The Liu and Nussenzweig groups identify the immediate precursor of CD1c+ and CD141+ dendritic cells in the circulation of healthy donors. These precursor cells (hpre-cDC) were detectable in cord blood, bone marrow, blood, and peripheral lymphoid organs.
Abstract
Two subsets of conventional dendritic cells (cDCs) with distinct cell surface markers and functions exist in mouse and human. The two subsets of cDCs are specialized antigen-presenting cells that initiate T cell immunity and tolerance. In the mouse, a migratory cDC precursor (pre-CDC) originates from defined progenitors in the bone marrow (BM). Small numbers of short-lived pre-CDCs travel through the blood and replace cDCs in the peripheral organs, maintaining homeostasis of the highly dynamic cDC pool. However, the identity and distribution of the immediate precursor to human cDCs has not been defined. Using a tissue culture system that supports the development of human DCs, we identify a migratory precursor (hpre-CDC) that exists in human cord blood, BM, blood, and peripheral lymphoid organs. hpre-CDCs differ from premonocytes that are restricted to the BM. In contrast to earlier progenitors with greater developmental potential, the hpre-CDC is restricted to producing CD1c+ and CD141+ Clec9a+ cDCs. Studies in human volunteers demonstrate that hpre-CDCs are a dynamic population that increases in response to levels of circulating Flt3L.
Conventional DCs (cDCs) induce immunity or tolerance by capturing, processing, and presenting antigen to T lymphocytes (Banchereau and Steinman, 1998). In the mouse, cDCs are short-lived cells, whose homeostasis in lymphoid and nonlymphoid tissues is critically dependent on continual replenishment from circulating pre-CDC (Liu et al., 2007; Liu and Nussenzweig, 2010). Murine pre-CDCs are BM-derived cells that are present in very small numbers in the blood but increase in response to Flt3L injection (Liu et al., 2007, 2009). pre-CDCs have a very short dwell time in the blood, 65% of these cells leave the circulation within 1 min after leaving the BM (Liu et al., 2007, 2009). Upon leaving the circulation, pre-CDCs seed tissues where they differentiate to cDCs, which divide further under the control of Flt3L (Liu et al., 2007, 2009). Thus, in addition to the BM and blood, mouse pre-CDCs are also found in peripheral lymphoid organs and nonlymphoid tissues (Naik et al., 2006; Bogunovic et al., 2009; Ginhoux et al., 2009; Liu et al., 2009; Varol et al., 2009).
Mouse cDCs can be divided into two major subsets, CD11b+ DCs and CD8+/CD103+ DCs that differ in their microanatomic localization, cell surface antigen expression, antigen-processing activity, and ability to contribute to immune responses to specific pathogens (Merad et al., 2013; Murphy, 2013). Despite these important differences, both CD11b+ and CD8+/CD103+ cDC subsets of mouse DCs are derived from the same immediate precursor (pre-CDC) that expresses CD135 (Flt3), the receptor for Flt3L, a cytokine that is critical to DC development in vivo (McKenna et al., 2000; Waskow et al., 2008).
Similar to the mouse, humans have two major subsets of cDCs. CD141 (BDCA3)+Clec9a+ DCs (CD141+ cDC herein) appear to be the human counterpart of mouse CD8+/CD103+ DCs, expressing XCR1, Clec9a, IRF8, and TLR3 and producing IL-12 (Robbins et al., 2008; Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Poulin et al., 2010; Haniffa et al., 2012). CD1c (BDCA1)+ cDCs appear to be more closely related to mouse CD11b+ DCs, expressing IRF4, inducing Th17 differentiation upon A. fumigatus challenge, and imprinting intraepithelial homing of T cells (Robbins et al., 2008; Crozat et al., 2010; Schlitzer et al., 2013; Yu et al., 2013). In the mouse, the superior ability of CD8+/CD103+ DCs to cross-present exogenous antigens to CD8+ T cells is attributed to both differential antigen uptake (Kamphorst et al., 2010) and to increased expression of proteins and enzymes that facilitate MHC class I presentation (Dudziak et al., 2007). Human CD141+ cDCs are more efficient than CD1c+ cDCs in cross-presentation (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Poulin et al., 2010), but this difference appears to result from differences in antigen uptake and cytokine activation rather than a specialized cell-intrinsic program (Segura et al., 2012; Cohn et al., 2013; Nizzoli et al., 2013).
