Expression of NLR family apoptosis inhibitory proteins (NAIPs) protect against the development of tumors in two models of colorectal cancer. Protection appeared to be independent of NLRC4 inflammasome activation
Abstract
NLR family apoptosis inhibitory proteins (NAIPs) belong to both the Nod-like receptor (NLR) and the inhibitor of apoptosis (IAP) families. NAIPs are known to form an inflammasome with NLRC4, but other in vivo functions remain unexplored. Using mice deficient for all NAIP paralogs (Naip1-6Δ/Δ), we show that NAIPs are key regulators of colorectal tumorigenesis. Naip1-6Δ/Δ mice developed increased colorectal tumors, in an epithelial-intrinsic manner, in a model of colitis-associated cancer. Increased tumorigenesis, however, was not driven by an exacerbated inflammatory response. Instead, Naip1-6Δ/Δ mice were protected from severe colitis and displayed increased antiapoptotic and proliferation-related gene expression. Naip1-6Δ/Δ mice also displayed increased tumorigenesis in an inflammation-independent model of colorectal cancer. Moreover, Naip1-6Δ/Δ mice, but not Nlrc4-null mice, displayed hyper-activation of STAT3 and failed to activate p53 18 h after carcinogen exposure. This suggests that NAIPs protect against tumor initiation in the colon by promoting the removal of carcinogen-elicited epithelium, likely in a NLRC4 inflammasome-independent manner. Collectively, we demonstrate a novel epithelial-intrinsic function of NAIPs in protecting the colonic epithelium against tumorigenesis.
Inflammatory bowel disease (IBD) is an important risk factor that favors the development and progression of colitis-associated cancer (CAC; Eaden et al., 2001; Terzić et al., 2010; Rubin et al., 2013). Even in the absence of overt inflammatory disease in colorectal cancer (CRC), loss of barrier function in the tumor epithelium enables translocation of microbial products into tumor tissue. This triggers the activation of lamina propria immunocytes and colonic epithelial cells via pattern-recognition receptors (PRRs) to produce cytokines and chemokines. Those factors then promote tumor growth and mediate recruitment of further immune cells (Grivennikov et al., 2012; Mueller, 2012). Alternatively, epithelial innate immune components could be subverted during tumorigenesis and influence tumor growth independently. Although cytokine/chemokine-mediated modulation of tumor growth has been described, the role of epithelial-intrinsic, innate immune components still remains elusive.
Several Nod-like receptors (NLRs) have previously been implicated in colon inflammation and tumorigenesis, mostly in protective roles (Allen et al., 2010; Hu et al., 2010; Chen et al., 2011; Elinav et al., 2011; Zaki et al., 2011; Carvalho et al., 2012). In some cases, this has been attributed to reduced inflammasome-mediated release of IL-18, which is protective for the colonic epithelium (Allen et al., 2010; Dupaul-Chicoine et al., 2010). In other cases, noninflammasome-mediated factors were found to protect mice against CAC development. For example, NLRP12 was protective against colonic inflammation and tumorigenesis by dampening NF-κB and ERK activation in macrophages (Zaki et al., 2011). However, several discrepancies also exist, as illustrated by Caspase-1–deficient mice, which display increased colon tumorigenesis. In one study, this was dependent on NLRC4 and was epithelial intrinsic rather than inflammation mediated (Hu et al., 2010), whereas, in another study, increased tumorigenesis involved NLRP3 and was inflammation and hematopoietic cell–dependent (Allen et al., 2010). Such discrepancies are suggested to arise from differences in microbiota between facilities or use of WT mice from external sources (Ubeda et al., 2012), but could also arise from opposing functions of inflammasome components in different tissues, which has been demonstrated in a skin tumorigenesis model (Drexler et al., 2012).
The physiological function of the NLR protein NAIP (NLR family apoptosis inhibitory protein, previously known as neuronal apoptosis inhibitory protein) is not fully characterized, mainly because mice have several possibly redundant Naip paralogs (e.g., 4 functional and 2 noncoding Naip genes in the C57BL/6 genome; Yaraghi et al., 1998; Endrizzi et al., 2000; Growney and Dietrich, 2000). Humans also have several NAIP genes, only one of which is full length (Schmutz et al., 2004; Romanish et al., 2009). NAIPs are intracellular, cytosolic proteins with a tripartite structure; three N-terminal baculovirus inhibitor of apoptosis (IAP) protein repeat (BIR) domains, a central NACHT domain and C-terminal leucine rich-repeat (LRR) domains. The latter two domains group NAIPs to the NLR family of proteins. Indeed, NAIPs are best characterized for their inflammasome function. Mouse and human NAIPs are involved in the detection of intracellular pathogens, such as Salmonella, and activation of the NLRC4 inflammasome, inducing pyroptosis and caspase-1–mediated cleavage of IL-1β and IL-18 (Kofoed and Vance, 2011; Zhao et al., 2011; Rayamajhi et al., 2013; Yang et al., 2013). In mice, NAIP paralogs provide specificity to different bacterial components (Kofoed and Vance, 2011; Zhao et al., 2011). In vivo, the NAIP5-NLRC4 inflammasome was required for sepsis-induced mortality by an Escherichia coli pathobiont or by systemic delivery of intracellular-targeted flagellin, although partial redundancy to other Naip paralogs was apparent (Ayres et al., 2012; von Moltke et al., 2012).
NAIPs also belong to the IAP family due to three N-terminal BIR domains; but whether they actually function as inhibitors of apoptosis is controversial. Some studies show direct binding and inhibition of caspase-3 and -9 (Maier et al., 2002; Davoodi et al., 2004, 2010), but others do not (Roy et al., 1997). Also, NAIPs lack certain caspase-interaction residues within the BIR domains that would be necessary for direct inhibition of caspases, raising concern about whether NAIP can inhibit caspases in physiological settings (Scott et al., 2005; Eckelman and Salvesen, 2006; Eckelman et al., 2006). Additionally, NAIPs mediate inflammasome-induced caspase-1 activation and induction of pyroptosis via NLRC4, which is contrary to the suggested inhibitor of apoptosis function (Kofoed and Vance, 2012). BIR domains, however, can mediate a broad range of protein–protein interactions and therefore could be implicated in diverse cellular functions in addition to inhibition of caspases. In NAIPs, the BIR domains appeared to be necessary for NLRC4 inflammasome formation and activation of caspase-1 (Kofoed and Vance, 2011).
A mouse model lacking all Naip paralogs has not been available, preventing definitive analysis of NAIPs physiological function. In this study, we describe the first complete Naip1-6 knockout mice and demonstrate a crucial role for NAIPs in preventing colonic tumor initiation and progression.
RESULTS
Naip1-6 knockout mice develop normally
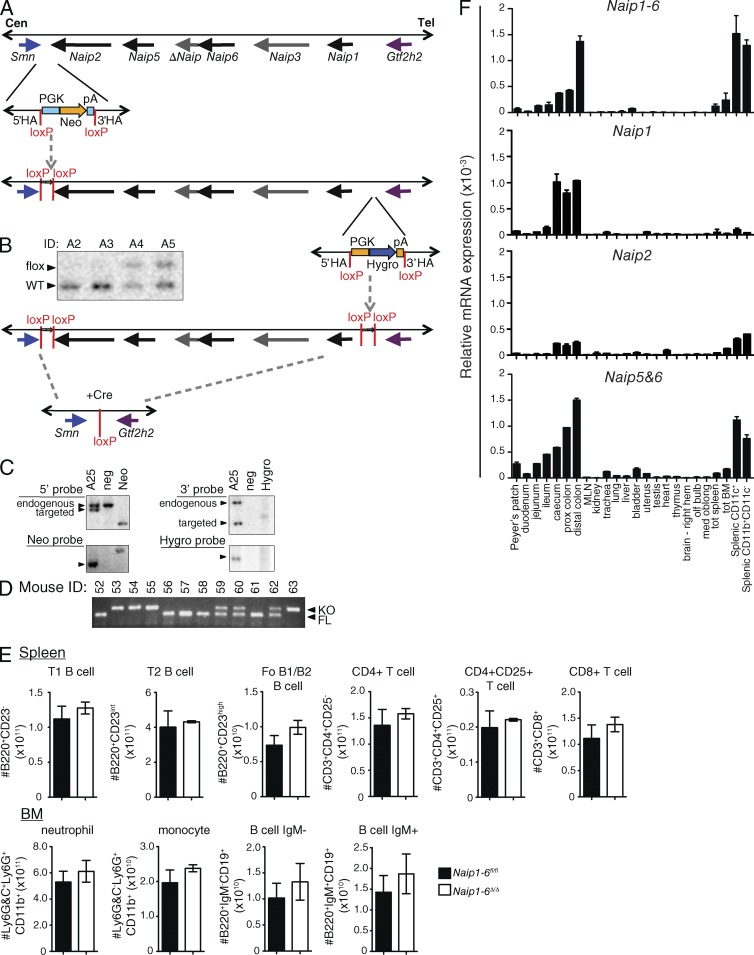
C57BL/6 mice have four functional copies of Naip (1, 2, 5, and 6) and two noncoding (nc) copies (Δ and 3; Fig. 1 A). Using a two-step targeting strategy, we generated C57BL/6 mice containing loxP sites flanking the Naip locus (Naip1-6fl/fl; Fig. 1, A–D). These mice were crossed with CMV-cre deleter mice (10 generations on C57BL/6 background) to generate full-body knockout of the Naip locus (Naip1-6Δ/Δ). Naip1-6Δ/Δ, and Naip1-6fl/fl mice bred as concurrent nonlittermate homozygous lines were used in most experiments, except where littermates are indicated. Naip1-6Δ/Δ mice were indistinguishable from their WT Naip1-6fl/fl counterparts when housed under specific pathogen–free conditions for up to one year. FACS analysis of immune cell subsets in the spleen and BM showed normal T cell, B cell, monocyte, and granulocyte populations (Fig. 1 E).
Figure 1.
