Jay and colleagues show that TREM2 deficiency reduces the number of macrophages infiltrating the brain and is protective against disease pathogenesis in mouse models of Alzheimer’s disease.
Abstract
Variants in triggering receptor expressed on myeloid cells 2 (TREM2) confer high risk for Alzheimer’s disease (AD) and other neurodegenerative diseases. However, the cell types and mechanisms underlying TREM2’s involvement in neurodegeneration remain to be established. Here, we report that TREM2 is up-regulated on myeloid cells surrounding amyloid deposits in AD mouse models and human AD tissue. TREM2 was detected on CD45hiLy6C+ myeloid cells, but not on P2RY12+ parenchymal microglia. In AD mice deficient for TREM2, the CD45hiLy6C+ macrophages are virtually eliminated, resulting in reduced inflammation and ameliorated amyloid and tau pathologies. These data suggest a functionally important role for TREM2+ macrophages in AD pathogenesis and an unexpected, detrimental role of TREM2 in AD pathology. These findings have direct implications for future development of TREM2-targeted therapeutics.
It has historically been debated whether inflammation plays an active role in Alzheimer’s disease (AD) pathogenesis or is ancillary to other AD pathologies (Wyss-Coray, 2006). However, recent genome-wide association studies linked polymorphisms in inflammation-related genes to increased AD risk, supporting the conclusion that inflammation can be a causative factor in disease pathology (Karch and Goate, 2015). Notably, variants of the immune cell–specific triggering receptor expressed on myeloid cells 2 (TREM2) confer dramatically elevated risk for developing AD (R. Guerreiro et al., 2013; Jonsson et al., 2013). TREM2 variants are also the genetic basis of Nasu-Hakola disease (Bird et al., 1983) and confer increased risk for frontotemporal dementia (FTD; R.J. Guerreiro et al., 2013), Parkinson’s disease (Rayaprolu et al., 2013), and amyotrophic lateral sclerosis (Cady et al., 2014). These data suggest that TREM2 may serve a common function that modifies risk for neurodegenerative disorders.
TREM2 is an important modulator of immune cell function. In the brain, the TREM2 receptor is expressed exclusively by myeloid cells (Colonna, 2003). TREM2, along with its obligate intracellular adaptor DAP12, was identified as a hub gene in systems biology analyses in AD (Forabosco et al., 2013; Zhang et al., 2013), highlighting the central importance of these myeloid cell signaling elements in neurodegeneration. In vitro, TREM2 promotes phagocytosis, suppresses toll-like receptor–induced inflammatory cytokine production and enhances antiinflammatory cytokine transcription (Neumann and Takahashi, 2007; Paradowska-Gorycka and Jurkowska, 2013). In contrast, Trem2−/− mice have less inflammation and enhanced phagocytosis in models of stroke and lung infection (Sieber et al., 2013; Sharif et al., 2014). These findings suggest that the role of TREM2 in modulating inflammation may be more complex than previously appreciated and may be dependent on the cell type in which it is expressed and the inflammatory context in which it is studied. In the AD brain, the role of TREM2 is poorly understood. In this study, we identify disease-relevant cell types that express TREM2 and examine the effects of loss of TREM2 function on AD pathologies.
RESULTS AND DISCUSSION
TREM2-expressing myeloid cells associate with β amyloid (Aβ) deposits
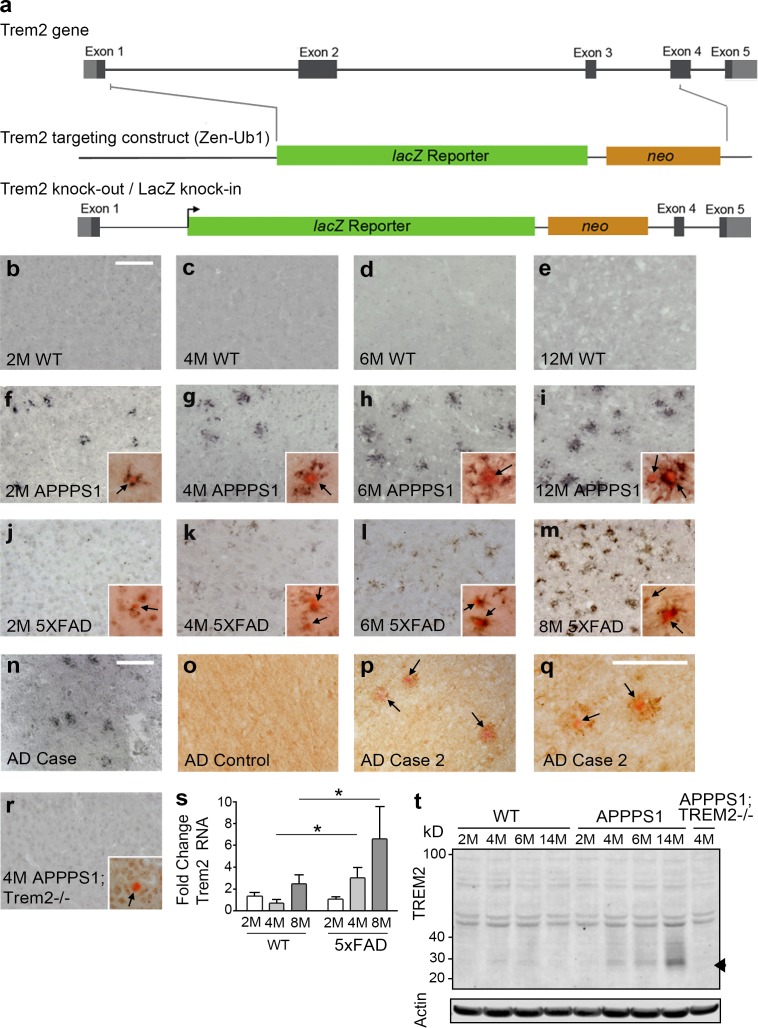
We defined the cellular localization and kinetics of TREM2 expression in tissues from two transgenic AD mouse models that exhibit age-related Aβ plaque deposition. TREM2 immunohistochemistry (IHC) revealed an age-dependent increase in TREM2-expressing cells around Congo red–positive plaques in both APPPS1 and 5XFAD mice (Fig. 1, f–m). We observed similar expression patterns in two neuropathologically confirmed AD cases (Fig. 1, n–q). Despite reports of TREM2 expression in microglia (Frank et al., 2008; Hickman et al., 2013), we did not detect TREM2 staining in nontransgenic mice (Fig. 1, b–e) or in nonplaque-associated cells (Fig. 1 o and see Fig. 4 l). Thus, TREM2 is expressed in parenchymal microglia at levels below the limit of detection using IHC. TREM2 antibody specificity was confirmed in APPPS1;Trem2−/− mice (Fig. 1 r). TREM2 RNA (Fig. 1 s) and protein (Fig. 1 t) analyses confirmed an age-dependent increase in TREM2 expression in the brains of AD mice.
Figure 1.
