathwa | ||
| Respiration | -Phenformin | IDHs | Lipid | -AGI-5198 (Xcessbio) | |
| GLUT1 | Glycolysis | -WZB117 | Biosynthesis | -AGI-6780 (Xcessbio) | |
| GLS1 | Glutamine | -968 | MGLL | Lipid | -JZL184 |
| Metabolism | -BPTES | Biosynthesis | |||
| Hexokinases | Glycolysis | -2-DG | PGAM1 | Pentose phosphate pathway | -PGMI-004A |
| -3-BP | PKM2 | Pentose | -TEPP-46 | ||
| -Lonidamine | Phosphate | -SAICAR | |||
| -Methyl | Pathway | -Serine | |||
| -Jasmonate |
|
||||
| MCT1 | Kreb’s cycle | -AR-C155858 | Targeting Other Metabolism
|
||
| -AR-C117977 | Targets | Pathway | Agents or approaches | ||
| -AZD3965 (AstraZeneca) |
|
||||
| -CHC | HIF1 | Hypoxic | -Acriflavine | ||
| PDK1 | Kreb’s cycle | -DCA | Responses | -PX-478 | |
| PKM2 | Glycolysis | -TLN-232(Thallion) | mTOR | Cell growth autophagy | -Rapalogues |
| -Torins | |||||
| PTGS2 | Cell growth | -Aspirin | |||
| AMPK | Autophagy | ||||
However, with the exception of glucose, the energy sources of cancer cells were largely unclear. Under these circumstances, the glucose level within the tumor is very low because of the lack of blood supply. Therefore, other resources such as fatty acids and amino acids may provide important energy supplies to the rapidly growing cancer cells. In fact, deletion of the fatty acid binding protein 4 gene, which encodes a known fatty acid transporter, led to strong tumor growth suppression (Nieman et al., 2011). Fatty acid oxidation appears to be an efficient method for energy generation in cancer cells because glycolysis inefficiently produces small amounts of ATPs in comparison with fatty acid oxidation (glycolysis generates 2 ATPs from one glucose versus 132 ATPs from one palmitate). Glycolysis effectively supports the biosynthesis of building blocks instead of generating energy in cancers. The exact primary source of energy in cancer cells remains under study.
Targeting bio-anabolism
There are two viewpoints regarding the targeting of anabolism. The popular view states that the characteristic cancer phenotype is rapid growth that requires a continuous supply of biomass. Anabolic metabolism in cancer cells is coordinately increased to supply biomasses, including protein, lipids, and nucleotides. Therefore, blocking the supply of biomasses is a good strategy for regulating cancer metabolism. The other view is the unusual slow growth of tumors that depend on fatty acid synthesis via acetate uptake for survival, such as hepatocellular carcinoma. Cancer cell survival via acetate-dependent fatty acid synthesis can be reversed by inhibiting fatty acid synthesis. However, it is not clear how to induce apoptosis followed by fatty acid synthesis inhibition in this type of cancer cell (Yun et al., 2009).
Several metabolic pathways have been suggested as targets for the inhibition of rapidly growing cancer cells, including the pentose phosphate pathway (PPP), serine synthesis pathway, and glutaminolysis (Fig. 3). PPP generates ribose-5-phosphate (a precursor of purines and pyrimidines) and NADPH. In non-small-cell lung cancer, approximately 40% of NADPH synthesis is supplied by the PPP; the remainder might be supplied by folate metabolism via the serine synthesis pathway (Fig. 3) (Fan et al., 2014). Therefore, simultaneous inhibition of the PPP and serine synthesis pathway might be a good strategic approach to block bio-anabolism. Glutamine plays an important role in cancer cell anabolic metabolism. TCA cycle intermediates provide not only a source of energy but also building blocks such as lipids, carbohydrates, and amino acids. Owing a lack of pyruvate conversion into the TCA cycle, cancer cells may use glutamine as a carbon source for the TCA cycle in a process called glutaminolysis. Glutaminolysis involves two steps: the first is catalyzed by glutaminase (GLS) and converts glutamine to glutamate, and the second is catalyzed by glutamate dehydrogenase (GDH) and converts glutamate to α-ketoglutarate (α-KG). 13C-tracking metabolic flux experiments have shown that cancer cells exhibiting Warburg-like metabolism do not stop utilizing the TCA cycle; instead these cells begin to rely on glutamine as a carbon source for the TCA cycle (DeBerardinis et al., 2007). A GLS inhibitor, Bis-2-[5-phenylacetamido-1,2,4-thiadiazol-2-yl] ethyl sulfide (BPTES), was shown to reduce aerobic cell proliferation (Robinson et al., 2007). GLS inhibition via BPTES slowed the growth of glioblastoma cells harboring an isocitrate dehydrogenase 1 (IDH1) mutation (Seltzer et al., 2010). Furthermore, GLS activity inhibition via BPTES decreases the levels of glutamate and α-ketoglutarate (Lottmann et al., 2012), resulting in increased levels of glycolytic intermediates. This suggests that the simultaneous inhibition of GLS and glycolysis might be a better strategy for treating cancer patients who harbor a mutant form of IDH1.