Generation of Naip1-6 knockout mice and Naip tissue expression. Naip1-6 knockout mice were generated at Ozgene Pty. Ltd. using C57BL/6 material (DNA, blastocysts and ES cells). (A) Schematic of the targeting strategy (Naip locus schematic adapted from; Endrizzi et al., 2000). First, the region between the Smn and Naip2 genes was targeted with a vector containing neomycin selection flanked by loxP sites (HA, homology arms). Targeted ES cells were selected using neomycin (Neo) and germline transmission of the mutation was achieved. (B) Southern blot demonstrating the presence of WT or floxed allele (A2 to A5 represent different mice). ES cells were then isolated from the resulting heterozygous mice and used for the second targeting of the 3′ end of the Naip locus, between Naip1 and Gtf2h2. Successfully double-targeted ES cells were used to generate the double-floxed Naip mice (Naip1-6fl/fl). (C) Southern blot confirmation of double-targeted Naip allele (A25, mouse sample ID A25; neg, negative control; Neo, neomycin control; Hygro, hygromycin control). Naip1-6fl/fl mice were crossed with general deleter CMV-Cre recombinase mice to generate the full knockout mice (Naip1-6Δ/Δ). (D) PCR detection of floxed or KO alleles. (E) Spleen from Naip1-6fl/fl and Naip1-6Δ/Δ mice were stained for B220, CD23, CD3, CD4, CD8 and CD25 and analyzed by flow cytometry. BM cells were stained for Ly6G&C, Ly6G, CD11b, B220, CD19, and IgM and analyzed by flow cytometry. Specific cell populations are indicated above each graph. n = 3 mice per group. Data are representative of two experiments. (F) Tissues were dissected from C57BL/6 mice, immune cell subsets were FACS-sorted from spleens, RNA was isolated from these samples, and qPCR analysis for Naips was performed using pan Naip1-6, as well as Naip1, Naip2, and Naip5-6–specific primers. Data were normalized to 18S rRNA expression. Data are representative of tissues isolated from three individual mice at independent time points. Spleen subsets were obtained from pooled samples and are representative of two independent experiments.
Naips are highly expressed in the colon and innate immune cells
We observed the highest expression of Naips in the large intestine, with increasing levels from the cecum to the distal colon (Fig. 1 F). Naips are also expressed in innate immune cells such as macrophages, dendritic cells, and neutrophils (Fig. 1 F; and BioGPS gene annotation portal). Similarly, high expression of human NAIP was reported in the colonic epithelium and in innate immune cells (Diez et al., 2000; see also BioGPS and the Human Protein Atlas). Expression of Naips in the colon would be congruent with their role in detecting enteric pathogens such as Salmonella (Kofoed and Vance, 2011; Zhao et al., 2011; Sellin et al., 2014); but it also raises the question of whether NAIPs play any other physiological role in this organ.
TLR and inflammasome responses in Naip1-6Δ/Δ mice
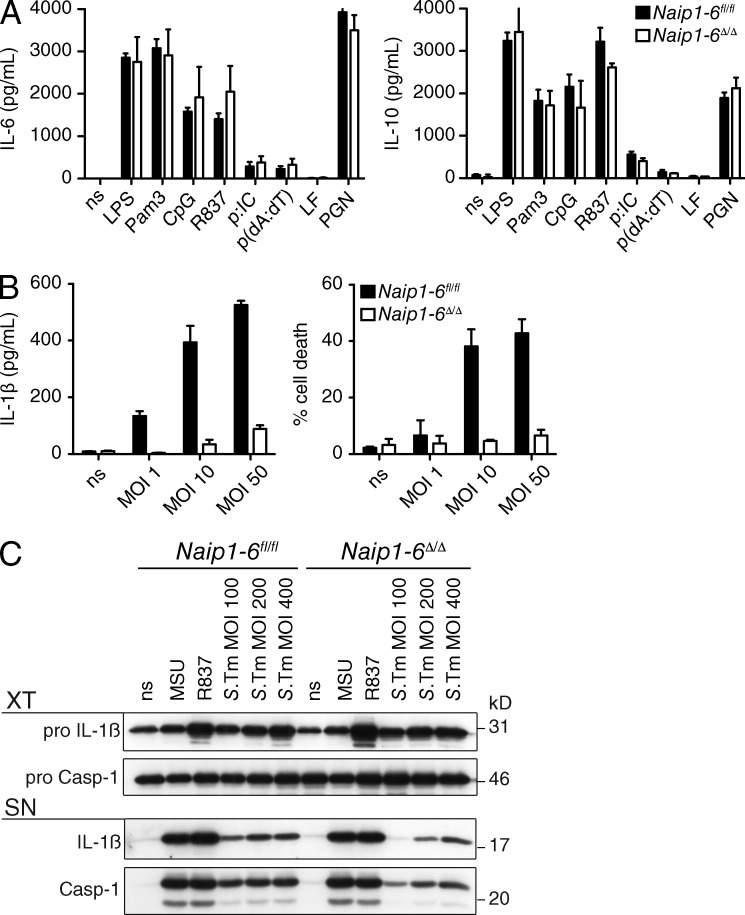
Naip1-6Δ/Δ BM-derived macrophages (BMDMs) responded normally to a range of Toll-like receptor (TLR) ligands with regard to production of the cytokines IL-6 and IL-10 (Fig. 2 A). However, consistent with NAIP’s role in NLRC4 inflammasome activation (Kofoed and Vance, 2011; Zhao et al., 2011; Yang et al., 2013), upon infection of Naip1-6Δ/Δ BMDMs with Salmonella typhimurium (S.Tm), IL-1β production and pyroptosis were severely attenuated (Fig. 2, B and C). Priming with LPS resulted in normal induction of pro–IL-1β, further confirming intact TLR responses in Naip1-6Δ/Δ BMDMs (Fig. 2 C). NLRP3 inflammasome activators (Fig. 2 C), and AIM2 inflammasome activator poly(dA:dT; not depicted), induced similar levels of caspase-1 activation and IL-1β production in both Naip1-6fl/fl and Naip1-6Δ/Δ BMDMs, indicating that other inflammasomes are intact in the Naip1-6Δ/Δ mouse. We observed residual activation of Caspase-1 and production of IL-1β in Naip1-6Δ/Δ BMDMs stimulated with high titers of S.Tm (Fig. 2, B and C), in line with previous observations of NLRC4-independent, NLRP3-mediated detection of S.Tm (Broz et al., 2010).
Figure 2.
TLR and inflammasome activation in Naip1-6Δ/Δ BMDMs. (A) Naip1-6fl/fl and Naip1-6Δ/Δ BMDMs were stimulated with a range of TLR ligands for 6 h. Supernatants were analyzed by ELISA for IL-6 and IL-10 production. Data are representative of two experiments. (B) BMDMs from Naip1-6fl/fl and Naip1-6Δ/Δ mice were prestimulated with LPS (20 ng/ml), and then infected with Salmonella (S.Tm; SB300 WT) at the indicated multiplicity of infection (MOI). Cell supernatants were collected after 2 h for analysis. IL-1β production was measured by ELISA. Cell death was measured by lactate dehydrogenase (LDH) release and results were normalized to a positive control for cell death. Data are representative of greater than three experiments. (C) Naip1-6fl/fl and Naip1-6Δ/Δ BMDMs were primed with LPS and stimulated with inflammasome activators; MSU (300 µg/ml), R837 (20 µg/ml), and S.Tm at the indicated MOI. Expression of pro-IL-1β, IL-1β, pro-caspase-1, and caspase-1 was assessed by Western blot. (top) Cell extract (XT); (bottom) cell supernatant (SN). Data are representative of three independent experiments, conducted on BMDM obtained from one or two mice for each experiment.