TREM2 expression is increased around Aβ plaques. (a) A novel Trem2 knockout with a LacZ reporter under the control of the Trem2 promoter was generated. (b–m) TREM2 IHC was performed in WT mice at 2 (b), 4 (c), 6 (d), and 12 mo (e) of age, in APPPS1 mice at 2 (f), 4 (g), 6 (h), and 12 mo (i) of age, and in 5XFAD mice at 2 (j), 4 (k), 6 (l), and 8 mo (m) of age (n = 4–5). Insets show Congo red costaining with arrows indicating plaques. (n–q) TREM2 was also observed around plaques (n, p, and q) but not in regions lacking Aβ deposition (o) in human AD cases (n = 2). Arrows point to Congo red–positive plaques. (r) TREM2 immunoreactivity was not observed in APPPS1;Trem2−/− mice (n = 7). Insets show Congo red costaining with arrows indicating plaques. (s and t) TREM2 expression was also assessed by qRT-PCR (s; two-way ANOVA, age P = 0.083, genotype P = 0.0036, interaction P = 0.185; Bonferroni-corrected Student’s t tests shown; n = 6–8 per group) and Western blot (t; n = 2–3). Arrowhead indicates position of TREM2-specific band. Error bars indicate SEM. *, P < 0.05. At least three independent experiments were performed for all analyses. Bars, 100 µm (bars in b and n apply to b–p and r).
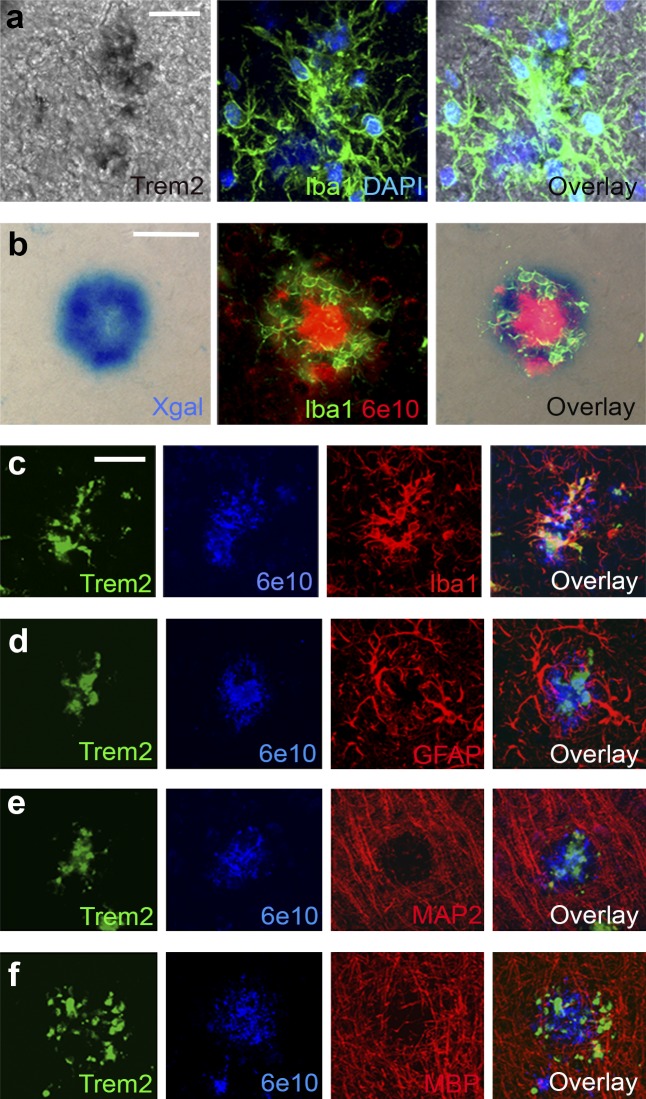
We next wanted to assess the cellular localization of TREM2. We demonstrated that TREM2 transcripts colocalized with plaque-associated Iba1+ myeloid cells in APP/PS1 mice using in situ hybridization (Fig. 2 a). This was confirmed in mice in which exons 2–4 of the Trem2 gene were replaced with LacZ (Fig. 1 a) as a reporter for TREM2 gene expression (Fig. 1 a). X-gal staining in APPPS1;Trem2LacZ/+ mice visualized LacZ expression in Iba1+ cells around Aβ deposits (Fig. 2 b). Triple fluorescent IHC confirmed that TREM2 protein colocalized with plaque-associated Iba1+ cells (Fig. 2 c), but not with markers of reactive astrocytes (GFAP, Fig. 2 d), neurons (MAP2, Fig. 2 e), oligodendrocytes (MBP, Fig. 2 f), or with parenchymal Iba1+ cells that were not associated with plaques (see Fig. 4 l). Collectively, these results demonstrate that TREM2 is selectively up-regulated by myeloid cells surrounding Aβ deposits.
Figure 2.
TREM2 is expressed in plaque-associated myeloid cells. (a) In situ hybridization with TREM2 probes colocalized with Iba1 (n = 2). (b) X-gal staining of brain tissue from 4-mo-old APPPS1;Trem2LacZ/+ mice colocalized with fluorescent IHC for Iba1 and 6E10 (n = 3). (c–f) Confocal microscopy was used to assess TREM2 colocalization with 6E10+ plaque-associated myeloid cells (c; Iba1), astrocytes (d; GFAP), neurons (e; MAP2), or oligodendrocytes (f; MBP; n = 8). At least two independent experiments were performed for all analyses. Bars: (a) 20 µm; (b–f) 50 µm.
TREM2+ plaque–associated myeloid cells express markers characteristic of monocyte-derived macrophages
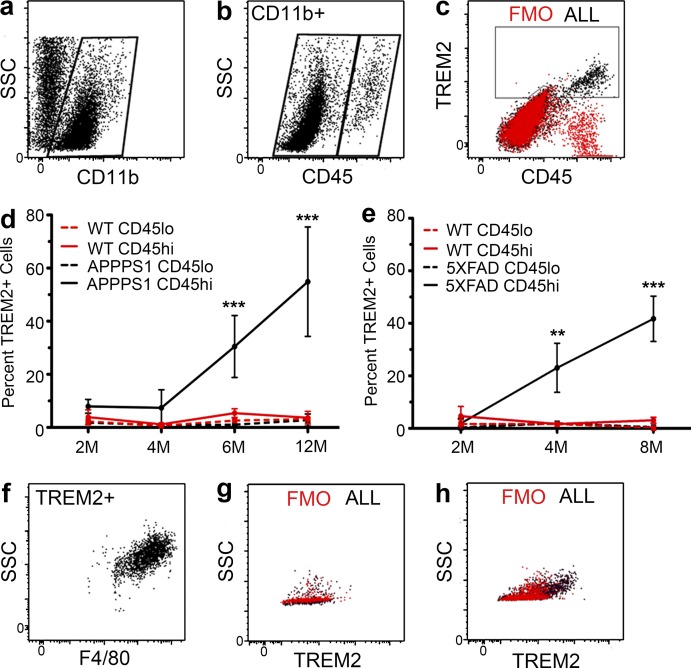
The identity of TREM2-expressing cells in AD mouse models was further examined using flow cytometry. The myeloid cell population was selected using CD11b (Fig. 3 a). Although there is no universally accepted marker to definitively distinguish macrophages derived from microglia and those derived from infiltrating monocytes, differences in levels of CD45 expression have been used extensively in flow cytometry to distinguish these two cell populations (Sedgwick et al., 1991; Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014). In these studies, CD45hi was used to identify cells that originate from bone marrow–derived monocytes, whereas CD45lo identified resident microglia. We found that CD11b+TREM2+ cells were exclusively CD45hi (Fig. 3 c). There was also a striking age-dependent increase in the percentage of brain TREM2+CD11b+CD45hi cells in two different AD mouse models (Fig. 3, d and e), whereas TREM2 expression by CD11b+CD45lo cells did not differ from that seen in wild-type mice at any time point. TREM2+ cells were also uniformly F4/80+, confirming their macrophage lineage (Fig. 3 f). TREM2 antibody specificity was validated in APPPS1;Trem2−/− mice (Fig. 3, g and h). Collectively, these results show that the TREM2+ macrophages that surround the Aβ deposits in the transgenic mouse models of AD are CD45hi, a canonical marker of peripherally derived macrophages.