Both CD1c+ cDCs and CD141+ cDCs are present in human blood and peripheral tissues. Each subset in the blood resembles its tissue counterpart in gene expression but appears less differentiated (Haniffa et al., 2012; Segura et al., 2012; Schlitzer et al., 2013). These observations are consistent with the idea that less differentiated human cDCs travel through the blood to replenish the cDC pool in the peripheral tissues (Collin et al., 2011; Segura et al., 2012; Haniffa et al., 2013). Others have postulated the existence of a less differentiated circulating DC progenitor based on absence of CD11c, expression of CD123, and response to Flt3L (O’Doherty et al., 1994; Pulendran et al., 2000), but the progenitor potential of these putative precursors that produced large amounts of IFN-α was never tested directly and they appear to correspond at least in part to plasmacytoid DCs (Grouard et al., 1997; Siegal et al., 1999). Thus, whether there is an immediate circulating precursor restricted to human immature and mature CD1c+ and CD141+ cDCs is not known.
Here, we report the existence of a migratory pre-CDC in humans (hpre-CDC) that develops from committed DC progenitors (hCDPs) in the BM (Lee et al., 2015) and is the immediate precursor of both CD1c+ and CD141+ cDCs, but not pDCs or monocytes. hpre-CDCs are present in BM, cord, and peripheral blood, as well as peripheral lymphoid tissues. In studies of human volunteers, Flt3L injection induces expansion of hpre-CDCs in the circulation. Thus, human cDC precursors constitute a dynamic circulating population whose homeostasis is regulated by Flt3L, a cytokine that is responsive to inflammatory and infectious agents.