Targeting autophagy
In normal tissues, starvation induces autophagy as a survival mechanism. Autophagy is a self-degradation process whereby cytosolic components and organelles are sequestered in autophagosomes and delivered to lysosomes for degradation and recycling. Interestingly, as mentioned earlier with regard to autophagy induction via mTOR inhibition, this phenomenon has also observed in cancer cells under treatment with tyrosine kinase inhibitors such as imatinib (Gleevec) (Ertmer et al., 2007) and erlotinib (Tarceva) (Gorzalczany et al., 2011). A similar induction of autophagy via anti-cancer drugs such as histone deacetylase inhibitors (SAHA) (Sooparb et al., 2004), and DNA alkylating agents (Kanzawa et al., 2004) has been reported to promote cancer cell survival. Therefore, combination therapy with autophagy inhibition appears necessary to improve the survival rate after conventional chemotherapy. A review article discussing autophagy as a cancer target introduced findings that autophagy inhibition exerts anti-cancer effects in both cancer cell lines and xenograft models (Janku et al., 2011). Chloroquine derivatives are the most commonly used autophagy inhibitors in clinical trials. Chloroquine, an anti-malarial drug, is known to inhibit lysosomal acidification and impair the fusion of autophagosomes with lysosomes and subsequent degradation (Kimura et al., 2013). In addition to chloroquine derivatives, the anti-tumor effects of various potential autophagy inhibitors, including 3-methyladenine, bafilomycin A1, and monensin, have been studied in vitro and in vivo (Cheong et al., 2012). Clinical trials of autophagy inhibition for cancer therapy are listed at http://www.clinicaltrial.gov.
CONCLUSION
We have reviewed the panorama of anti-cancer drug discovery in the past, present, and future. Our philosophy of the cancer process taught us that cancer cell metabolism might be affected by cancer driver gene-mediated cell signaling. Is cancer metabolism a passive environmental stress response that is unrelated to cell death or survival? Hypoxia induces hypoxia inducible factor-1alpha (HIF-1α) expression in normal cells. Without oxygen, normal cells that continue to respire via the mitochondrial oxidative phosphorylation system will be killed by an acute increase in the levels of oxygen free radicals generated by blocked mitochondrial electron transfer. Clearly, one duty of HIF-1α must be inhibiting the mitochondrial respiration system by inducing PDK1, which re-directs energy resources to glycolysis. The metabolic destiny appears to be determined immediately by the expression of the oncoprotein HIF-1α before cell signaling has fully adapted to the environment. HIF-1α may be induced constitutively in cancer cells under either hypoxic or normoxic conditions as a result of the three-dimensional tumor structure of tumor or mutation of a negative HIF-1α regulator such as Von Hippel-Lindau tumor suppressor or prolyl hydroxylase, which drives aerobic glycolysis for biomass production while intact mitochondria are present. Therefore, the cancer cell has a survival advantage resulting from increased glycolysis and lactate production and reduced ROS generation. DCA-mediated PDK inhibition showed good therapeutic potential (Michelakis et al., 2008). Importantly, this suggests that PDK inhibition or PDH induction suggests a good therapeutic approach, although PDK overexpression or PDH knockout is not sufficient for tumor induction. This is the beginning of a deductive science process because cancer biologists target common metabolic pathways used by both normal and cancer cells but on which only cancer cells depend universally. For another example, therapeutic inhibition of mutant IDH1 is being investigated in a clinical trial against glioblastoma multiforme (GBM). Inhibition of mutant IDH1 resulted in a huge increase in the survival of mice bearing mIDH1 tumors, although mIDH1 transgenic mice did not exhibit the same GBM phenotype. This result suggests that scientists identified a therapeutic approach to target one common metabolic signature pathway of GBM, rather than targeting innumerable cancer targets identified in a genomics study. Again, this is a deductive science method. In conclusion, a paradigm shift in cancer research has already begun, and I hope that we will obtain good results regarding therapeutic strategies for patients with resistant cancers.