Naip1-6Δ/Δ mice have increased colon tumorigenesis
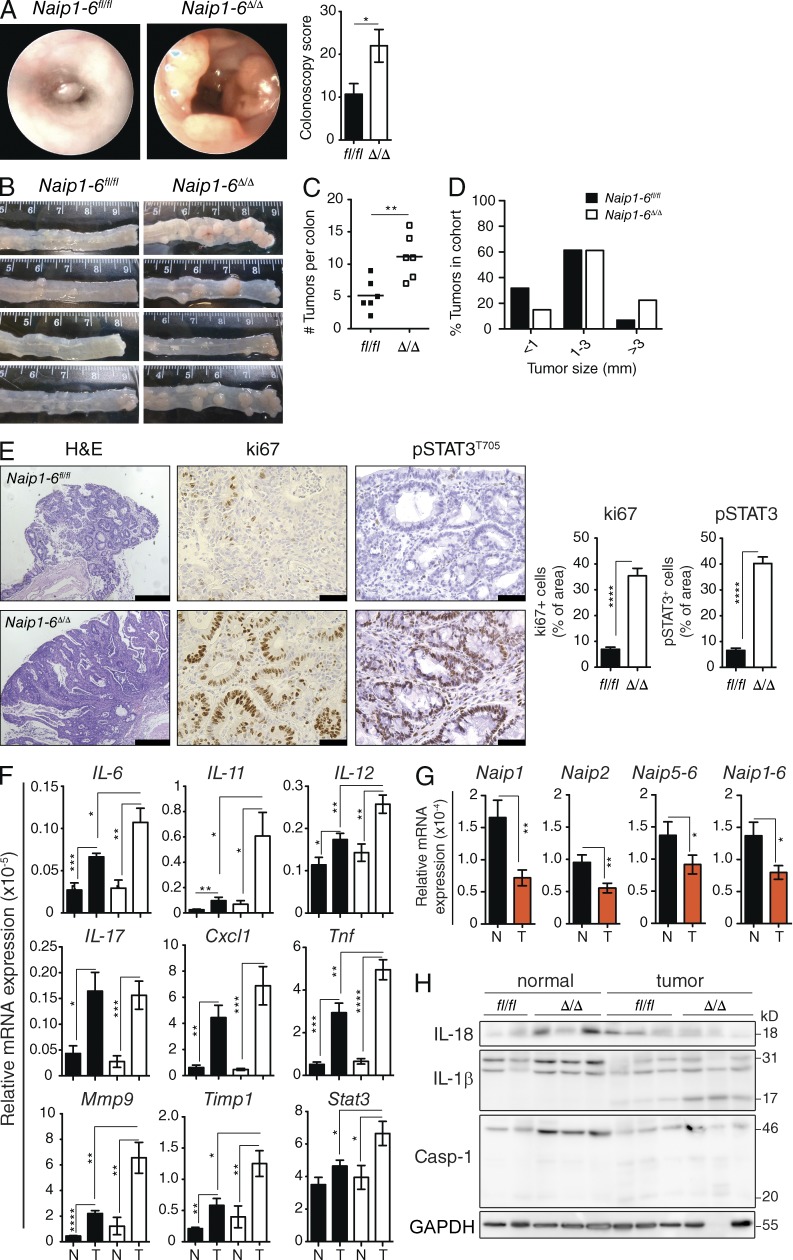
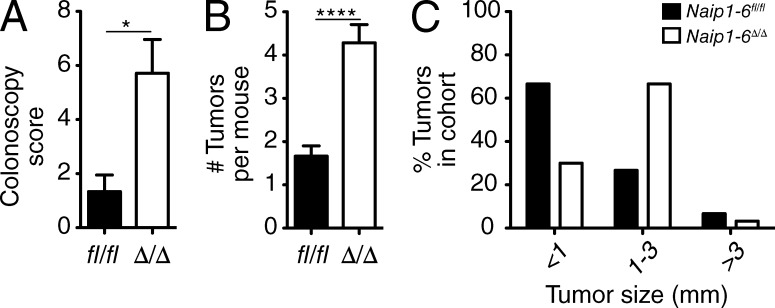
NAIPs have been suggested to act as tumor promoters because they are IAP family members. However, whether they act as IAPs is controversial. Additionally, NAIPs belong to the NLR family, and several NLRs are protective in colonic inflammation and tumorigenesis, through a variety of mechanisms (Allen et al., 2010; Hu et al., 2010; Chen et al., 2011; Elinav et al., 2011; Zaki et al., 2011; Carvalho et al., 2012). Naip1-6fl/fl and Naip1-6Δ/Δ mice (nonlittermates) were challenged in a CAC model with administration of the carcinogen azoxymethane (AOM; 10 mg/kg) followed by three cycles of dextran sulfate sodium (DSS; 2.5% wt/vol) to induce colitis and accelerate tumorigenesis. Colonic endoscopy assessment after the last dose of DSS revealed greater tumor burden in Naip1-6Δ/Δ mice compared with Naip1-6fl/fl (Fig. 3 A). Autopsy confirmed increased tumor burden, and also increased tumor size, in Naip1-6Δ/Δ mice (Fig. 3, B–D). Histological analysis of tumors demonstrated development of tubular adenomas with high-grade dysplasia in Naip1-6Δ/Δ mice, whereas Naip1-6fl/fl mice mostly had low-grade dysplasia, with only a few cases of high-grade dysplasia (Fig. 3 E). Naip1-6Δ/Δ tumors showed increased staining for ki67 compared with Naip1-6fl/fl tumors (Fig. 3 E), indicating increased proliferation, which is consistent with the increased tumor size. In accordance with this, STAT3 was highly phosphorylated and located in the nucleus of Naip1-6Δ/Δ tumor epithelium (as well as in infiltrating leukocytes; Fig. 3 E). However, tumors in Naip1-6fl/fl showed very little pSTAT3 staining in epithelial cells (but did have pSTAT3-expressing infiltrating leukocytes; Fig. 3 E). STAT3 activation is an important stimulus for tumor growth (Levy and Darnell, 2002; Bollrath et al., 2009; Terzić et al., 2010).
Figure 3.
Increased colon tumorigenesis in Naip1-6Δ/Δ mice AOM/DSS CAC model. Naip1-6fl/fl and Naip1-6Δ/Δ (nonlittermates) mice were injected i.p. with AOM (10 mg/kg) on day −1 and on day 0 mice were treated DSS (2.5% wt/vol) in the drinking water for 7 d, followed by 14 d of normal drinking water. DSS treatment was repeated twice. Mice were sacrificed after last DSS exposure on day 56. (A) Representative endoscopic view at day 55 and colonoscopy score. (B) Macroscopic appearance of colons at autopsy (day 56) shows tumor development in distal colon, (C) tumor burden, and (D) tumor size, expressed as percent of tumors in the entire cohort observed in the indicated size range. (E) Immunohistology staining for H&E showing representative tumor sections (bars, 500 µm) and ki67 and pSTAT3T705 (showing tumors of relatively similar development to enable comparison of proliferation; bars, 50 µm). Bar graphs on the right show quantification of ki67 and pSTAT3 expression. All tumors imaged were in the distal 1–2 cm of colon. (F) Relative expression levels of indicated mRNAs isolated from tumors T or the adjacent normal tissue N (normalized to 18S rRNA). (G) Same as F for the indicated Naips in Naip1-6fl/fl mice. (H) Western blot analysis for IL-18, Caspase-1, and IL-1β in normal and tumor tissue. Data are representative of two independent experiments with six to eight male mice per group. Data are shown as mean ± SEM. *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P < 0.0001.
Analysis of transcripts within tumor and in adjacent nontumoral tissue revealed an up-regulation of many proinflammatory cytokines within tumors of both genotypes (Fig. 3 F), which would be congruent with decreased barrier function within tumor epithelium (Grivennikov et al., 2012). Expression of IL-6, IL-11, IL-12, and Tnf were all significantly higher in Naip1-6Δ/Δ tumors compared with Naip1-6fl/fl (Fig. 3 F). Because IL-6 and IL-11 can activate STAT3 (Bollrath et al., 2009; Putoczki et al., 2013), increased pSTAT3 observed in Naip1-6Δ/Δ tumors might be related to the observed up-regulation of those cytokines. Also consistent with STAT3 activation, downstream targets Mmp9 and Timp1, which can act to enhance tumor growth or invasion (Kim et al., 2012; Shuman Moss et al., 2012), as well as Stat3 itself, were elevated (Fig. 3 F). In contrast, transcripts for each of the Naips were significantly down-regulated in tumor tissue, compared with surrounding nontumoral tissue of WT mice (Fig. 3 G). This suggests that reduced expression of all Naip paralogs could be associated with tumor progression, and therefore that they may all play a role in this phenotype.
Analysis of inflammasome activation in tumoral and nontumoral tissues of Naip1-6fl/fl and Naip1-6Δ/Δ mice revealed reduced mature IL-18, but increased IL-1β levels in tumors of both genotypes (Fig. 3 H). Because this was performed on whole-tissue homogenates, it is not possible to distinguish between colonic epithelium and hematopoietic-derived cytokine, which may account for the different regulation of IL-18 and IL-1β. Cleaved caspase-1 was also similarly detected at a low level, in both genotypes, with slight increase in tumor tissue (Fig. 3 H). These results make it unlikely that tumor progression in Naip1-6Δ/Δ would be caused by differences in inflammasome-derived cytokines.
Collectively, this data shows increased tumor development in Naip1-6Δ/Δ mice compared with Naip1-6fl/fl. Both genotypes showed induction of an inflammatory milieu in developed tumors, but this was more marked in Naip1-6Δ/Δ mice. In addition, Naip1-6Δ/Δ tumors had increased levels of proliferation markers (ki67) and STAT3 activation, which might, at least in part, account for the increased tumor size. No major differences in IL-18 and IL-1β were observed between genotypes, indicating normal activation of canonical inflammasomes in Naip1-6Δ/Δ mice. In addition, the down-regulation of Naips in WT tumors further suggests a role for Naips in tumor suppression.
Decreased colitis in Naip1-6Δ/Δ mice
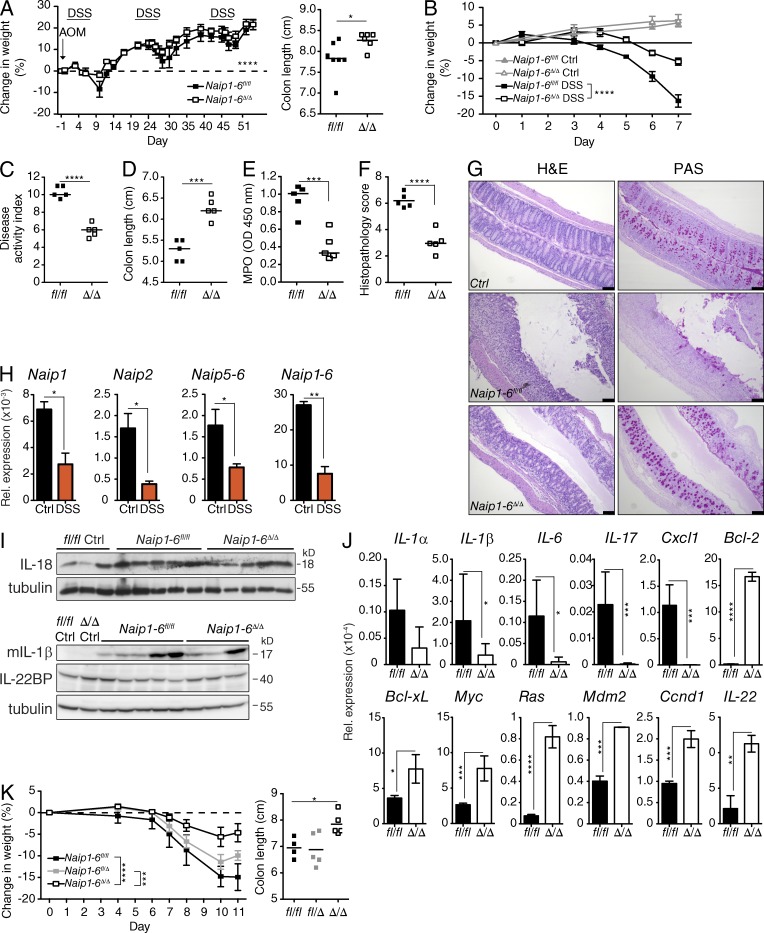
The inflammatory response to DSS-induced tissue damage is an important tumor promoter in the CAC model (Terzić et al., 2010). However, we observed reduced signs of colitis in Naip1-6Δ/Δ mice during the AOM/DSS protocol, including reduced weight loss and reduced colon shortening (Fig. 4 A). To further assess the colitis phenotype, we analyzed mice after 7 d of oral DSS exposure (acute colitis). The severity of colitis in Naip1-6Δ/Δ mice was reduced when compared with Naip1-6fl/fl mice, with reduced weight loss, disease activity index, and colon shortening (Fig. 4, B–D). There was also diminished myeloperoxidase activity, indicative of reduced neutrophil accumulation in the colon (Fig. 4 E). Histological analysis also demonstrated reduced immune cell infiltrates and protection from mucosal damage and loss of mucous-producing goblet cells in Naip1-6Δ/Δ mice (Fig. 4, F and G). mRNA levels of all Naip paralogs were reduced in the distal colon after 7 d of DSS (Fig. 4 H), although expression was not altered in the proximal or mid colon (not depicted). Intriguingly, the distal colon is the site of greatest tumorigenesis in this model. This might suggest that Naips are down-regulated to enable epithelial repair, or might reflect loss of NAIP-expressing cells during colon damage.