RESULTS
Identification of an immediate precursor to human cDCs
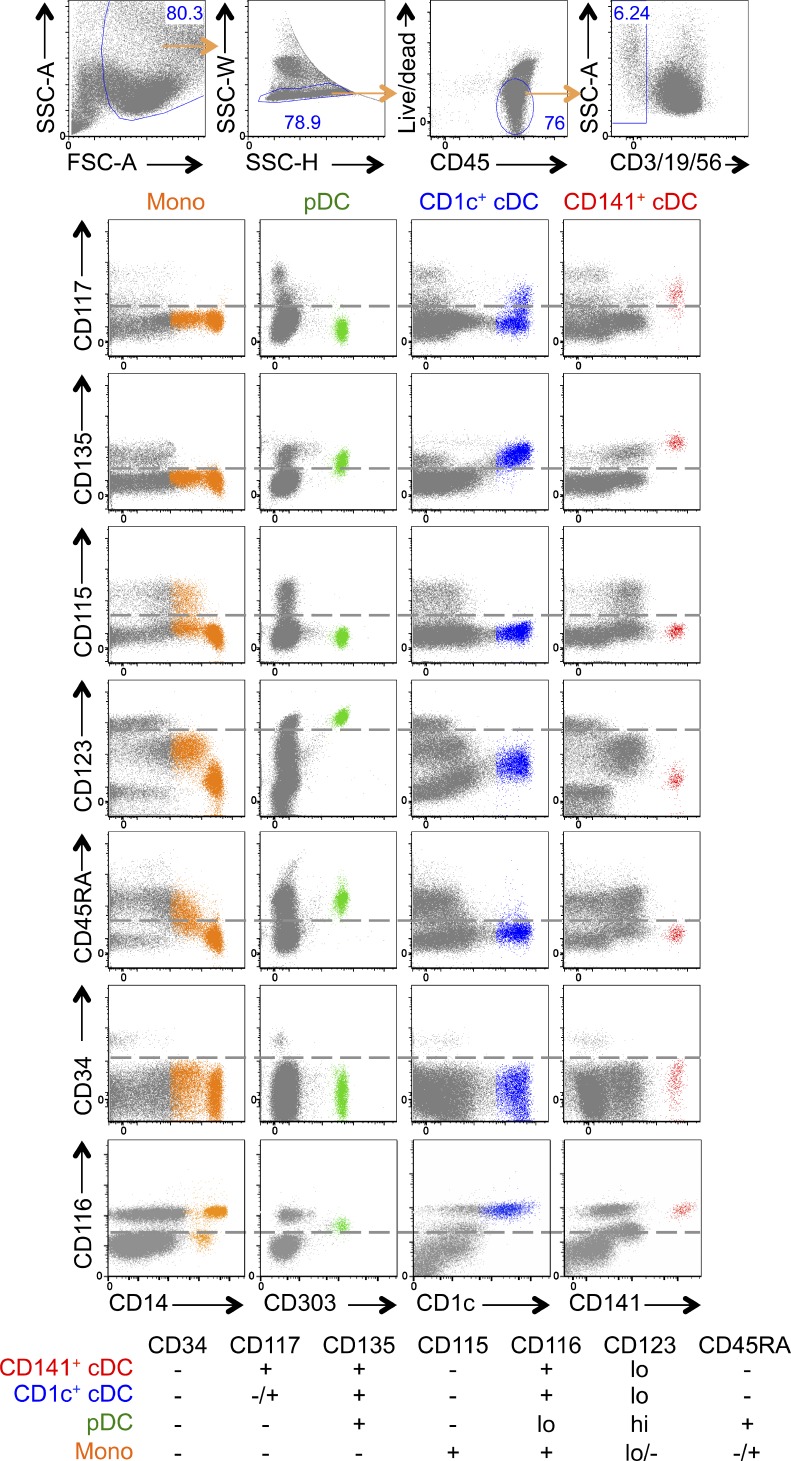
To define cell surface markers that might be expressed by circulating hpre-CDCs, we examined DCs and monocytes for expression of cytokine receptors that are found on BM DC progenitors (Lee et al., 2015). Whereas CD116 (GM-CSF receptor) is expressed on differentiated monocytes and DCs, CD135 (Flt3L receptor) is restricted to DCs, CD115 (M-CSF receptor) is expressed only on monocytes, and CD117 (SCF receptor) is found on the surface of CD141+ cDCs and most CD1c+ cDCs, but not on pDCs or monocytes (Fig. 1). CD45RA, a marker expressed by several different hematopoetic cell progenitors including granulocyte monocyte progenitors (GMP) and common lymphoid progenitors (CLP; Chicha et al., 2004; Doulatov et al., 2010), is highly expressed only on pDCs (Fig. 1). Finally, CD34 is expressed on early DC progenitors (Chicha et al., 2004; Doulatov et al., 2010; Lee et al., 2015), but not on fully differentiated DCs or monocytes, suggesting that loss of CD34 would precede cDC development (Fig. 1). Collectively, the data suggests that if a hpre-CDC exists in human blood it may be found in the CD34−CD115−CD116+CD117+CD135+ fraction.
Figure 1.
Expression of cytokine receptors on DCs and monocytes. Flow cytometry plots show expression of CD117, CD135, CD115, CD123, CD45RA, CD34, and CD116 on gated CD3−CD19−CD56− cells (gray), differentiated monocytes (CD14+ orange), pDCs (CD303+ green), CD1c+ cDCs (blue), and CD141+ cDCs (red) from peripheral blood. Table summarizes results of flow cytometry.