Figure 3.
TREM2 is specifically expressed on CD11b+CD45hiF4/80+ macrophages in AD mice. (a and b) Isolated brain myeloid cells were gated on CD11b (a) and divided into CD45lo and CD45hi populations (b). (c and f) TREM2+ cells were exclusively CD45hi (c) and F4/80+ (f; n = 41). (d and e) TREM2 expression was quantified on CD45lo and CD45hi populations in APPPS1 mice (d; two-way ANOVA, age P < 0.0001; genotype/cell type P < 0.0001, interaction P < 0.0001; Bonferroni-corrected Student’s t tests shown; n = 2–8), 5XFAD mice (e; two-way ANOVA, age P = 0.025, genotype/cell type P < 0.0001, interaction P = 0.0003; Bonferroni-corrected Student’s t tests shown; n = 5–9), and WT controls (d and e; n = 4–14). Error bars indicate SEM. **, P < 0.01; ***, P < 0.001. (g and h) Flow cytometry on APPPS1;Trem2−/− mice (g) revealed a lack of TREM2+ cells compared with APPPS1;Trem2+/+ mice (h; n = 7). At least two independent experiments were performed for all analyses.
TREM2-deficient mice have reduced Aβ plaque–associated macrophages
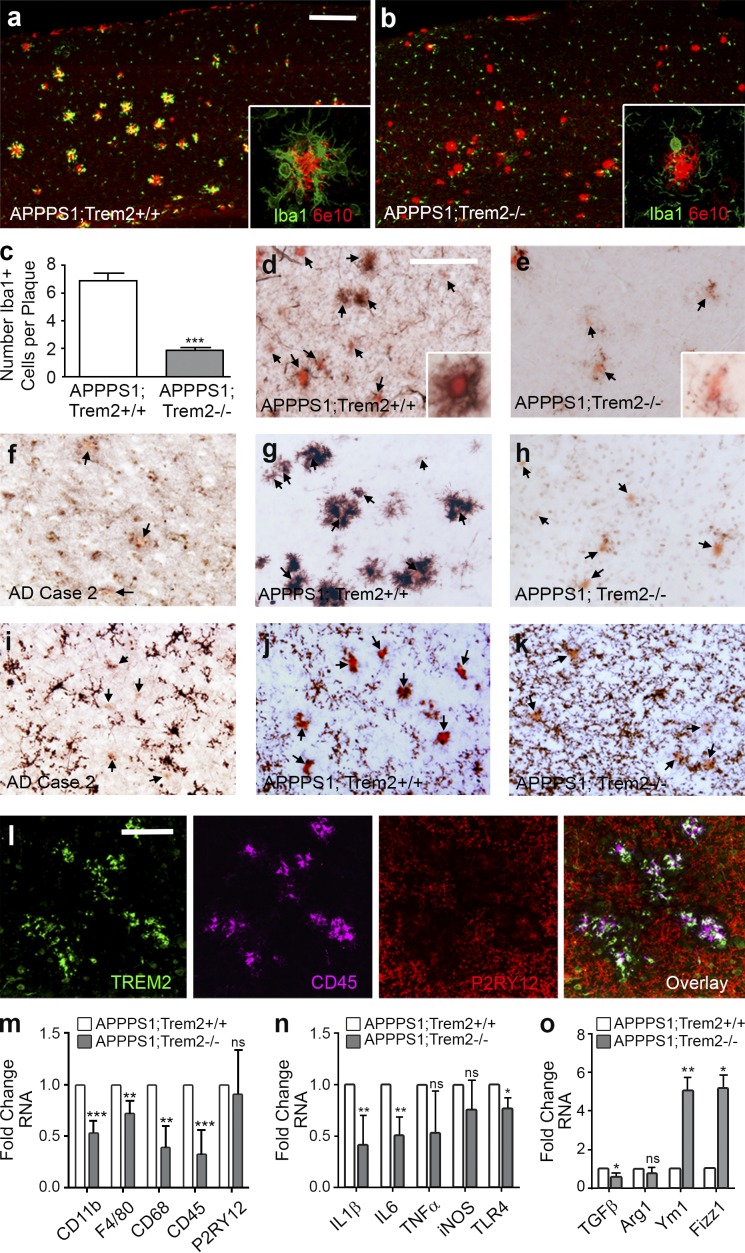
To determine the role of TREM2 in AD-like pathology, APPPS1 mice were crossed with Trem2−/− mice (Fig. 1 a). Although APPPS1;Trem2+/+ mice exhibit robust accumulation of Iba1+ myeloid cells around plaques (Fig. 4 a), APPPS1;Trem2−/− mice have a fivefold decrease in plaque-associated Iba1+ cells (Fig. 4, b and c), approximately twice the effect size previously observed in APPPS1;Trem2+/− mice (Ulrich et al., 2014). Cells expressing high levels of the peripheral monocyte marker Ly6C (Fig. 4 d) and CD45 (Fig. 4 g) are exclusively associated with Congo red–positive plaques in AD mouse models and in human AD tissue (Fig. 4 f), whereas TREM2-deficient animals revealed a near absence of plaque-associated CD45+Ly6C+ macrophages (Fig. 4, e and h). Consistent with these data, transcript levels of the myeloid cell markers CD11b, CD68, F4/80, and CD45 were all significantly decreased in brain lysates from APPPS1;Trem2−/− animals compared with APPPS1;Trem2+/+ controls (Fig. 4 m). Interestingly, P2RY12, a purinergic receptor selectively expressed in microglia (Hickman et al., 2013; Butovsky et al., 2014), was expressed by parenchymal, nonplaque-associated cells in APPPS1;Trem2+/+ mice (Fig. 4 j) and in human AD tissue (Fig. 4 i), in contrast to CD45 and TREM2 (Fig. 4 l). Unlike the CD45+Ly6C+ cells, the P2RY12+ cells were not eliminated in TREM2-deficient mice (Fig. 4 k), and there was no change in P2RY12 RNA in APPPS1;Trem2−/− mice compared with APPPS1;Trem2+/+ controls (Fig. 4 m). Collectively, these results demonstrate a dramatic reduction in Aβ plaque–associated CD45+Ly6C+ macrophages, but not in parenchymal P2RY12+ microglia in TREM2-deficient mice.