Acknowledgments
This work was supported by a research grant from the National Cancer Center of Korea (NCC1410671-2).
REFERENCES
- Arduino LJ, Mellinger GT. Clinical trial of busulfan (NSC-750) in advanced carcinoma of prostate. Cancer Chemother Rep. 1967;51:295–303. [PubMed] [Google Scholar]
- Armour AA, Watkins CL. The challenge of targeting EGFR: experience with gefitinib in nonsmall cell lung cancer. Eur Respir Rev. 2010;19:186–196. doi: 10.1183/09059180.00005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Sahra I, Laurent K, Loubat A, Giorgetti-Peraldi S, Colosetti P, Auberger P, Tanti JF, Le Marchand-Brustel Y, Bost F. The antidiabetic drug metformin exerts an antitumoral effect in vitro and in vivo through a decrease of cyclin D1 level. Oncogene. 2008;27:3576–3586. doi: 10.1038/sj.onc.1211024. [DOI] [PubMed] [Google Scholar]
- Bernards R. A missing link in genotype-directed cancer therapy. Cell. 2012;151:465–468. doi: 10.1016/j.cell.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Possemato R, Yilmaz OH, Koch CE, Chen WW, Hutchins AW, Gultekin Y, Peterson TR, Carette JE, Brummelkamp TR, Clish CB, Sabatini DM. MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat Genet. 2013;45:104–108. doi: 10.1038/ng.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook J, Bateman JR, Steinfeld JL. Evaluation of Melphalan (Nsc-8806) in Treatment of Multiple Myeloma. Cancer Chemother Rep. 1964;36:25–34. [PubMed] [Google Scholar]
- Buck E, Eyzaguirre A, Haley JD, Gibson NW, Cagnoni P, Iwata KK. Inactivation of Akt by the epidermal growth factor receptor inhibitor erlotinib is mediated by HER-3 in pancreatic and colorectal tumor cell lines and contributes to erlotinib sensitivity. Mol Cancer Ther. 2006;5:2051–2059. doi: 10.1158/1535-7163.MCT-06-0007. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabner BA, Roberts TG., Jr Timeline: Chemotherapy and the war on cancer. Nat. Rev. Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- Chan S. Targeting the mammalian target of rapamycin (mTOR): a new approach to treating cancer. Br. J. Cancer. 2004;91:1420–1424. doi: 10.1038/sj.bjc.6602162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson JH, Lagakos S, Stolbach L, Perlia CP, Bennett JM, Mansour EG, Horton J, Regelson W, Cummings FJ, Israel L, Brodsky I, Shnider BI, Creech R, Carbone PP. Mechlorethamine (NSC-762) plus CCNU (NSC-79037) in the treatment of inoperable squamous and large cell carcinoma of the lung. Cancer Treat Rep. 1976;60:625–627. [PubMed] [Google Scholar]
- Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, Elsasser HP, Schatzl HM. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–942. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausel C. Targeted chronic myeloid leukemia therapy: Seeking a cure. Am J Health Syst Pharm. 2007;64:S9–15. doi: 10.2146/ajhp070482. [DOI] [PubMed] [Google Scholar]
- Fisher BK, Elliott GB. Triple drug therapy with actinomycin D (Nsc-3053), chlorambucil (Nsc-3088), and methotrexate (Nsc-740) in metastatic solid tumors in children. Cancer Chemother Rep. 1965;45:45–51. [PubMed] [Google Scholar]
- Foley JF, Kennedy BJ. Effect of cyclophosphamide (Nsc-26271) on far-advanced neoplasia. Cancer Chemother Rep. 1964;34:55–58. [PubMed] [Google Scholar]
- Gilman A. The initial clinical trial of nitrogen mustard. Am J Surg. 1963;105:574–578. doi: 10.1016/0002-9610(63)90232-0. [DOI] [PubMed] [Google Scholar]