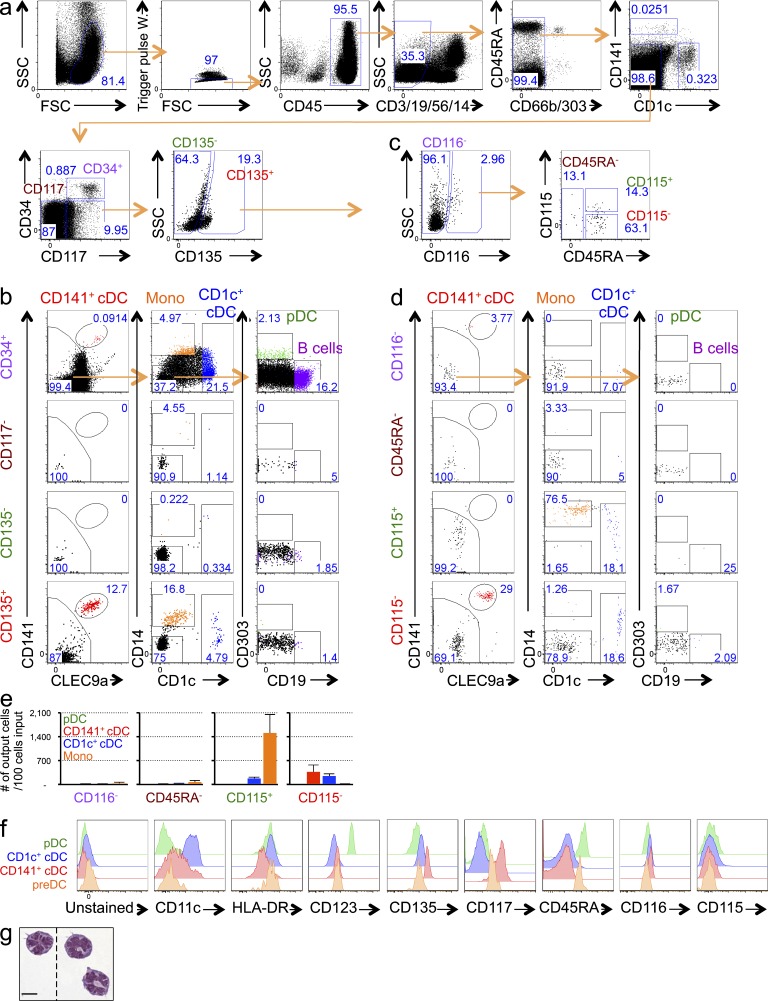
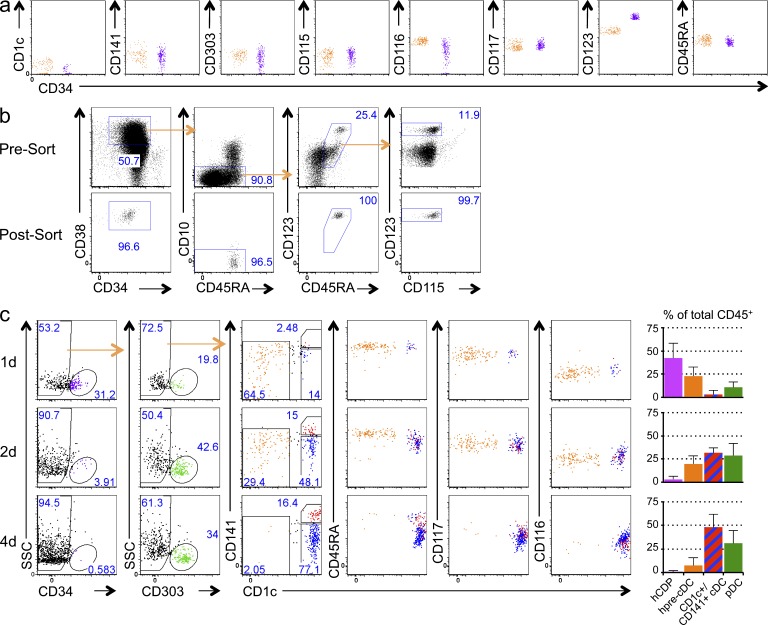
To try to identify hpre-CDCs in cord blood, we excluded terminally differentiated lymphoid cells (CD19+CD3+CD56+), granulocytes (CD66b+), monocytes (CD14+), pDCs (CD303/BDCA2+), and cDCs (CD1c+ and CD141+). The remaining CD45+ cells were subdivided into 4 groups by expression of CD34, CD117, and CD135, as follows: CD34+CD117+ (CD34+), CD34−CD117− (CD117−), CD34−CD117+CD135− (CD135−), and CD34−CD117+CD135+ (CD135+; Fig. 2 a). Pools of 1,000 purified cells were cultured on MS5 stromal cells with Flt3L, SCF, and GM-CSF cytokines (MS5+FSG herein) under conditions that support DC development in vitro (Lee et al., 2015). Only CD34+CD117+ and CD34−CD117+CD135+ cells produced identifiable progeny (Fig. 2 b). As expected, CD34+CD117+ cells, which should contain the least committed progenitors because they retain CD34 expression, developed into monocytes, granulocytes, pDCs, CD1c+ and CD141+ cDCs, and B cells, indicating that they are indeed multipotent (Fig. 2 b and not depicted). In contrast, CD34−CD117+CD135+ cells were restricted to CD1c+ and CD141+ cDCs and monocytes, suggesting that this pool of cells contains the immediate precursor to human cDCs and monocytes (Fig. 2 b). To further purify the hpre-CDC, we fractionated the CD34−CD117+CD135+ pool based on expression of CD116, CD115, and CD45RA (Chicha et al., 2004; Doulatov et al., 2010) and assayed their potential in MS5+FSG cultures (Fig. 2 c). Whereas CD116+CD45RA+CD115+ cells produced primarily CD14+CD1c− monocytes, a small number of CD1c+CD14+ cDCs (Patterson et al., 2005; Granelli-Piperno et al., 2006), and no CD141+ cDCs; CD116+CD45RA+CD115− cells yielded primarily CD1c+ and CD141+ cDCs and only rare monocytes and pDCs (Fig. 2 d). These results were reproducible in multiple different human donors (Fig. 2 e). Therefore, CD116+CD45RA+CD115− cells show the restricted developmental