Figure 4.
Plaque-associated myeloid cells are reduced in TREM2-deficient mice. (a–c) Confocal microscopy was used to examine Iba1 and 6E10 expression in 4-mo-old APPPS1;Trem2−/− mice (b) and APPPS1;Trem2+/+ controls (a; quantified in c). (d and e) IHC and Congo red costaining (arrows) was performed for Ly6C in APPPS1;Trem2+/+ mice (d) and APPPS1;Trem2−/− animals (e). (a, b, d, and e) Insets show higher-magnification images. (f–h) CD45 and Congo red staining (arrows) was performed in human AD tissue (f; n = 2), APPPS1;Trem2+/+ mice (g), and APPPS1;Trem2−/− mice (h). (i–k) P2RY12 and Congo red staining (arrows) was performed in human AD tissue (i; n = 2), APPPS1;Trem2+/+ mice (j), and APPPS1;Trem2−/− animals (k). (l) TREM2 colocalized with CD45 but not P2RY12. (m–o) qRT-PCR was performed on whole brain lysates to examine transcript levels of myeloid cell markers (m), proinflammatory cytokines (n), and antiinflammatory markers (o). All experiments used n = 7–8 mice per group unless otherwise noted, and at least two independent experiments were performed for all analyses. Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars: (a and b) 50 µm; (d–l) 20 µm.
TREM2 deficiency reduces neuroinflammation
Because plaque-associated myeloid cells have been shown to play an important role in modulating neuroinflammation in AD (Akiyama et al., 2000), we assessed inflammatory markers in APPPS1 mice lacking TREM2. Transcript levels of inflammatory cytokines IL-1β and IL-6 were reduced (Fig. 4 n), whereas antiinflammatory markers chitinase-like 3/Ym1 and resistin B–like/Fizz1 were substantially increased in TREM2-deficient animals (Fig. 4 o). These data show that TREM2 deficiency reduces transcripts for two cardinal inflammatory mediators and increases transcripts for selected antiinflammatory markers.
TREM2 deficiency reduces hippocampal Aβ deposition
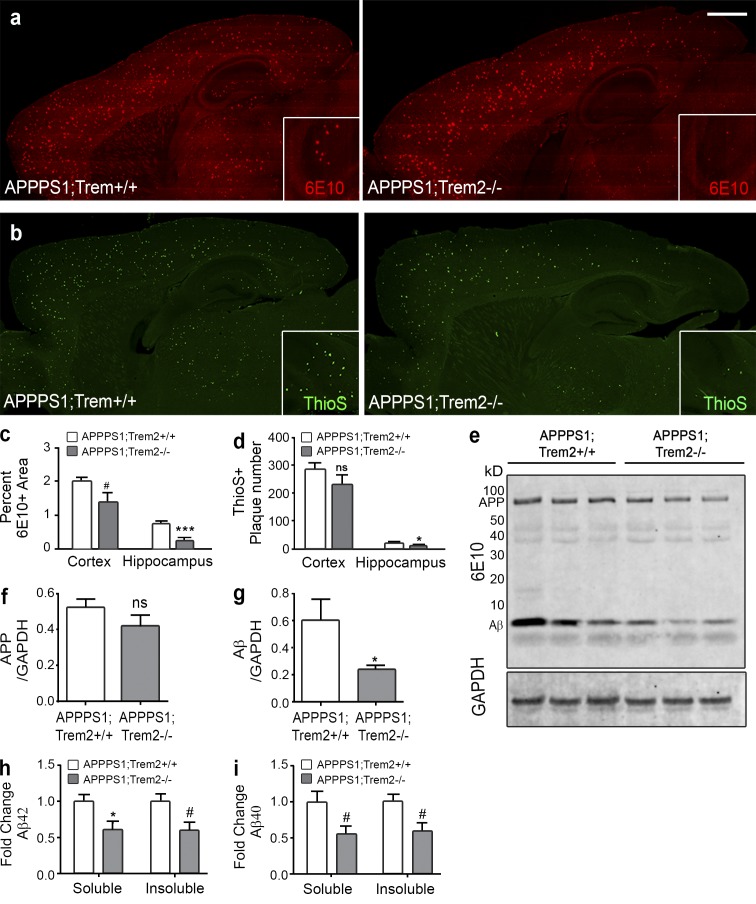
Given that TREM2 deficiency had robust effects on macrophage accumulation and neuroinflammation, factors associated with Aβ deposition, we assessed resultant changes in amyloid pathology in these mice. At 4 mo of age, APPPS1;Trem2−/− mice displayed significantly reduced 6E10 area in the hippocampus (Fig. 5, a and c), as well as other brain regions including the olfactory bulb and thalamus (not depicted), compared with APPPS1;Trem2+/+ controls. Interestingly, the effect of TREM2 deficiency on amyloid accumulation was not consistent across brain regions, as we observed only modest, nonsignificant effects in the cortex (Fig. 5, a and c) and brainstem (not depicted). Thioflavin S staining of dense core plaques revealed similar results (Fig. 5, b and d). Although the mechanism underlying this region specificity requires further investigation, similar effects have been observed in other contexts (Riddell et al., 2007). The histological results were confirmed by Western blot analysis (Fig. 5 e), which indicated no significant changes in APP (Fig. 5 f) but lower Aβ levels in APPPS1;Trem2−/− mice (Fig. 5 g). To assess which Aβ species were affected by TREM2 deficiency, we performed ELISAs on brain lysates. These results demonstrated reduced levels of insoluble Aβ42 (Fig. 5 h) and trends toward a reduction in soluble Aβ42 (Fig. 5 h) and soluble and insoluble Aβ40 (Fig. 5 i) in APPPS1;Trem2−/− mice compared with APPPS1;Trem2+/+ mice. These data demonstrate that TREM2 deficiency ameliorates amyloid pathology in a region-specific manner.
Figure 5.
TREM2 deficiency reduces Aβ accumulation. (a) IHC with 6E10 was performed on brain slices from 4-mo-old APPPS1;Trem2+/+ and APPPS1;Trem2−/− mice (quantified in c; n = 7–8). (b) Analysis of ThioS plaque number revealed similar results (quantified in d; n = 7–8). (a and b) Insets show a higher magnification of the hippocampus. Bar, 1 mm. (e) APP and Aβ levels were assessed by Western blot using 6E10. (f and g) Quantified relative to GAPDH, there was no significant change in APP protein levels (f) but a significant reduction in Aβ (g) in APPPS1;Trem2−/− mice (n = 3–4). (h and i) ELISAs on brain lysates also showed a significant reduction in insoluble Aβ42 and a trend toward a reduction in soluble Aβ42 (h) and a trend toward a reduction in soluble and insoluble Aβ40 (i; n = 3–4). #, P < 0.10; *, P < 0.05; ***, P < 0.001. At least two independent experiments were performed for all analyses.