- Goldman JM, Melo JV. Chronic myeloid leukemia--advances in biology and new approaches to treatment. N Engl J Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- Gorzalczany Y, Gilad Y, Amihai D, Hammel I, Sagi-Eisenberg R, Merimsky O. Combining an EGFR directed tyrosine kinase inhibitor with autophagy-inducing drugs: a beneficial strategy to combat non-small cell lung cancer. Cancer Lett. 2011;310:207–215. doi: 10.1016/j.canlet.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Haugrud AB, Zhuang Y, Coppock JD, Miskimins WK. Dichloroacetate enhances apoptotic cell death via oxidative damage and attenuates lactate production in metformin-treated breast cancer cells. Breast Cancer Res Treat. 2014;147:539–550. doi: 10.1007/s10549-014-3128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Jackson RC. Unresolved issues in the biochemical pharmacology of antifolates. NCI Monogr. 1987:9–15. [PubMed] [Google Scholar]
- Jacobs EM, Peters FC, Luce JK, Zippin C, Wood DA. Mechlorethamine HCl and cyclophosphamide in the treatment of Hodgkin’s disease and the lymphomas. JAMA. 1968;203:392–398. doi: 10.1001/jama.1968.03140060016005. [DOI] [PubMed] [Google Scholar]
- Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol. 2009;27:3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73:3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lottmann HB, Margaryan M, Bernuy M, Rouffet MJ, Bau MO, El-Ghoneimi A, Aigrain Y, Stenberg A, Lackgren G. The effect of endoscopic injections of dextranomer based implants on continence and bladder capacity: a prospective study of 31 patients. J Urol. 2002;168:1863–1867. doi: 10.1016/S0022-5347(05)64431-X. [DOI] [PubMed] [Google Scholar]
- Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br. J. Cancer. 2008;99:989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, Lengyel E. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelson W, Holland JF, Frei E, 3rd, Gold GL, Hall T, Krant M, Miller SO. Comparative clinical toxicity of 6-mercaptopurine (Nsc-755)-1 and 6-mercaptopurine ribonucleoside (Nsc-4911)-2 administered intravenously to patients with advanced cancer. Cancer Chemother Rep. 1964;36:41–48. [PubMed] [Google Scholar]
- Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, Hansen JC, Curthoys NP. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausville EA, Johnson JI. Molecules for the millennium: how will they look? New drug discovery year 2000. Br. J. Cancer. 2000;83:1401–1404. doi: 10.1054/bjoc.2000.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, Dang CV, Riggins GJ. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Copetti T, De Saedeleer CJ, Vegran F, Verrax J, Kennedy KM, Moon EJ, Dhup S, Danhier P, Frerart F, Gallez B, Ribeiro A, Michiels C, Dewhirst MW, Feron O. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PloS One. 2012;7:e33418. doi: 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, Kelley MJ, Gallez B, Wahl ML, Feron O, Dewhirst MW. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. doi: 10.1016/S1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Warmuth M, Sellers WR, Dorsch M. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin Pharmacol Ther. 2010;87:543–552. doi: 10.1038/clpt.2009.297. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Simpson AJ, Old LJ, Riggins GJ. Oncogenomics and the development of new cancer therapies. Nature. 2004;429:469–474. doi: 10.1038/nature02627. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature reviews. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7:3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PubMed] [Google Scholar]
- Yun M, Bang SH, Kim JW, Park JY, Kim KS, Lee JD. The importance of acetyl coenzyme A synthetase for 11C-acetate uptake and cell survival in hepatocellular carcinoma. J Nucl Med. 2009;50:1222–1228. doi: 10.2967/jnumed.109.062703. [DOI] [PubMed] [Google Scholar]
- Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–10273. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Liu H, Riker AI, Fodstad O, Ledoux SP, Wilson GL, Tan M. Emerging metabolic targets in cancer therapy. Front Biosci. 2011;16:1844–1860. doi: 10.2741/3826. [DOI] [PMC free article] [PubMed] [Google Scholar]