potential expected of hpreDCs and CD116+CD45RA+CD115+ cells of premonocytes. hpre-CDCs express high HLA-DR, an intermediate level of CD123, and a low level of CD11c (Fig. 2 f). Morphologically, hpre-CDCs are mononuclear cells that present a distinctive formation of multilobulated nucleus and a few veiled processes but do not possess the membrane extensions typically associated with cDCs (Fig. 2 g).
Figure 2.
Screening of populations in the cord blood for committed cDC lineage potential. (a) Flow cytometry plots show gating of CD45+CD3−CD19−CD56−CD14−CD66b−CD1c−CD141−CD303− cells in human cord blood can be divided into four populations based on CD117, CD34, and CD135: CD34+CD117+ (CD34+), CD34−CD117− (CD117−), CD34−CD117+CD135− (CD135−), and CD34−CD117+CD135+ (CD135+). Numbers indicate the frequency of respective gates. (b) Differentiation potential of 1,000 purified cells from each of 4 populations indicated in (a) in MS5+FSG cultures for 7 d. Flow cytometry plots show gated live human CD45+ cells. (c) Flow cytometry plots show CD135+ cells indicated as in (a) can be further separated into 4 populations based on CD116, CD115, and CD45RA: CD135+CD116− (CD116−), CD135+CD116+CD45RA−(CD45RA−), CD135+CD116+CD45RA+CD115+ (CD115+), and CD135+CD116+CD45RA+ CD115− (CD115−). (d and e) Differentiation potential of 100 purified cells from each of 4 populations indicated in (c) in MS5+FSG cultures for 7 d. (d) Flow cytometry plots of CD45+ cells gated as in (c), showing expression of CD141, CLEC9a, CD1c, and CD19. (e) Graphs show mean output of pDC, CD1c+ cDC, CD141+ cDC and monocytes from each population from three independent experiments. (f) Histograms show expression of CD11c, HLA-DR, CD123, CD135, CD117, CD45RA, CD116, and CD115 on indicated cell-type. (g) Morphology of purified cord blood hpre-CDCs by Giemsa staining of cytospin preparations (100×). Dotted lines indicate cropping out of white space between cells. Bar, 10 µm.