TREM2 deficiency reduces astrocytosis and microtubule-associated protein τ (MAPT) pathology
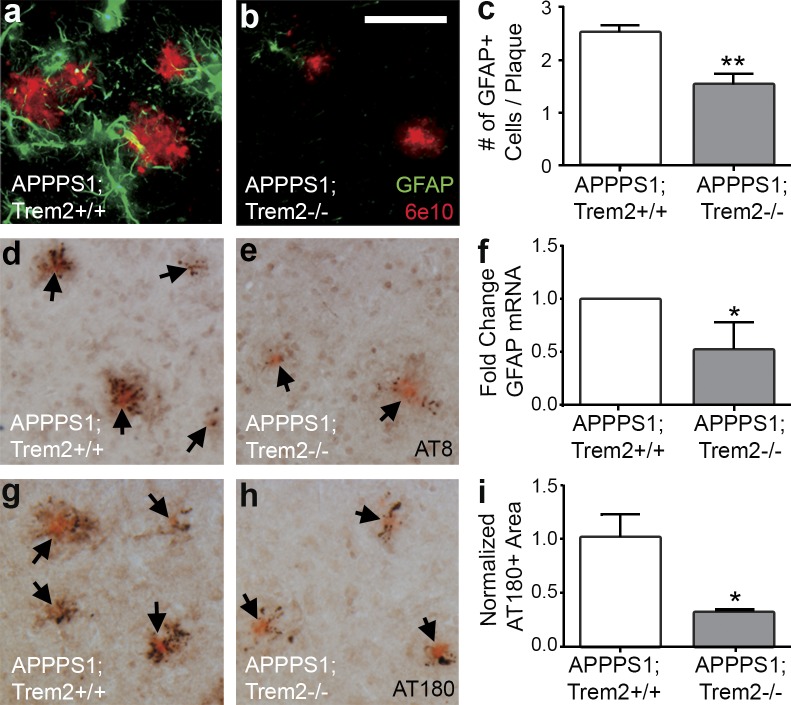
In addition to amyloid deposition, APPPS1 mice also exhibit astrocytosis and accumulation of hyperphosphorylated MAPT in dystrophic neurites around amyloid plaques. We examined the effects of TREM2 deficiency on these pathological hallmarks. TREM2-deficient APPPS1 mice had decreased numbers of GFAP immunoreactive astrocytes around Aβ deposits (Fig. 6, a–c) and a corresponding decrease in GFAP mRNA (Fig. 6 f). Additionally, there was a dramatic reduction in phosphorylated MAPT staining associated with Aβ deposits in APPPS1;Trem2−/− mice when compared with APPPS1;Trem2+/+ controls, as detected with AT8 (Fig. 6, d and e) and AT180 (Fig. 6, g–i) antibodies. Further investigation will be required to determine whether these changes in MAPT phosphorylation are a result of reduced amyloid deposition or whether they are regulated independently in this context. However, TREM2 deficiency not only ameliorated amyloid pathology but also reduced reactive astrocyte accumulation and MAPT hyperphosphorylation.
Figure 6.
TREM2 deficiency reduces astrocytosis and MAPT phosphorylation. (a and b) Astrocytosis was assessed in 4-mo-old APPPS1;Trem2+/+ (a) and APPPS1;Trem2−/− mice (b) using IHC for GFAP and 6E10 (n = 7–8). (c and f) The number of GFAP+ cells surrounding plaques was quantified (c; n = 3–4), and results were confirmed by qRT-PCR (f; n = 7–8). (d, e, g, and h) Hyperphosphorylated MAPT was detected in APPPS1;Trem2−/− (e and h) and APPPS1;Trem2+/+ mice (d and g) with AT8 (d and e) and AT180 antibodies (g and h; n = 7–8). Arrows indicate Congo red–positive plaques. (i) Quantification of the area of AT180 immunoreactivity revealed significant decreases in APPPS1;Trem2−/− mice (n = 3–4). At least two independent experiments were performed for all analyses. Error bars indicate SEM. *, P < 0.05; **, P < 0.01. Bar, 50 µm.
In this study, we demonstrated several surprising results regarding the expression and function of TREM2 in AD. First, we found that TREM2+ cells in AD mouse models expressed high levels of CD45 and Ly6C, canonical markers of macrophages derived from peripheral monocytes. The potential role of peripherally derived macrophages in AD has been controversial (Prinz and Priller, 2014), but it is well accepted that CD45hiLy6C+CCR2+ monocytes enter the central nervous system (CNS) and modulate pathology in other disease contexts (Mildner et al., 2011). Although our experiments identify a marker signature on TREM2+ cells consistent with this peripherally derived population, further investigation will be required to conclusively demonstrate the functional relevance of peripherally derived cells in AD and the ontogeny of the TREM2+ cell population in the AD brain.
If TREM2+ cells are derived from the periphery, as their marker expression would suggest, then TREM2 deficiency could result in the striking reduction in macrophage accumulation observed in this study through several mechanisms: impaired transmigration of TREM2-negative cells across the blood–brain barrier or alterations directly to the brain vasculature that alter cell trafficking, reduced chemotaxis of TREM2-deficient cells to parenchymal Aβ deposits, or shortened survival of these cells within the CNS. An alternative interpretation of the data in this study is that TREM2 is necessary for large-scale changes in microglial phenotype, including the up-regulation of CD45 and Ly6C and coincident down-regulation of P2RY12. In this case, TREM2 deficiency could reduce macrophage accumulation by eliminating microglial recognition of Aβ as a relevant stimulus, impairing migration of microglia to plaques, or preventing these phenotypic changes. Additional studies will be required to determine which mechanisms are responsible for the changes in TREM2-deficient AD mice in the current study.
Regardless of mechanism, our results demonstrate that TREM2 deficiency is protective against disease pathogenesis in AD mouse models. These results are surprising given similar findings in AD mice in which TREM2 was overexpressed (Jiang et al., 2014), although in this study TREM2 expression was not restricted to the cell types in which TREM2 is expressed physiologically. Our findings are also counterintuitive based on the human genetic data that demonstrate that presumed loss-of-function variants in TREM2 promote AD pathogenesis. TREM2 variants such as the missense mutation Q33X (R. Guerreiro et al., 2013) and the FTD-related variants T66M and Y38C which impair protein maturation (Kleinberger et al., 2014), almost certainly impair TREM2 function. However, the primary AD risk allele, R47H, harbors a change within the putative ligand-binding domain of TREM2, and alterations of its surface trafficking and function were less dramatic than the FTD variants (Kleinberger et al., 2014). Thus, it is possible that this variant could confer both loss- and gain-of-function phenotypes, which would be consistent with our findings. Going forward, it will be important to generate mice with knock-in TREM2 risk alleles to asses this possibility.