Figure 4.
Decreased colitis severity in Naip1-6Δ/Δ mice. (A) Change in body weight was assessed at the indicated time points and the length of colons at autopsy of mice undergoing the AOM/DSS CAC model (as per Fig. 3). (B–I) Mice were treated with DSS (2.5% wt/vol) in the drinking water for 7 d, and were sacrificed and tissues analyzed on day 7. n = 5 for each group. (B) The percent change in weight of Naip1-6fl/fl and Naip1-6Δ/Δ mice at the indicated time points; n = 5 mice per group (n = 3 for untreated controls [Ctrl]). (C) Disease activity index on day 6 of the treatment. (D) Colon length. (E) Myeloperoxidase measurements of colon homogenates, expressed as optical density at 450 nm. (F) Histopathology scores performed blinded on H&E-stained colon sections. (G) Representative sections of colons stained with H&E and periodic acid-Schiff stain (PAS). Bars, 100 µm. (H) Expression of Naips mRNA in colon homogenate of control and DSS-treated Naip1-6fl/fl mice at day 7 were measured by qPCR (normalized to GAPDH). (I) Western blot analysis of IL-18, IL-1β, and IL-22BP in colon homogenates from DSS-treated mice on day 7 (genotypes indicated, samples pooled for IL-1β and IL-22BP). (J) Male mice were injected i.p. with AOM (10 mg/kg) and the following day given DSS (2.5% wt/vol) in the drinking water for 7 d, followed by 2 d normal water. Relative expression levels of indicated mRNAs isolated from colons (normalized to 18S rRNA). n = 5 for Naip1-6fl/fl and n = 3 for Naip1-6Δ/Δ mice (data are shown as mean ± SEM). Data are representative of two independent experiments. (K) Weight loss and colon lengths of Naip1-6fl/fl (n = 4), fl/Δ (n = 5), and Δ/Δ (n = 5) littermates undergoing acute DSS (2.5% wt/vol) induced colitis (7 d of DSS followed by normal water until day 11). Data representative of three independent experiments in B–F and of 2 independent experiments in G–K. Data shown were acquired with male mice. Two-way ANOVA was used for weight loss statistics and two-way Student’s t test for was used for other statistics. Data are shown as mean ± SEM. *, P < 0.05; **, P ≤ 0.01; ***, P < 0.001; ****, P ≤ 0.0001.
Inflammasome activation during DSS-induced colitis has previously been shown to be protective, particularly through the production of IL-18. IL-18 and IL-1β were induced at the protein level in colon lysates of both genotypes after DSS (Fig. 4 I). IL-22bp mRNA was shown to be negatively regulated during DSS colitis in a time-dependent manner and also after biopsy-induced damage. In the biopsy model, this regulation was absent in NLRP3 and NLRP6 inflammasome-deficient mice (Huber et al., 2012). We observed no change in IL-22BP in DSS exposed animals, with no difference between genotypes (Fig. 4 I). This is in contrast to Huber et al. (2012); however, the DSS dosing and timing, and line susceptibility, may affect the timing of IL-22BP protein and mRNA regulation. This data indicates that inflammasomes other than NAIP/NLRC4 are activated during acute colitis, but neither IL-18 and IL-1β, nor IL-22 BP levels, play a role in the Naip1-6Δ/Δ phenotype.
We then compared gene expression after AOM and DSS exposure in Naip1-6fl/fl and Naip1-6Δ/Δ mice. Analysis of mRNA expression in colon homogenates revealed a reduction of IL-1β, IL-6, IL-17, and Cxcl1 transcript levels in Naip1-6Δ/Δ mice (Fig. 4 J), consistent with reduced disease severity. In contrast, antiapoptotic, proliferation, or survival-related genes Bcl-2, Bcl-xL, Myc, Ras, Mdm2, Ccnd1 (Cyclin D1), and IL-22 were all increased in Naip1-6Δ/Δ mice (Fig. 4 J). This indicates that reduced colitis was not a result of failure to react to DSS-induced damage, but rather that tissue protective and regenerative responses were elicited.
Colitis was also assessed among littermates to rule out any effect of microbiota drift between the Naip1-6Δ/Δ and Naip1-6fl/fl mice, which were bred as concurrent homozygous lines. In litters of Naip1-6fl/fl, Naip1-6fl/Δ, and Naip1-6Δ/Δ, the Naip1-6Δ/Δ mice still displayed less severe colitis, with reduced weight loss and less reduction in colon length compared with Naip1-6fl/fl and Naip1-6fl/Δ1-6 (Fig. 4 K). Naip heterozygotes showed slightly less severe weight loss but a similar degree of colon shortening as Naip1-6fl/fl (Fig. 4 K). This demonstrates that the phenotype of Naip1-6Δ/Δ mice is not a result of microbiota drift between parental lines.
Altogether, this data suggests that Naip1-6Δ/Δ mice are protected from severe DSS-induced epithelial damage because increased survival and proliferation maintains epithelial integrity. By keeping an intact epithelial barrier, the Naip1-6Δ/Δ colon would be protected from the ensuing inflammation that follows epithelial barrier disruption. However, the prosurvival and proliferative response could also act to promote tumor progression.
Increased tumorigenesis is epithelium intrinsic
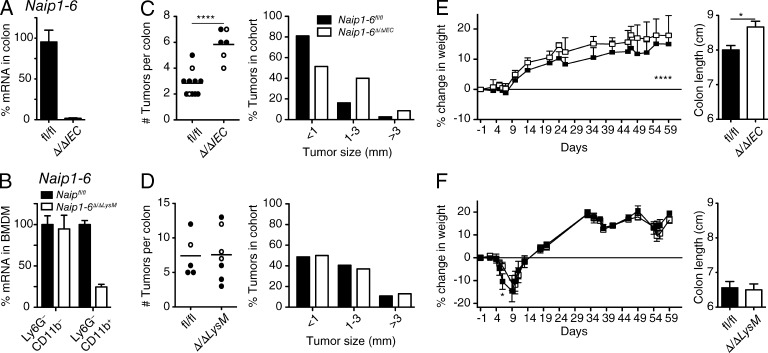
Next, we wanted to determine which tissue was responsible for NAIP protection against tumorigenesis. Because NAIPs are highly expressed in the colon and in the innate immune myeloid cell compartment, we generated epithelial (Naip1-6Δ/ΔIEC) and myeloid (Naip1-6Δ/ΔLysM) cell–specific Naip KO mice by crossing Naip1-6fl/fl mice to Villin-cre or LysM-cre, respectively (both are on the C57BL/6 background), and performed the AOM/DSS CAC model. Loss of Naips via Villin-cre deletion almost completely removed Naip expression from the colon, indicating the high expression of Naips within colonic epithelial cells (Fig. 5 A). Naips knockout in LysM-cre-deleted mice was around 75% in BM Ly6G−CD11b+ macrophages (Fig. 5 B). Deletion efficiency by LysM-cre has been shown to vary across the myeloid lineage (Clausen et al., 1999).
Figure 5.
Increased tumorigenesis in the absence of Naips is epithelium-intrinsic. Epithelial-specific (Naip1-6Δ/ΔIEC), or myeloid-specific (Naip1-6Δ/ΔLysM) Naip knockouts were exposed to the AOM/DSS CAC model as described in Fig. 3, except where indicated. Mice were sacrificed on day 62. Littermates of both sexes were used (filled circles, males; open circles, females). (A) Naip1-6 mRNA in colon of Naip1-6fl/fl and Naip1-6Δ/ΔIEC mice (relative to 18S RNA and expressed as percentage of transcript in Naip1-6fl/fl). (B) Naip1-6 mRNA in Ly6G−CD11b+ (macrophage) and Ly6G−CD11b− (nonmyeloid) BM populations derived from Naip1-6fl/fl and Naip1-6Δ/ΔLysM mice (relative to 18S RNA and expressed as percentage of transcript in Naip1-6fl/fl). (C) Tumor burden and tumor size in Naip1-6fl/fl (n = 11) and Naip1-6Δ/ΔIEC (n = 6) littermates. Data were pooled from two independent experiments (in C, mice were treated with DSS [2.5%] for 5 d and normal water for 16 d for 3 cycles). (D) Tumor burden and tumor size in Naip1-6fl/fl (n = 5) and Naip1-6Δ/ΔLysM (n = 7) littermates, representative of two independent experiments. (E and F) Weight loss and colon length of mice treated as in C and D, respectively. Data are shown as mean ± SEM. *, P < 0.05; ****, P ≤ 0.0001.
Similar to the full Naip1-6Δ/Δ mice, Naip1-6Δ/ΔIEC mice showed increased tumorigenesis compared with littermate Naip1-6fl/fl mice, with increased tumor burden and tumor size (Fig. 5 C). Naip1-6Δ/ΔLysM mice, however, showed the same tumor burden as littermate controls (Fig. 5 D). Colitis assessed during the AOM/DSS protocol showed a significant difference in Naip1-6Δ/ΔIEC mice with higher weight and less colon shortening compared with Naip1-6fl/fl mice (Fig. 5 E). In this experiment, mice received DSS for only 5 d, explaining why the impact on weight loss, colon shortening and tumor burden was less prominent than in other experiments. Naip1-6Δ/ΔLysM mice displayed a slight reduction of colitis in the early phase but no protection after 9 d, and colons exhibited equivalent shortening compared with WT littermates (Fig. 5 F). A role for Naips in myeloid cells in the initial phase of colitis could reflect activation of myeloid cells (macrophages or neutrophils) after barrier disruption. Together, this data indicates that NAIP deficiency in the colonic epithelium, and not in resident or infiltrating myeloid cells, drives the increased tumorigenesis.