hpre-CDCs in adult blood and lymphoid organs
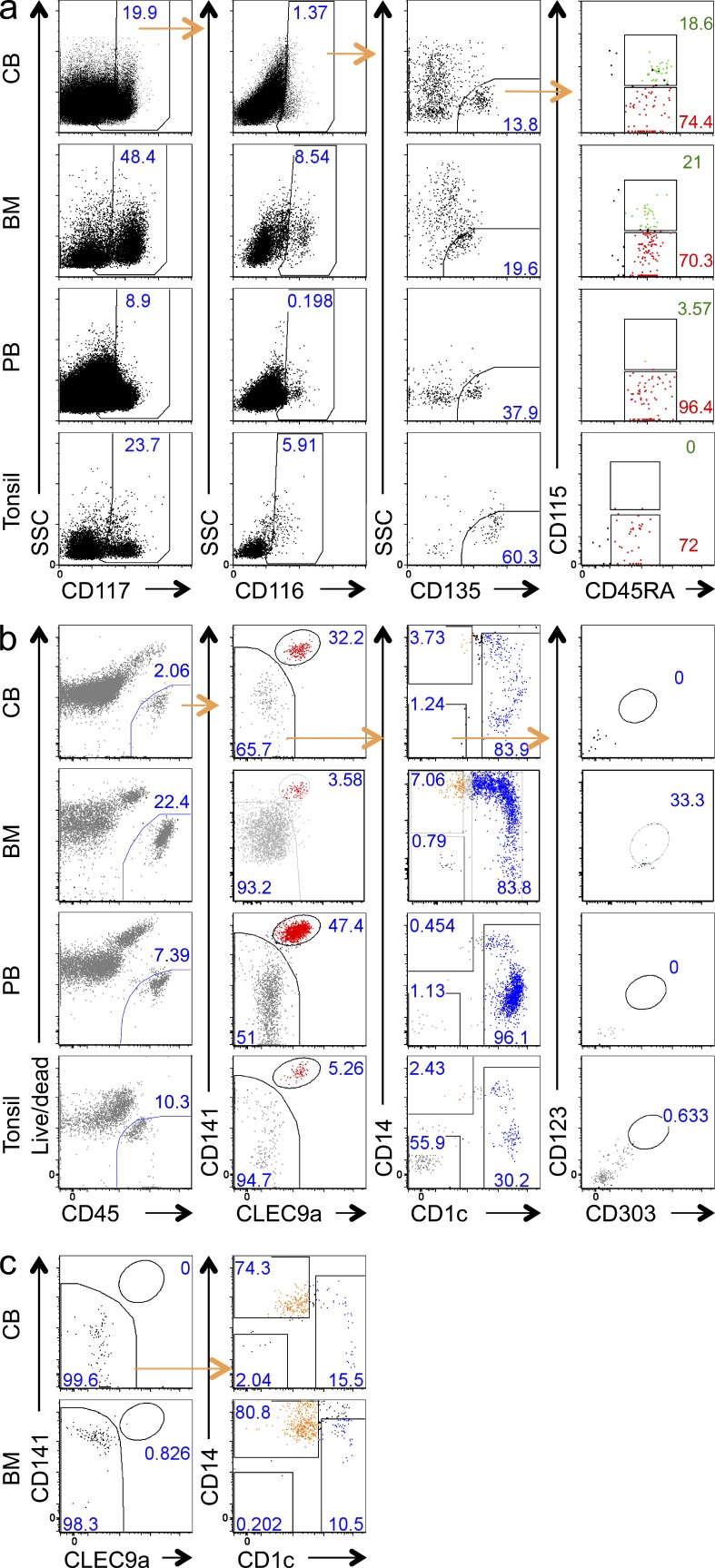
In the mouse, pre-CDCs are found in circulation and in tissues. To determine the physiological distribution of hpre-CDCs in adult humans, we examined BM, peripheral blood, and tonsils using the same cocktail of surface markers used to analyze cord blood. Lin−CD34−CD117+CD135+ cells in the BM resembled those in the cord blood and contained CD116+CD115− hpre-CDC-like cells and CD116+CD115+ premonocyte-like cells (Fig. 3 a). These hpre-CDC-like (Lin−CD34−CD117+CD135+CD116+CD115−) cells were also detected in the peripheral blood and the tonsil. In contrast, the premonocyte-like (Lin−CD34−CD117+CD135+CD116+CD115+) cells are lacking in the peripheral blood and tonsil (Fig. 3 a). To determine the developmental potential of the hpre-CDC-like cells in different tissues, we purified them and cultured them under conditions that support DC development. Although occasional hpre-CDCs remained undifferentiated, all cultures, irrespective of hpre-CDC origin from cord blood, BM, blood, or tonsils, predominantly produced CD1c+ and CD141+ cDCs and few, if any, CD14+CD1c− monocytes (Fig. 3 b). CD1c+ cDCs produced in our culture system can express some CD14, but gene array data show that these CD1c+ CD14+ cells are cDCs and not monocytes (Lee et al., 2015). In contrast, the CD115+CD116+ premonocytes in the BM, which are absent from the blood and tonsils, produced primarily monocytes and no CD141+ cDCs (Fig. 3 c). Cells showing the