Although TREM2 has perhaps received the most attention for the high risk it confers for developing AD, TREM2 variants also confer risk for developing other neurodegenerative diseases, including FTD, amyotrophic lateral sclerosis, and Parkinson’s disease (R.J. Guerreiro et al., 2013; Rayaprolu et al., 2013; Borroni et al., 2014; Cady et al., 2014). Thus, it is plausible that TREM2 plays a common role in modifying risk for developing these diverse CNS pathologies. It will be important to determine whether TREM2 is expressed on a common macrophage subset and promotes similar changes in neuroinflammation in these other models. The results from the present study will help inform the future research agenda related to TREM2 biology in these other disease contexts.
MATERIALS AND METHODS
Mice.
Two amyloid mouse models of AD were analyzed. APPPS1-21 (termed APPPS1) mice (provided by M. Jucker, German Center for Neurodegenerative Diseases [DZNE], Tubingen, Germany) express human APP with the Swedish (K670M/N671L) and PSEN1 L166P mutations under control of the Thy1 promoter (Radde et al., 2006). This mouse was maintained on the B6 background. 5XFAD mice (The Jackson Laboratory B6SJL-Tg(APPSwFlLon,PSEN1*M146*l286V)) express mutant human APP(695) with Swedish (K670N/M671L), Florida (I716V), and London (V717I) mutations and human PSEN1 with M146L and L286V mutations under control of the Thy1 promoter (Oakley et al., 2006). This mouse was maintained on a mixed B6/SJL background.
We also used a novel Trem2−/− mouse model (Trem2tm1(KOMP)Vlcg), which has a LacZ reporter cassette knocked into the endogenous Trem2 locus in place of exons 2 and 3 and most of exon 4, resulting in a loss of TREM2 function as well as expression of the LacZ reporter under the control of the TREM2 promoter (Fig. 1 a). This mouse was generated by the trans-NIH Knock-Out Mouse Project (KOMP). These mice were maintained on a B6 background. These mice were crossed with APPPS1 mice to yield APPPS1;Trem2+/LacZ and APPPS1;Trem2LacZ/LacZ genotypes (also termed throughout the paper APPPS1;Trem2+/− and APPPS1;Trem2−/−, respectively).
Mice were housed in the Cleveland Clinic Biological Resources Unit, Case Western Reserve University Animal Resource Center and The Jackson Laboratory, facilities fully accredited by the Association and Accreditation of Laboratory Animal Care. All experimental procedures were approved by the Institutional Animal Care and Use Committee at each respective institution.
Human tissue.
Human AD tissue was obtained from two neuropathologically confirmed AD patients. Tissue labeled “AD Case 1” is hippocampal tissue from an 88-yr-old female with a clinical diagnosis of Braak stage VI-C AD. Postmortem interval was 6 h, and tissue was immediately frozen. Tissue labeled “AD Case 2” is from the hippocampus of a 78-yr-old male with an 8-yr-history of cognitive decline. The patient’s son and power-of-attorney provided written permission for release for research purposes of any autopsy tissue not needed for diagnosis. Patient identifying information is housed in an Institutional Review Board–approved database. Postmortem interval was 36 h. The tissue was stored at 4°C in 4% paraformaldehyde for 2 d, followed by cryoprotection solution (20% glycerol in 0.08 M phosphate buffer, pH 7.6) until use. Human experiments were approved by The Cleveland Clinic and University of Washington Institutional Review Boards.
In situ hybridization.
Mice were perfused with 4% PFA, and 25-µm-thick frozen brain sections were prepared. A digoxigenin (DIG)-labeled riboprobe for TREM2 was transcribed from a cDNA clone (MMM1013-202767203; Thermo Fisher Scientific). The plasmid was digested with Sal1, and in vitro transcription was performed with T7 polymerase. A TREM2 sense probe was digested with a Not1 restriction enzyme and transcribed by SP6 RNA polymerase. Colorimetric detection of hybridized mRNA was performed using an anti-DIG conjugated to alkaline phosphatase and was developed using NBT-BCIP substrate (Roche).
Quantitative RT-PCR (qRT-PCR).
Mice were perfused with PBS, and their brains were removed, snap frozen, and kept at −80°C until use. Tissue was homogenized in 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1:100 protease inhibitor cocktail in PBS. RNA was isolated using chloroform extraction and was purified using Purelink RNA Mini kit (Life Technologies) and treated with DNase Purelink (Life Technologies). cDNA was prepared from 1.5 µg RNA using a QuantiTect Reverse Transcription kit (QIAGEN), and real-time PCR was performed for 40 cycles with the StepOne Plus Real Time PCR system (Life Technologies). All primers and TaqMan probes were purchased from the Life Technologies database. Relative gene expression was determined using the ΔΔCT method. A two-way ANOVA was performed, and significance between ΔCT values was determined using a Bonferroni post-hoc test for the TREM2 qPCR and a Student’s t test used for qPCR assays comparing APPPS1;Trem2+/+ and APPPS1;Trem2−/− animals.
Western blotting.
Tissue was extracted and processed as described above for qRT-PCR. After sonication and centrifugation, protein concentration was determined using a BCA kit (Thermo Fisher Scientific). Proteins were denatured for 15 min at 95°C in 35% denaturing buffer containing LDS sample buffer (Life Technologies) and reducing agent (Life Technologies). 30–50 µg of protein per sample was loaded along with 5 µl Magic Mark XP protein ladder (Life Technologies) onto Novex 4–12% Bis-Tris gels (Life Technologies), run at 160 V for 30–45 min, and transferred onto PVDF membranes (EMD Millipore) in 1× TAE buffer at 100 mA overnight at room temperature. After transfer, membranes were blocked using Odyssey Blocking Buffer in PBS (LI-COR Biosciences) for 1 h at room temperature and incubated in the appropriate primary antibodies in blocking buffer with 0.1% Tween 20 overnight at 4°C, 6E10 (1:5,000; Signet), TREM2 (1:500; R&D Systems), CT15 (1:10,000; a gift from E.H. Koo, University of California, San Diego, La Jolla, CA), and GAPDH (1:10,000; Thermo Fisher Scientific). Membranes were washed in PBST (0.1% Tween 20), incubated in the appropriate IR dye–conjugated secondary antibody (Thermo Fisher Scientific) in blocking buffer/0.1% Tween 20, and imaged using an Odyssey IR Scanner (LI-COR Biosciences) system. ImageJ software (National Institutes of Health) was used for densitometric analysis, and each experimental sample was normalized to GAPDH.
ELISA.