Increased tumorigenesis in AOM-only model of CRC
Typically, decreased colonic inflammation is associated with decreased tumorigenesis. However, Naip1-6Δ/Δ mice developed significantly more tumors than controls (Fig. 3, A–D), despite decreased inflammation (Fig. 4, A–G). This lack of correlation between tumorigenesis and inflammation led us to check whether Naip1-6 deficiency could also lead to increased tumorigenesis in an inflammation-independent setting. To test this, we used a model of AOM-induced CRC that is free of DSS or other inflammatory challenges (Schwitalla et al., 2013). Mice were injected with AOM (10 mg/kg) once per week for 6 wk and were assessed for tumor burden after 24 wk. Naip1-6Δ/Δ mice had increased tumor burden, assessed by colonoscopy and at autopsy, compared with those of Naip1-6fl/fl mice (Fig. 6, A and B). Naip1-6Δ/Δ tumors also tended to be larger compared with Naip1-6fl/fl mice; however, both genotypes had a similar percentage of tumors greater than 3 mm (Fig. 6 C). These data demonstrate that Naip deficiency can drive increased tumorigenesis in an inflammation-independent manner, and point toward a role in tumor initiation.
Figure 6.
Increased tumorigenesis in Naip1-6Δ/Δ mice in AOM-induced CRC. Naip1-6fl/fl (n = 9) and Naip1-6Δ/Δ (n = 7) mice were injected with AOM (10 mg/kg) once per week for 6 wk and were analyzed at week 24. (A) Colonoscopy score assessed at week 24, (B) tumor burden (data are shown as mean ± SEM), and (C) tumor size assessed at 25 wk, expressed as percentage of tumors in the indicated size range, calculated on the entire cohort. In all panels, data are representative of two independent experiments. Data from male mice are shown (similar results were obtained in female mice). *, P < 0.05; ****, P < 0.0001.
Altogether, this data suggests that the initiation of tumorigenesis by AOM is sufficient for increased tumorigenesis in Naip1-6Δ/Δ mice. Furthermore, in the setting of AOM/DSS CAC, the tissue-protective response observed in Naip1-6Δ/Δ mice in response to DSS, in addition to the induction of an inflammatory milieu in developed tumors, could act to promote tumor growth.
The early response to AOM is altered in Naip1-6Δ/Δ mice
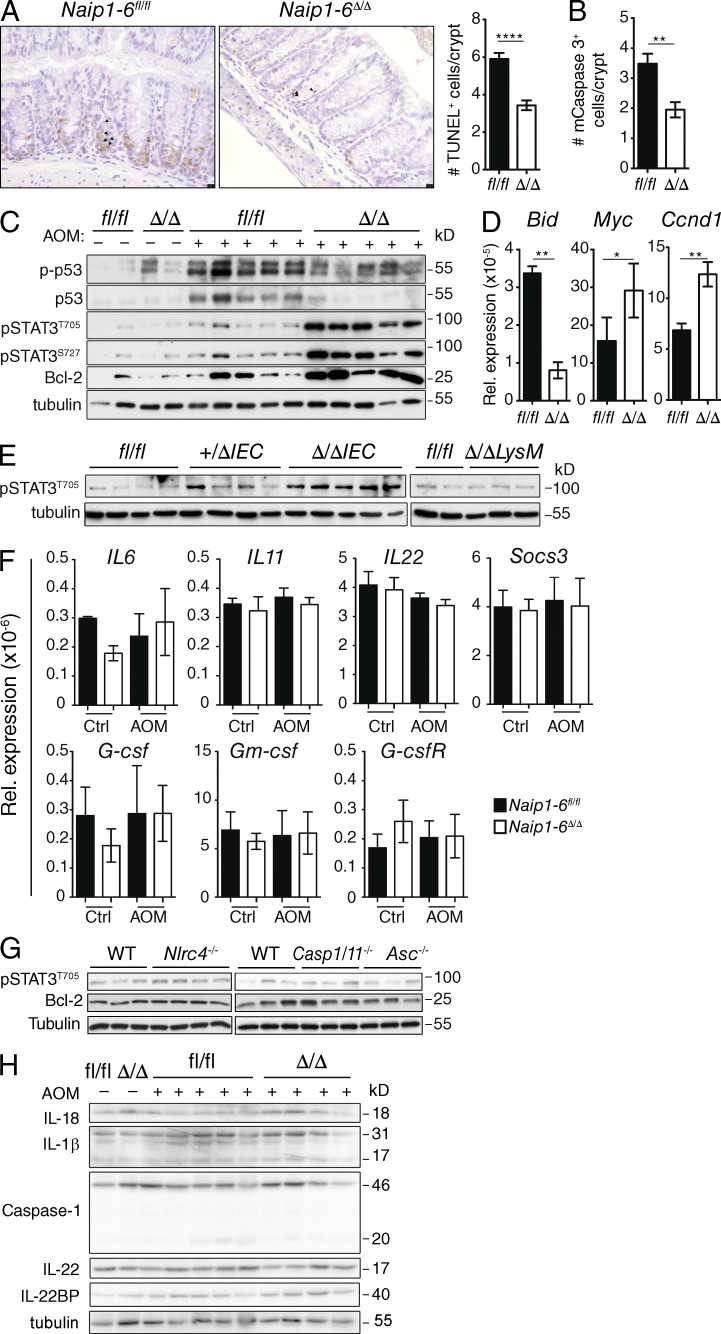
AOM induces O-6 methylguanine adducts on DNA guanine, causing G-to-A base changes, which usually elicits a wave of p53-mediated apoptosis that can be detected early after AOM injection (Hu et al., 2002; Kerr et al., 2013; Schwitalla et al., 2013). Failure to repair damaged DNA, or to eliminate AOM-mutated cells, leads to increased tumor burden (Schwitalla et al., 2013). Because Naip1-6Δ/Δ mice developed increased tumor burden in the AOM-only model of CRC, we assessed the role of NAIPs in tumor initiation. Mice were injected with AOM (10 mg/kg) and the colons were analyzed 18 h later. Naip1-6fl/fl mice showed induction of apoptosis in the colon, with increased immunoreactivity for TUNEL and active caspase 3, whereas this was significantly reduced in Naip1-6Δ/Δ colons (Fig. 7, A and B). This illustrates a failure to induce apoptosis upon AOM administration in the absence of NAIPs, which is contrary to their suggested IAP role. Consistent with this, Western blot analysis showed reduced activation of p53 (phosphorylation on S15 and stabilization of total p53) in Naip1-6Δ/Δ compared with Naip1-6fl/fl mice (Fig. 7 C). Additionally, mRNA analysis revealed increased induction of Bid, the pro-apoptotic p53 target, in Naip1-6fl/fl but not Naip1-6Δ/Δ mice (Fig. 7 D). In contrast, Naip1-6Δ/Δ mice showed up-regulation of Bcl2, Myc and Ccnd1 (cyclin D1; Fig. 7, C and D), which is consistent with increased survival and proliferation. This suggests that the increased tumor burden observed in Naip1-6Δ/Δ mice results from an altered response to AOM.
Figure 7.
The early response to AOM is altered in Naip1-6Δ/Δ mice. Naip1-6fl/fl and Naip1-6Δ/Δ mice were injected with AOM (10 mg/kg) and colons were analyzed 18 h later. (A) Representative sections of TUNEL staining on colon sections and quantification. 10 crypts in 5 independent fields of view per mouse were counted. Data are expressed as number of TUNEL+ cells per crypt. (B) Quantification of active caspase-3+ cells in colon sections, performed as in A. (C) Western blot analysis of the indicated proteins in lysates prepared from whole-colon homogenates. (D) Relative expression of indicated mRNA from whole colon homogenates (normalized to 18S rRNA). Five mice per group, representative of three independent experiments (A–D and F). Data shown as mean ± SEM. (E) Levels of pSTAT3 in colon homogenates of Naip1-6Δ/ΔIEC, Naip1-6Δ/ΔLysM, or Naip1-6fl/fl littermates after 18 h AOM exposure. Representative of two independent experiments. (F) Relative expression of indicated STAT-3–inducing mRNA targets from Naip1-6fl/fl and Naip1-6Δ/Δ whole colon samples as described for D. Data shown as mean ± SEM. (G) Nlrc4−/−, Caspase-1/11−/−, and Asc−/− mice were injected with AOM (10 mg/kg) and colons were analyzed 18 h later. Expression of pSTAT3 and Bcl-2 in colon homogenates was determined by Western blot (representative of two independent experiments). (H) Western blot analysis of Caspase-1, IL-18, IL-1β, IL-22, and IL-22BP in whole-colon homogenates from AOM and control-treated Naip1-6fl/fl and Naip1-6Δ/Δ mice (representative of two independent experiments). *, P < 0.05; **, P ≤ 0.01; ****, P < 0.0001.
The phenotype of Naip1-6Δ/Δ mice, namely increased colon tumors in a context of lower inflammation, has been described before in gp130Y757F mice, which express hyperactivated STAT3 (Bollrath et al., 2009). Similarly, IL-6−/− mice display increased colitis severity but decreased tumorigenesis, largely due to defective STAT3 activation (Grivennikov et al., 2009). Although STAT3 involvement in tumor progression is well established (Levy and Darnell, 2002; Bollrath et al., 2009; Terzić et al., 2010), a role in tumor initiation is less studied. However, a few studies have demonstrated a role for STAT3 in initiation of a skin tumorigenesis model (Chan et al., 2004; Miyatsuka et al., 2006; Kim et al., 2009) and in Apcmin/+ intestinal tumor initiation (Musteanu et al., 2010). To determine if STAT3 was affected during the initiation phase of AOM exposure in Naip1-6Δ/Δ mice, we analyzed phosphorylation of STAT3 in colon homogenates 18 h after AOM injection. Strikingly, Naip1-6Δ/Δ, but not Naip1-6fl/fl mice, had abundant phosphorylation of STAT3 (T705 and S727) after AOM injection (Fig. 7 C). Accordingly, expression of downstream targets of STAT3 (Bcl-2, Myc, and Ccnd1) was also increased in colons from Naip1-6Δ/Δ, but not Naip1-6fl/fl mice (Fig. 7, C and D). Increased STAT3 activation was also observed in Naip1-6Δ/ΔIEC, but not Naip1-6Δ/ΔLysM mice (Fig. 7 E), demonstrating an epithelial-intrinsic role for NAIPs in the early response to AOM. Activation of STAT3 usually occurs via cytokine or growth factor signaling downstream of receptor tyrosine kinase activation (Levy and Darnell, 2002). Therefore, we checked expression of cytokines and growth factors IL-6, IL-11, IL-22, G-csf, and Gm-csf, as well as Socs3 (suppressor of cytokine signaling 3) by qRT-PCR; however none of these factors changed after administration of AOM, either between the genotypes or between AOM and control-treated mice (Fig. 7 F). This would indicate that AOM does not affect expression of cytokines or growth factors. Overall, this data suggests that STAT3 activation occurs in an epithelial-intrinsic manner in the absence of NAIPs and could be an important impetus of tumor initiation in the absence of NAIPs.