hpre-CDC phenotype constitute 0.008% (range, 0.001–0.016%) of the CD45+ cells in cord blood, 0.117% (range, 0.056–0.188%) in the BM, 0.001% (range of 0.001–0.002%) in the peripheral blood, and 0.001% (range, 0.001–0.002%) in the tonsil (Fig. 3 a). Thus, whereas hpre-CDCs originate in the BM and travel through the blood to seed lymphoid tissues, the monocyte precursor is retained in the BM.
Figure 3.
hpre-CDCs in human lymphoid organs and blood. (a) Representative flow cytometry plots of gated Lin(CD3/19/56/14/66b)−DC(CD1c/141/303)−CD34− cells show gating strategy and composition of hpre-CDCs (SSCloCD117+CD116+CD135+ CD45RA+CD115−, red) and premonocytes (SSCloCD117+CD116+CD135+ CD45RA+CD115+, green) in human cord blood (CB), BM, peripheral blood (PB), and tonsil. Numbers indicate frequency of cells of parent gate (CB, n = 5; BM, n = 4, PB, n = 5; Tonsil, n = 3; n, number of donors). (b) Differentiation potential of purified hpre-CDCs indicated in (a) in MS5+FSG cultures for 5 d. Flow cytometry plots of gated live human CD45+ cells show culture output of CD141+ cDC (red) and CD1c+ cDC (blue). Representative results of four (CB), three (BM), three (PB), and two (tonsil) independent experiments are shown. (c) Differentiation potential of purified premonocytes from cord blood (CB) and BM in MS5+FSG cultures for 5 d. FACs plots show phenotype of gated live human CD45+ culture-derived cells including monocytes (orange) and CD1c+ cDCs (blue). Representative of three independent experiments are shown.