Aβ extraction was performed on microdissected brain tissue enriched for cortex and hippocampus. Lysates were mixed with an equal volume of 0.4% diethylamine in NaCl and centrifuged for 13,500 g for 1 h at 4°C. The supernatant was collected and neutralized with 0.5 M Tris, pH 6.8, and analyzed as the soluble Aβ fraction. The pellet was sonicated with 70% formic acid and centrifuged at 105,000 g for 45 min at 4°C. The supernatant was neutralized and analyzed as the insoluble Aβ fraction. Samples were analyzed by sandwich ELISA using 6E10 as the capture antibody and Aβ1-42 as detection antibody (Covance) as previously described (Cramer et al., 2012).
IHC.
Mice were deeply anesthetized with avertin and perfused with PBS. Brains were drop-fixed in 4% paraformaldehyde in PBS and cryoprotected in 30% sucrose. Brains were embedded in OCT, and free-floating 30-µm sagittal sections were collected and stored at 4°C in PBS. For 3,3′-diaminobenzidine (DAB) staining, endogenous peroxidases were quenched by incubating sections in 1% H2O2 in PBS for 30 min. Sections were blocked in 5% NGS/0.3% Triton X-100 in 1× PBS for 1 h. The following primary antibodies were added overnight at 4°C: Iba1 (1:1,000; Wako Pure Chemical Industries), AT8 (1:500; Thermo Fisher Scientific), AT180 (1:500; Thermo Fisher Scientific), CD11b (1:500; EMD Millipore), CD45 (1:500; AbD Serotec), CD68 (1:500; AbD Serotec), TREM2 (1:100; R&D Systems), and P2RY12 (1:2,000; a gift from H. Weiner, Brigham and Women’s Hospital, Boston, MA). To block nonspecific staining with antibodies generated in mouse and rat, Mouse on Mouse Blocking Reagent (Vector Laboratories) was used at 1 µl/ml of block. Slices were incubated with appropriate biotinylated secondary antibodies (1:200; Vector Laboratories) and VECTASTAIN Elite ABC kit (Vector Laboratories) and developed with DAB with nickel chloride. Indicated slices were counterstained with Congo red to visualize dense core Aβ plaques. Slices were mounted with Permount (Thermo Fisher Scientific).
TREM2 immunofluorescence was performed as described above for DAB staining except incubation with ABC was followed with incubation in the TSA Biotin System kit (PerkinElmer) and incubation with SA-488 (1:200; Life Technologies). Slices were then incubated with Iba1 (1:500; Wako Pure Chemical Industries), 6E10 (1:500; Covance), CT15 (1:250; a gift from E.H. Koo), GFAP (1:500; Sigma-Aldrich), MAP2 (1:500; EMD Millipore), or MBP (1:200; Abcam), followed by species-specific Alexa secondary antibodies (1:1,000; Life Technologies) and mounted using Vectashield Hard Set mounting media (Vector Laboratories).
β-Galactosidase activity was assessed in 30 µm free-floating sections with an X-gal staining solution (1 mg/ml X-gal, 5 mM potassium ferricyanate, 5 mM potassium ferrocyante, and 25 µM sodium deoxycholate). Slices were incubated in the solution overnight at 37°C, washed in PBS, costained, and mounted with Permount.
Brightfield images were taken on a DMLS microscope (Leica), using QImaging camera (QImaging) using QCapture Software (QImaging). For quantification of plaque area, whole brain sections were imaged on the SCN400F slide scanner (Leica) with SCN Client Software (Leica) and analyzed using Image Pro Plus Software (Media Cybernetics). Confocal images were taken on a LSM 510 META microscope (Carl Zeiss). 12–20 slices, 1 µm apart, were imaged and z-stacks were reconstructed in ImageJ.
Flow cytometry.
Myeloid cells were isolated from APPPS1 and 5XFAD mice along with age-matched controls. Mice were perfused with HBSS. Brains were chopped and digested in papain (Roche) at 37°C with shaking for 30 min. Homogenates were passed through 18- and 22-gauge needles and transferred into 20% FBS. Isotonic Percoll was added to create a 30% solution, and 1 ml of 70% Percoll was underlaid. This was centrifuged at 800 g for 30 min, and 2–3 ml of the 70%/30% interface was collected and washed. Samples were resuspended in FACS buffer (PBS, 1% BSA, 0.1% NaN3, and 5 mM EDTA) and blocked with a CD16/CD32 antibody (1:200; BD) for 10 min. Pooled portions of samples were used for unstained and single-stained control, whereas individual samples were stained with a master mix of CD45/AX700 (1:500; BioLegend), CD11b/BV605 (1:500; BioLegend), F4/80/PEefluor610 (1:125; eBioscience), and Ly6C/PECy7 (1:50,000; BioLegend). Each sample was divided in two tubes, a fluorescence minus one “FMO” control and an “ALL” sample to which TREM2/APC (1:125; R&D Systems) was added. Events were acquired on a Fortessa SORP (BD) and analyzed using FlowJo. For analysis, events were gated on single cells and CD11b-positive events. Samples with >5,000 CD11b-positive events were used for analysis. This population was divided into CD45lo and CD45hi subsets. TREM2+ events were assessed by overlaying FMO and ALL plots for each sample and gating on the population that was present only in the ALL sample. Significant differences between ages and genotypes were determined using a two-way ANOVA and Bonferroni-corrected Student’s t tests between all groups.
Statistics.
Statistical analyses were performed using Prism (GraphPad Software). Two-way ANOVAs and significance between individual groups were determined using a Bonferroni post-hoc test for analyses with multiple comparisons. Two-sided, unpaired Student’s t tests were used to determine statistical difference between samples in analyses that required only single comparisons. Although the data were not formally tested, based on previous results (Lee et al., 2014), we assumed they conform to a Gaussian distribution and that variance between groups was comparable. Biological replicates were used to define each n. Statistical outliers were excluded from all datasets. The mean of each group is graphed, and the error bars represent the SEM. Degree of significance between groups is represented as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. No tests were performed a priori to determine the sample size; however, the sample sizes used here are similar to those used in a previous study (Lee et al., 2014). 3–12 mice from at least two cohorts were included in each group.
Acknowledgments
We thank Rebecca Achey and Jiayang Li and the flow cytometry and imaging cores for their technical support.