NAIPs regulate STAT3 phosphorylation independent of the inflammasome axis
Because NAIPs can act in concert with NLRC4 to induce inflammasome activity upon detection of intracellular pathogens, we tested whether NLRC4, Caspase-1, or ASC had any effect on STAT3 phosphorylation status. We injected Nlrc4−/−, Caspase-1/11−/−, Asc−/−, or WT mice with AOM (10 mg/kg) and checked colon homogenates 18 h later. No differences in STAT3 phosphorylation, or downstream target Bcl-2, were observed between any of the genotypes (Fig. 7 G). Immunoblots for IL-18, IL-1β, and caspase-1 revealed no detectable activation of caspase-1 or downstream cleavage of IL-18 or IL-1β beyond basal levels in Naip1-6fl/fl or Naip1-6Δ/Δ colons after AOM administration (Fig. 7 H). IL-22 protein levels also did not change (Fig. 7 H), which is in line with the mRNA levels (Fig. 7 F). There was a modest increase in IL-22BP in Naip1-6Δ/Δ colons, the significance of which is unknown. Collectively, this data suggests that IL-22 signaling does not play a role in activation of STAT3 after AOM exposure in Naip1-6Δ/Δ mice. The mechanism of how STAT3 is activated in Naip1-6Δ/Δ colons after AOM administration remains to be determined. Altogether, these results demonstrate that NAIPs suppress STAT3 activation after carcinogen exposure independent of the inflammasome axis and cytokine production, and in an epithelium-intrinsic manner.
In summary, in the absence of NAIPs, we observed that colonic epithelial cells failed to induce apoptosis in response to AOM exposure. Instead, there was an induction of STAT3 phosphorylation and signaling. This response to AOM was sufficient to drive tumorigenesis in an AOM-only model of CRC. In the colitis-associated cancer model, additional elements were at play. During acute colitis, similar to the altered response to AOM, we observed a change in gene expression indicative of a proliferative and protective epithelial response. However, by the stage of polyp induction, we could see greater induction of some inflammatory cytokines in Naip1-6Δ/Δ compared with Naip1-6fl/fl mice. This is consistent with loss of barrier function within tumors as they develop (Grivennikov et al., 2012). Naip1-6Δ/Δ mice also had increased pSTAT3 and ki67 within tumors, which would account for the increase in tumor size in this model.
DISCUSSION
In this study, we unveil a novel function for NAIPs in the suppression of colon tumorigenesis. Additionally, we also confirm the known role of NAIPs in the innate immune response of macrophages to Salmonella typhimurium and activation of inflammasome activity. In contrast to NAIP’s proposed role as inhibitors of apoptosis, NAIPs favored cell death at the expense of cell proliferation in the colonic epithelium under stress conditions. Under normal conditions, mice deficient for NAIPs do not have any abnormal phenotype. After insult with a chemical mutagen, Naip1-6Δ/Δ mice failed to activate p53 compared with control Naip1-6fl/fl mice, which demonstrated p53 stabilization, activation, and downstream induction of pro-apoptotic targets such as Bid (Fig. 7, A–D).
NAIPs also dampened the generation of pSTAT3 and its downstream effects on cell survival and proliferation (Bcl2 and Ccnd1 expression; Fig. 7, C and D). It remains to be determined how STAT3 is hyperphosphorylated in the absence of NAIPs. None of the cytokines or growth factors known to activate STAT3, that we checked, were altered at the mRNA level (Fig. 7 F). IL-22 and IL-22BP protein levels were also not affected in a way that would enable increased IL-22 signaling. The increase in STAT3 phosphorylation was also observed in IEC-specific Naip-deficient mice, which would indicate an epithelial cell–intrinsic mechanism. Whether and how direct or indirect loss of inhibition of STAT3 occurs in the absence of Naips remains to be elucidated. It is also unclear whether there is a causal link between reduced p53 activation and apoptosis in Naip1-6Δ/Δ mice and STAT3 phosphorylation.
We determined that NAIP’s effect on the early activation of STAT3 was independent of the NLRC4 inflammasome, because Nlcr4−/−, Asc−/−, and Casp-1/11−/− mice did not exhibit a similar dysregulation of STAT3 (Fig. 7 G). However it is uncertain as to whether the phenotype of Naip1-6Δ/Δ mice in the AOM/DSS CAC model is independent of NLRC4. Caspase-1−/− and Nlrc4−/− mice have also been reported to have increased tumorigenesis in the AOM/DSS CAC model (Hu et al., 2010); however, another study reported no role for NLRC4 (Allen et al., 2010). The study by Hu et al. (2010), demonstrated that, similar to Naip1-6Δ/Δ mice, Nlrc4−/− and Caspase-1−/− mice display an epithelial-intrinsic increase in tumorigenesis that was not dependent on IL-1β or IL-18. Their results differ to the Naip1-6Δ/Δ phenotype in that Nlrc4−/− and Caspase-1−/− mice have equivalent levels of DSS-induced colitis compared with WT counterparts (which we saw similarly for Nlrc4−/− in our laboratory; unpublished data), whereas Naip1-6Δ/Δ mice have decreased susceptibility. Therefore, it appears that there are some differences between Naip1-6Δ/Δ mice and Nlrc4−/− mice. It is likely that some epithelial-intrinsic functions of NAIPs are mediated via NLRC4, such as NAIP/NLRC4 inflammasome-mediated extrusion of S.Tm-infected enterocytes (Sellin et al., 2014). However, it is also apparent that there are some NLRC4-independent functions of NAIPs. This is not surprising because NAIPs contain three BIR domains, which can mediate a variety of functions.
We propose that increased tumorigenesis observed in Naip-deficient mice challenged with a chemical carcinogen results from decreased cell death of damaged/mutated cells during the initiation stage with AOM and from a growth advantage throughout tumor development. Further, similar mechanisms may be at play upon stress imposed by DSS, where damage is being compensated by a robust proliferation and survival response in the absence of NAIPs. Naip deficiency and/or constitutive STAT3 activation may confer tumor cells with survival and proliferation signals usually provided by an inflammatory context and provide a rational explanation for the observed dissociation between tumorigenesis and inflammation in these mice. Finally, although inflammation does promote tumor growth—hence, development of colorectal tumors within a relatively short period of time in the AOM/DSS model—the level of inflammation does not necessarily directly correlate with tumor burden, as shown in Naip1-6Δ/Δ mice. Instead, it would appear that different types of tissue responses to stressors could have the capacity to drive tumorigenesis. This has implications for the identification of populations at risk of developing CRC.
As Naip1-6Δ/Δ mice experienced deletion of a large genomic fragment, a possible contribution of intergenic elements to the phenotype should be kept in mind. Although currently available genome browsers did not detect micro RNAs or other elements predicted to be important in this region.
A previous study (Endo et al., 2004) found that NAIP mRNA expression was decreased in well and moderately differentiated colon adenocarcinoma, compared with adjacent tissue, in line with our observation that, in mice, Naips are down-regulated in colon tumor versus normal colon tissue (Fig. 3 G). This supports that down-regulation of NAIPs could play a role in human CRC. Additionally, data available on the Human Protein Atlas shows strong staining for NAIP in the human colon but weak staining in samples of colon adenocarcinoma. These results should prompt further interest into the role of NAIP in human CRC.
MATERIALS AND METHODS
General and specific deletion of Naip1-6 in mice
Mice were generated by Ozgene Pty. Ltd. as described in Fig. 1, on a C57BL/6 background. To create full-body knockout of Naip1-6, the double “floxed” targeted mice were crossed with cre-deleter (JAX, B6.C-Tg(CMV-cre)1Cgn/J) mice to remove the ∼250-kb Naip locus. The CMV-cre line within our facility was backcrossed 10 generations to C57BL/6 background, which should preclude contamination with non-B6 alleles. Resulting Naip1-6fl/fl or Naip1-6Δ/Δ mice were maintained as concurrent WT (FL) or deficient (KO) lines (which were regularly renewed by intercrossing to avoid microbiota drift). Where indicated, mice were bred as heterozygote lines to use littermates. Tissue-specific deletion of Naip1-6 was achieved using Villin-Cre (JAX, B6.SJL-Tg (Vil-cre)997Gum/J) and LysM-Cre mice (JAX, B6.129P2-Lyz2tm1(cre)Ifo/J), which were also backcrossed in our facility at least 10 times on a C57BL/6 background. Genotyping of Naip1-6–deficient mice by PCR was performed on ear biopsy lysates using the following primers: 5′-TTGCTGTACTGACATCTGG-3′ (fwd), 5′-TCATACAATTCAGGATGGAA-3′ (rev), 5′-TGCTGACAGGACAGATATGA-3′ (fwd), and 5′-TAGAATTAATTCGCCAGGC-3′ (rev). Nlrc4−/−, Asc−/− (Mariathasan et al., 2004), and Caspase-1/11−/− (Kuida et al., 1995) mice were described previously. Nonlittermate WT control mice for these lines were bred and maintained in the same facility. Mice were handled according to Swiss Federal Veterinary Office guidelines, and protocols were approved by the Office Vétérinaire Cantonal du Canton de Vaud. Mice are now maintained at RIKEN.
Models of CRC and colitis
Colitis and colitis-associated cancer was induced as previously described (Wirtz et al., 2007). In brief:
AOM/DSS CAC.
Mice were injected i.p. on day −1 with AOM (10 mg/kg; Sigma-Aldrich). On day 0 dextran sodium sulfate (DSS; MW 36,000–50,000; MP Biochemicals) was given in the drinking water (2.5% wt/vol) for 7 d (unless otherwise specified), followed by 14 d normal water; DSS treatment was repeated twice. Mice were sacrificed and tissue was analyzed between days 52 and 62. Colons were excised and washed and tumors were counted using a dissecting microscope. Colons were either fixed in formalin and paraffin-embedded for histological analysis or tumor and normal tissue was dissected and frozen immediately for later analysis.
Colitis.
Acute colitis was induced by administering 2.5% (wt/vol) DSS in the drinking water for 7 d. Mice were weighed every day and percent of body weight change for each mouse was calculated. Clinical scores are a combination of weight loss, rectal bleeding, and stool consistency (between 0 and 4, with 0 being normal and 1–4 being diarrhea) as described previously (Cooper et al., 1993). Mice were sacrificed at day 7. Colons were removed, measured, weighed, washed, and fixed in 10% buffered formalin. A section of the colon was taken for MPO or RNA analyses.
AOM-induced CRC.
AOM (Sigma-Aldrich) was injected i.p. (10 mg/kg) once per week for 6 wk. After 24 wk, mice were assessed by colonoscopy for development of tumors. For analysis of the early response to AOM, mice were sacrificed 18 h after AOM injection.
Colonoscopy procedures
Colonoscopy was performed using a Coloview miniendoscopic system, consisting of a rigid Hopkins II miniature endoscope (0° direct vision, 30 cm length, 2 mm outer diameter) coupled to a Xenon 175 light source, an Endovision SLB Telecam camera (all from Karl Storz) and a low-pressure air pump (Rena Air 200; Rena). Polyps were classified and the colitis-associated cancer severity index calculated according to previously published parameters (Becker et al., 2005).
Histology
Colons were dissected, washed with PBS, rolled, and fixed in formalin. Tissues were then paraffin embedded, cut, and stained using standard protocols for H&E and PAS after rehydration of sections. For IHC, pSTAT3 (CST; 1:100 dilution) and anti-caspase-3 (CST, 1:200 dilution) were used after blocking with normal goat serum and were detected using EnVision anti–rabbit and DAB (DAKO). TUNEL staining of sections was performed using Terminal Transferase (TdT; Roche) as per manufacturer’s instructions.
Histology image acquisition and analysis
Images were acquired with a Leica DMI6000B and associated software, and a DFC450 camera. Quantification of ki67 and pSTAT3 IHC was performed using ImageJ (National Institutes of Health) software.
Colitis histolopathology assessment
Colitis histological damage was scored using the following system. Cell infiltration: 0, no infiltrate; 1, infiltrate around crypt base; 2, infiltrate reaching lamina; 3, confluence of infiltrate extending into submucosa; 4, transmural extension of the infiltrate. Epithelium (0–4): 0, normal, intact; 1, loss of goblet cells; 2, loss of goblet cells in large areas; 3, loss of crypts; 4, loss of crypts in large areas. Maximum total score, 8.
RNA isolation and qPCR
RNA was isolated using nucleospin RNA isolation kit (Macherey-Nagel) as per the manufacturer’s instructions. Tissues were lysed as per the manufacturer’s instructions using a QIAGEN TissueLyser. Reverse transcription was performed using standard protocols, using an M-MLV reverse transcription (Promega). Quantitative real-time PCR was performed with the LightCycler 480 Real-Time PCR System (Roche) and SYBR Green detection reagent. Gene-specific primers (Microsynth) are listed in Table S1.
Colon tissue analysis
For protein isolation and Western blot, colons were homogenized in tissue lysis buffer (Zaki et al., 2010) using a TissueLyser (QIAGEN). Protein was quantified, denatured with SDS buffer and boiled for 5 min. Proteins were separated by PAGE and blotted onto nitrocellulose membranes. Membranes were then incubated with the indicated primary antibodies overnight and detected with a relevant secondary antibody conjugated to horseradish peroxidase. Blots were revealed with regular or high-fidelity ECL solution (Thermo Fisher Scientific). Myeloperoxidase was measured in colon homogenates as previously described (Vieira et al., 2005).
Antibodies
Fluorescently labeled antibodies used for flow cytometry were all purchased from eBioscience. Antibodies used for Western blot were as follows: Bcl-2, p-p53, STAT3, and pSTAT (T705 and S727; CST), p53 (Vector Laboratories) and tubulin (Sigma-Aldrich).
FACS analysis
Single-cell suspensions of BM or spleen were obtained and cells were stained with indicated antibodies (eBioscience) conjugated to fluorescent dyes. Cells were analyzed on FACSCanto (BD) and data analyzed using FlowJo (Tree Star). To check the deletion efficiency of the Naip locus in Naip1-6ΔLysM mice, FACS sorting was performed for BM cells using Ly6G and CD11b antibodies (eBioscience), to isolate Ly6G−CD11b+ macrophages.
BM-derived macrophage culture
BM was flushed from the tibia and femur, red blood cells were lysed, and cells were cultured in BMDM medium (DMEM, 10% FCS, 20% supernatant from L929 cell-conditioned medium, 1% Penicillin-Streptomycin (10 000 U/ml; Invitrogen) in 10 cm culture Petri dishes for 6–7 d. Adherent cells were lifted from the Petri dish using Accutase (Invitrogen).
In vitro TLR and inflammasome stimulation
Naip1-6fl/fl or Naip1-6Δ/Δ BMDM (2.5 × 105 cells per stimulation) were prestimulated with LPS (20 ng/ml), and then infected with S.Tm (SB300 WT) at the indicated MOI for 20 min (Salmonella strains were supplied by W.-D. Hardt, ETH, Zurich, Switzerland). Cells were then washed to remove extracellular bacteria and incubated for a further 2 h with medium containing gentamycin. Supernatants were then removed for ELISA and LDH analysis. For TLR and other inflammasome stimulation (shown by Western blot) cells were incubated for 6 h with the indicated stimuli.
ELISA and lactate dehydrogenase measurements
ELISA (eBioscience) and LDH assays (Cayman Chemical) were performed as per manufacturer’s instruction using 50 µl of cell culture supernatants.
Statistical analysis
Data were analyzed using GraphPad Prism Software. Two-tailed Student’s t test was used for comparison between two groups, and two-way ANOVA was used when two independent variables were being assessed. A P-value equal or less than P = 0.05 was considered statistically significant. Data shown are mean ± SEM, unless otherwise stated.
Supplementary Material
Acknowledgments
We acknowledge the late Professor Jurg Tschopp (University of Lausanne) for initiating this project. Unfortunately he did not get to oversee the project. We thank Nicolas Fasel (University of Lausanne) for his support and discussions, and enabling the continuation of the Tschopp laboratory until completion of these studies. Thanks also to Fabio Martinon (University of Lausanne), Wolf-Dietrich Hardt, and Mikael Sellin (ETH, Zurich) for advice and critical reading of the manuscript. Philippe Menu (University of Lausanne) did initial studies on inflammasome activation in Naip1-6Δ/Δ BMDMs. Jean-Christophe Stehle and Janine Horlbeck (Mouse Pathology Unit, University of Lausanne) provided expert assistance with histology. Aude De Gassart and Antonia Di Micco (University of Lausanne) provided assistance with some experiments.
This work was funded by grants from the Swiss National Science Foundation, the Institute of Arthritis Research, the National Center of Competence in Research Molecular Oncology, and the Louis-Jeantet foundation (to Jurg Tschopp). K.M. Maslowski was funded by an EMBO long-term fellowship an Australian NHMRC overseas post-doctoral fellowship and is currently supported by RIKEN’s Programs for Junior Scientists. C.W. Yu is currently employed by Roche.
The authors declare no additional competing financial interests.
Author contributions: K.M. Maslowski conceived the project, planned and performed experiments, analyzed data, and wrote the manuscript. R. Allam helped with experimental work and writing of the manuscript. A. Tardivel maintained the mouse lines. V. Chennupati, C.W. Yu, and H. Bega helped with experiments. M.H. Maillard and D. Velin performed colonoscopy and provided assistance with manuscript preparation. P. Schneider helped with laboratory matters, data analysis and writing of the manuscript.