This work was supported by an Alzheimer’s Association Multi-Center Program Grant, the Jane and Lee Seidman Fund, a generous donation from Chet and Jane Scholtz, National Institute on Aging grant AG023012, National Institute of Neurological Disorders and Stroke grants NS047804 and NS087298, Department of Defense grant W81XWH12-1-0629, BrightFocus Foundation grant A2013252F, and National Research Service Awards T32 NS067431 and T32 GM007250.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- Aβ
- β amyloid
- AD
- Alzheimer’s disease
- DAB
- 3,3′-diaminobenzidine
- FTD
- frontotemporal dementia
- IHC
- immunohistochemistry
- MAPT
- microtubule-associated protein τ
References
- Akiyama H., Barger S., Barnum S., Bradt B., Bauer J., Cole G.M., Cooper N.R., Eikelenboom P., Emmerling M., Fiebich B.L., et al. 2000. Inflammation and Alzheimer’s disease. Neurobiol. Aging. 21:383–421 10.1016/S0197-4580(00)00124-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird T.D., Koerker R.M., Leaird B.J., Vlcek B.W., and Thorning D.R.. 1983. Lipomembranous polycystic osteodysplasia (brain, bone, and fat disease): a genetic cause of presenile dementia. Neurology. 33:81–86 10.1212/WNL.33.1.81 [DOI] [PubMed] [Google Scholar]
- Borroni B., Ferrari F., Galimberti D., Nacmias B., Barone C., Bagnoli S., Fenoglio C., Piaceri I., Archetti S., Bonvicini C., et al. 2014. Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol. Aging. 35:934.e7–934.e10 10.1016/j.neurobiolaging.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. 2014. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat. Neurosci. 17:131–143 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady J., Koval E.D., Benitez B.A., Zaidman C., Jockel-Balsarotti J., Allred P., Baloh R.H., Ravits J., Simpson E., Appel S.H., et al. 2014. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol. 71:449–453 10.1001/jamaneurol.2013.6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu I.M., Morimoto E.T., Goodarzi H., Liao J.T., O’Keeffe S., Phatnani H.P., Muratet M., Carroll M.C., Levy S., Tavazoie S., et al. 2013. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 4:385–401 10.1016/j.celrep.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M.2003. TREMs in the immune system and beyond. Nat. Rev. Immunol. 3:445–453 10.1038/nri1106 [DOI] [PubMed] [Google Scholar]
- Cramer P.E., Cirrito J.R., Wesson D.W., Lee C.Y., Karlo J.C., Zinn A.E., Casali B.T., Restivo J.L., Goebel W.D., James M.J., et al. 2012. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science. 335:1503–1506 10.1126/science.1217697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forabosco P., Ramasamy A., Trabzuni D., Walker R., Smith C., Bras J., Levine A.P., Hardy J., Pocock J.M., Guerreiro R., et al. 2013. Insights into TREM2 biology by network analysis of human brain gene expression data. Neurobiol. Aging. 34:2699–2714 10.1016/j.neurobiolaging.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Burbach G.J., Bonin M., Walter M., Streit W., Bechmann I., and Deller T.. 2008. TREM2 is upregulated in amyloid plaque-associated microglia in aged APP23 transgenic mice. Glia. 56:1438–1447 10.1002/glia.20710 [DOI] [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. Alzheimer Genetic Analysis Group. 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R.J., Lohmann E., Brás J.M., Gibbs J.R., Rohrer J.D., Gurunlian N., Dursun B., Bilgic B., Hanagasi H., Gurvit H., et al. 2013. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 70:78–84 10.1001/jamaneurol.2013.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.C., Means T.K., and El Khoury J.. 2013. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16:1896–1905 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Tan L., Zhu X.C., Zhang Q.Q., Cao L., Tan M.S., Gu L.Z., Wang H.F., Ding Z.Z., Zhang Y.D., and Yu J.T.. 2014. Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology. 39:2949–2962 10.1038/npp.2014.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368:107–116 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch C.M., and Goate A.M.. 2015. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry. 77:43–51 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger G., Yamanishi Y., Suárez-Calvet M., Czirr E., Lohmann E., Cuyvers E., Struyfs H., Pettkus N., Wenninger-Weinzierl A., Mazaheri F., et al. 2014. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6:243ra86 10.1126/scitranslmed.3009093 [DOI] [PubMed] [Google Scholar]
- Lee S., Xu G., Jay T.R., Bhatta S., Kim K.W., Jung S., Landreth G.E., Ransohoff R.M., and Lamb B.T.. 2014. Opposing effects of membrane-anchored CX3CL1 on amyloid and tau pathologies via the p38 MAPK pathway. J. Neurosci. 34:12538–12546 10.1523/JNEUROSCI.0853-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A., Schlevogt B., Kierdorf K., Böttcher C., Erny D., Kummer M.P., Quinn M., Brück W., Bechmann I., Heneka M.T., et al. 2011. Distinct and non-redundant roles of microglia and myeloid subsets in mouse models of Alzheimer’s disease. J. Neurosci. 31:11159–11171 10.1523/JNEUROSCI.6209-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H., and Takahashi K.. 2007. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J. Neuroimmunol. 184:92–99 10.1016/j.jneuroim.2006.11.032 [DOI] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradowska-Gorycka A., and Jurkowska M.. 2013. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Hum. Immunol. 74:730–737 10.1016/j.humimm.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Prinz M., and Priller J.. 2014. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15:300–312 10.1038/nrn3722 [DOI] [PubMed] [Google Scholar]
- Radde R., Bolmont T., Kaeser S.A., Coomaraswamy J., Lindau D., Stoltze L., Calhoun M.E., Jäggi F., Wolburg H., Gengler S., et al. 2006. Aβ42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7:940–946 10.1038/sj.embor.7400784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayaprolu S., Mullen B., Baker M., Lynch T., Finger E., Seeley W.W., Hatanpaa K.J., Lomen-Hoerth C., Kertesz A., Bigio E.H., et al. 2013. TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson’s disease. Mol. Neurodegener. 8:19 10.1186/1750-1326-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell D.R., Zhou H., Comery T.A., Kouranova E., Lo C.F., Warwick H.K., Ring R.H., Kirksey Y., Aschmies S., Xu J., et al. 2007. The LXR agonist TO901317 selectively lowers hippocampal Aβ42 and improves memory in the Tg2576 mouse model of Alzheimer’s disease. Mol. Cell. Neurosci. 34:621–628 10.1016/j.mcn.2007.01.011 [DOI] [PubMed] [Google Scholar]
- Sedgwick J.D., Schwender S., Imrich H., Dörries R., Butcher G.W., and ter Meulen V.. 1991. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc. Natl. Acad. Sci. USA. 88:7438–7442 10.1073/pnas.88.16.7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif O., Gawish R., Warszawska J.M., Martins R., Lakovits K., Hladik A., Doninger B., Brunner J., Korosec A., Schwarzenbacher R.E., et al. 2014. The triggering receptor expressed on myeloid cells 2 inhibits complement component 1q effector mechanisms and exerts detrimental effects during pneumococcal pneumonia. PLoS Pathog. 10:e1004167 10.1371/journal.ppat.1004167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber M.W., Jaenisch N., Brehm M., Guenther M., Linnartz-Gerlach B., Neumann H., Witte O.W., and Frahm C.. 2013. Attenuated inflammatory response in triggering receptor expressed on myeloid cells 2 (TREM2) knock-out mice following stroke. PLoS ONE. 8:e52982 10.1371/journal.pone.0052982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J.D., Finn M.B., Wang Y., Shen A., Mahan T.E., Jiang H., Stewart F.R., Piccio L., Colonna M., and Holtzman D.M.. 2014. Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol. Neurodegener. 9:20 10.1186/1750-1326-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T.2006. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat. Med. 12:1005–1015. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., et al. 2013. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 153:707–720 10.1016/j.cell.2013.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]