Footnotes
Abbreviations used:
- AOM
- azoxymethane
- BIR
- baculoviral IAP repeat
- CAC
- colitis-associated cancer
- CRC
- colorectal cancer
- DSS
- dextran sulfate sodium
- IAP
- inhibitor of apoptosis
- IBD
- inflammatory bowel disease
- LRR
- leucine-rich repeat
- NAIP
- NLR family apoptosis inhibitory protein
- NLR
- Nod-like receptor
- S.Tm
- Salmonella Typhimurium
References
- Allen I.C., TeKippe E.M., Woodford R.M., Uronis J.M., Holl E.K., Rogers A.B., Herfarth H.H., Jobin C., and Ting J.P.. 2010. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207:1045–1056 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S., Trinidad N.J., and Vance R.E.. 2012. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 18:799–806 10.1038/nm.2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C., Fantini M.C., Wirtz S., Nikolaev A., Kiesslich R., Lehr H.A., Galle P.R., and Neurath M.F.. 2005. In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut. 54:950–954 10.1136/gut.2004.061283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., et al. 2009. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 15:91–102 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., and Monack D.M.. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F.A., Nalbantoglu I., Aitken J.D., Uchiyama R., Su Y., Doho G.H., Vijay-Kumar M., and Gewirtz A.T.. 2012. Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges. Mucosal Immunol. 5:288–298 10.1038/mi.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.S., Sano S., Kiguchi K., Anders J., Komazawa N., Takeda J., and DiGiovanni J.. 2004. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 114:720–728 10.1172/JCI200421032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.Y., Liu M., Wang F., Bertin J., and Núñez G.. 2011. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186:7187–7194 10.4049/jimmunol.1100412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Cooper H.S., Murthy S.N., Shah R.S., and Sedergran D.J.. 1993. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Invest. 69:238–249. [PubMed] [Google Scholar]
- Davoodi J., Lin L., Kelly J., Liston P., and MacKenzie A.E.. 2004. Neuronal apoptosis-inhibitory protein does not interact with Smac and requires ATP to bind caspase-9. J. Biol. Chem. 279:40622–40628 10.1074/jbc.M405963200 [DOI] [PubMed] [Google Scholar]
- Davoodi J., Ghahremani M.H., Es-Haghi A., Mohammad-Gholi A., and Mackenzie A.. 2010. Neuronal apoptosis inhibitory protein, NAIP, is an inhibitor of procaspase-9. Int. J. Biochem. Cell Biol. 42:958–964 10.1016/j.biocel.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Diez E., Yaraghi Z., MacKenzie A., and Gros P.. 2000. The neuronal apoptosis inhibitory protein (Naip) is expressed in macrophages and is modulated after phagocytosis and during intracellular infection with Legionella pneumophila. J. Immunol. 164:1470–1477 10.4049/jimmunol.164.3.1470 [DOI] [PubMed] [Google Scholar]
- Drexler S.K., Bonsignore L., Masin M., Tardivel A., Jackstadt R., Hermeking H., Schneider P., Gross O., Tschopp J., and Yazdi A.S.. 2012. Tissue-specific opposing functions of the inflammasome adaptor ASC in the regulation of epithelial skin carcinogenesis. Proc. Natl. Acad. Sci. USA. 109:18384–18389 10.1073/pnas.1209171109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaul-Chicoine J., Yeretssian G., Doiron K., Bergstrom K.S., McIntire C.R., LeBlanc P.M., Meunier C., Turbide C., Gros P., Beauchemin N., et al. 2010. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 32:367–378 10.1016/j.immuni.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Eaden J.A., Abrams K.R., and Mayberry J.F.. 2001. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 48:526–535 10.1136/gut.48.4.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelman B.P., and Salvesen G.S.. 2006. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J. Biol. Chem. 281:3254–3260 10.1074/jbc.M510863200 [DOI] [PubMed] [Google Scholar]
- Eckelman B.P., Salvesen G.S., and Scott F.L.. 2006. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 7:988–994 10.1038/sj.embor.7400795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Abe S., Seidlar H.B., Nagaoka S., Takemura T., Utsuyama M., Kitagawa M., and Hirokawa K.. 2004. Expression of IAP family proteins in colon cancers from patients with different age groups. Cancer Immunol. Immunother. 53:770–776 10.1007/s00262-004-0534-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi M.G., Hadinoto V., Growney J.D., Miller W., and Dietrich W.F.. 2000. Genomic sequence analysis of the mouse Naip gene array. Genome Res. 10:1095–1102 10.1101/gr.10.8.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., and Karin M.. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 15:103–113 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Wang K., Mucida D., Stewart C.A., Schnabl B., Jauch D., Taniguchi K., Yu G.Y., Osterreicher C.H., Hung K.E., et al. 2012. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 491:254–258 10.1038/nature11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Growney J.D., and Dietrich W.F.. 2000. High-resolution genetic and physical map of the Lgn1 interval in C57BL/6J implicates Naip2 or Naip5 in Legionella pneumophila pathogenesis. Genome Res. 10:1158–1171 10.1101/gr.10.8.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Martin J., Le Leu R., and Young G.P.. 2002. The colonic response to genotoxic carcinogens in the rat: regulation by dietary fibre. Carcinogenesis. 23:1131–1137 10.1093/carcin/23.7.1131 [DOI] [PubMed] [Google Scholar]
- Hu B., Elinav E., Huber S., Booth C.J., Strowig T., Jin C., Eisenbarth S.C., and Flavell R.A.. 2010. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc. Natl. Acad. Sci. USA. 107:21635–21640 10.1073/pnas.1016814108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Zenewicz L.A., Huber F.J., Bosurgi L., Hu B., Hedl M., Zhang W., O’Connor W. Jr, Murphy A.J., et al. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 491:259–263 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr C.A., Hines B.M., Shaw J.M., Dunne R., Bragg L.M., Clarke J., Lockett T., and Head R.. 2013. Genomic homeostasis is dysregulated in favour of apoptosis in the colonic epithelium of the azoxymethane treated rat. BMC Physiol. 13:2 10.1186/1472-6793-13-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.J., Kataoka K., Rao D., Kiguchi K., Cotsarelis G., and Digiovanni J.. 2009. Targeted disruption of stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 69:7587–7594 10.1158/0008-5472.CAN-09-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.H., Kang J.G., and Ko J.H.. 2012. Expression level and glycan dynamics determine the net effects of TIMP-1 on cancer progression. BMB Rep. 45:623–628 10.5483/BMBRep.2012.45.11.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2012. NAIPs: building an innate immune barrier against bacterial pathogens. NAIPs function as sensors that initiate innate immunity by detection of bacterial proteins in the host cell cytosol. BioEssays. 34:589–598 10.1002/bies.201200013 [DOI] [PubMed] [Google Scholar]
- Kuida K., Lippke J.A., Ku G., Harding M.W., Livingston D.J., Su M.S., and Flavell R.A.. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 267:2000–2003 10.1126/science.7535475 [DOI] [PubMed] [Google Scholar]
- Levy D.E., and Darnell J.E. Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651–662 10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- Maier J.K., Lahoua Z., Gendron N.H., Fetni R., Johnston A., Davoodi J., Rasper D., Roy S., Slack R.S., Nicholson D.W., and MacKenzie A.E.. 2002. The neuronal apoptosis inhibitory protein is a direct inhibitor of caspases 3 and 7. J. Neurosci. 22:2035–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Miyatsuka T., Kaneto H., Shiraiwa T., Matsuoka T.A., Yamamoto K., Kato K., Nakamura Y., Akira S., Takeda K., Kajimoto Y., et al. 2006. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev. 20:1435–1440 10.1101/gad.1412806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.2012. Danger-associated molecular patterns and inflammatory bowel disease: is there a connection? Dig. Dis. 30:40–46 10.1159/000342600 [DOI] [PubMed] [Google Scholar]
- Musteanu M., Blaas L., Mair M., Schlederer M., Bilban M., Tauber S., Esterbauer H., Mueller M., Casanova E., Kenner L., et al. 2010. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 138:1003–1011: e1–e5 10.1053/j.gastro.2009.11.049 [DOI] [PubMed] [Google Scholar]
- Putoczki T.L., Thiem S., Loving A., Busuttil R.A., Wilson N.J., Ziegler P.K., Nguyen P.M., Preaudet A., Farid R., Edwards K.M., et al. 2013. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell. 24:257–271 10.1016/j.ccr.2013.07.018 [DOI] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D.E., Chavarria-Smith J., Vance R.E., and Miao E.A.. 2013. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish M.T., Nakamura H., Lai C.B., Wang Y., and Mager D.L.. 2009. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS ONE. 4:e5761 10.1371/journal.pone.0005761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Deveraux Q.L., Takahashi R., Salvesen G.S., and Reed J.C.. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 16:6914–6925 10.1093/emboj/16.23.6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D.T., Huo D., Kinnucan J.A., Sedrak M.S., McCullom N.E., Bunnag A.P., Raun-Royer E.P., Cohen R.D., Hanauer S.B., Hart J., and Turner J.R.. 2013. Inflammation is an independent risk factor for colonic neoplasia in patients with ulcerative colitis: a case-control study. Clin. Gastroenterol. Hepatol. 11:1601–1608: e1–e4 10.1016/j.cgh.2013.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Martin J., Terry A., Couronne O., Grimwood J., Lowry S., Gordon L.A., Scott D., Xie G., Huang W., et al. 2004. The DNA sequence and comparative analysis of human chromosome 5. Nature. 431:268–274 10.1038/nature02919 [DOI] [PubMed] [Google Scholar]
- Schwitalla S., Ziegler P.K., Horst D., Becker V., Kerle I., Begus-Nahrmann Y., Lechel A., Rudolph K.L., Langer R., Slotta-Huspenina J., et al. 2013. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 23:93–106 10.1016/j.ccr.2012.11.014 [DOI] [PubMed] [Google Scholar]
- Scott F.L., Denault J.B., Riedl S.J., Shin H., Renatus M., and Salvesen G.S.. 2005. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 24:645–655 10.1038/sj.emboj.7600544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Shuman Moss L.A., Jensen-Taubman S., and Stetler-Stevenson W.G.. 2012. Matrix metalloproteinases: changing roles in tumor progression and metastasis. Am. J. Pathol. 181:1895–1899 10.1016/j.ajpath.2012.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzić J., Grivennikov S., Karin E., and Karin M.. 2010. Inflammation and colon cancer. Gastroenterology. 138:2101–2114: e5 10.1053/j.gastro.2010.01.058 [DOI] [PubMed] [Google Scholar]
- Ubeda C., Lipuma L., Gobourne A., Viale A., Leiner I., Equinda M., Khanin R., and Pamer E.G.. 2012. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209:1445–1456 10.1084/jem.20120504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira A.T., Pinho V., Lepsch L.B., Scavone C., Ribeiro I.M., Tomassini T., Ribeiro-dos-Santos R., Soares M.B., Teixeira M.M., and Souza D.G.. 2005. Mechanisms of the anti-inflammatory effects of the natural secosteroids physalins in a model of intestinal ischaemia and reperfusion injury. Br. J. Pharmacol. 146:244–251 10.1038/sj.bjp.0706321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N.J., Moayeri M., Kintzer A.F., Wang S.B., van Rooijen N., Brown C.R., Krantz B.A., Leppla S.H., Gronert K., and Vance R.E.. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 490:107–111 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S., Neufert C., Weigmann B., and Neurath M.F.. 2007. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2:541–546 10.1038/nprot.2007.41 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaraghi Z., Korneluk R.G., and MacKenzie A.. 1998. Cloning and characterization of the multiple murine homologues of NAIP (neuronal apoptosis inhibitory protein). Genomics. 51:107–113 10.1006/geno.1998.5378 [DOI] [PubMed] [Google Scholar]
- Zaki M.H., Boyd K.L., Vogel P., Kastan M.B., Lamkanfi M., and Kanneganti T.D.. 2010. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 32:379–391 10.1016/j.immuni.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Vogel P., Malireddi R.K., Body-Malapel M., Anand P.K., Bertin J., Green D.R., Lamkanfi M., and Kanneganti T.D.. 2011. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 20:649–660 10.1016/j.ccr.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.