Proliferative capacity and clonal potential of hpre-CDC
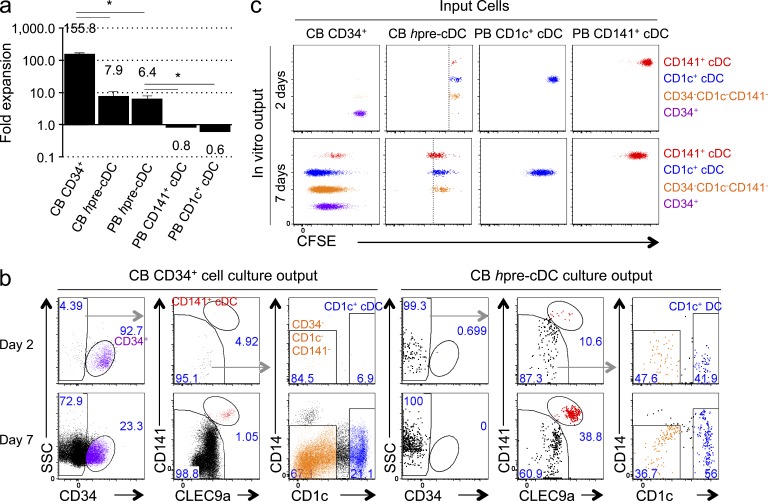
To examine the proliferative potential of hpre-CDC, we purified them from cord or peripheral blood and compared their developmental potential to CD34+ hematopoietic stem and progenitor cells (HSPCs) purified from cord blood in MS5+FSG culture (Lee et al., 2015). Whereas CD34+ progenitors expanded 156-fold during the 7-d culture period, hpre-CDCs from cord blood or peripheral blood expanded only 8- and 6-fold, respectively (Fig. 4 a). The relatively low level of hpre-CDC expansion could result from low proliferative capacity or poor clonal efficiency. To determine their proliferative capacity, we loaded purified hpre-CDCs or CD34+ HSPCs with CFSE and documented division by dye dilution at days 2 and 7 of culture. On day 2, HSPCs remained CD34+ and produced neither CD1c+ nor CD141+ cDCs, whereas hpre-CDCs differentiated into CD1c+ and CD141+ cDCs (Fig. 4, b and c). On day 7, both CD34+ HSPC and hpre-CDC cultures contained CD1c+ and CD141+ cDCs (Fig. 4 b). CD34+ HSPCs divided up to 9 times in 7 d, whereas hpre-CDCs underwent a maximum of 4 divisions (Fig. 4 c). Comparison of CFSE dilution revealed that hpre-CDC-derived CD1c+ cDCs and CD141+ cDCs underwent at least one more division than hpre-CDCs in the culture (Fig. 4 c), indicating that cDCs are also capable of cell division. Consistent with this notion, purified blood CD1c+ and CD141+ cDCs also divided 3–4 times in culture (Fig. 4 c). Thus, hpre-CDCs have a more limited proliferative capacity than CD34+ HSPCs in vitro. Nevertheless, a single hpre-CDC may be able to give rise to as many as 256 cDCs.
Figure 4.
Proliferative capacity of hpre-CDCs. (a) Expansion of purified hpre-CDCs or CD34+ HSPCs from peripheral blood (PB) or cord blood (CB) in MS5+FSG cultures for 7 d. Graph shows the mean fold change of live human CD45+ cells from 100 input cells from three independent experiments. *, P < 0.005, unpaired two-tailed Student’s t test. (b and c) CD34+ HSPCs and hpre-CDCs were purified from CB, labeled with CFSE, and cultured in MS5+FSG for 2 or 7 d; proliferation was assessed by flow cytometry. FACs plots show (b) gated CD45+ culture-derived cells, including CD34+ cells (purple), CD34−CD1c−CD141− cells (orange), CD141+ cDCs (red), and CD1c+ cDCs (blue) and (c) their CFSE dilution. Dotted lines mark last division by hpre-CDCs. Plots are representative of three independent experiments.
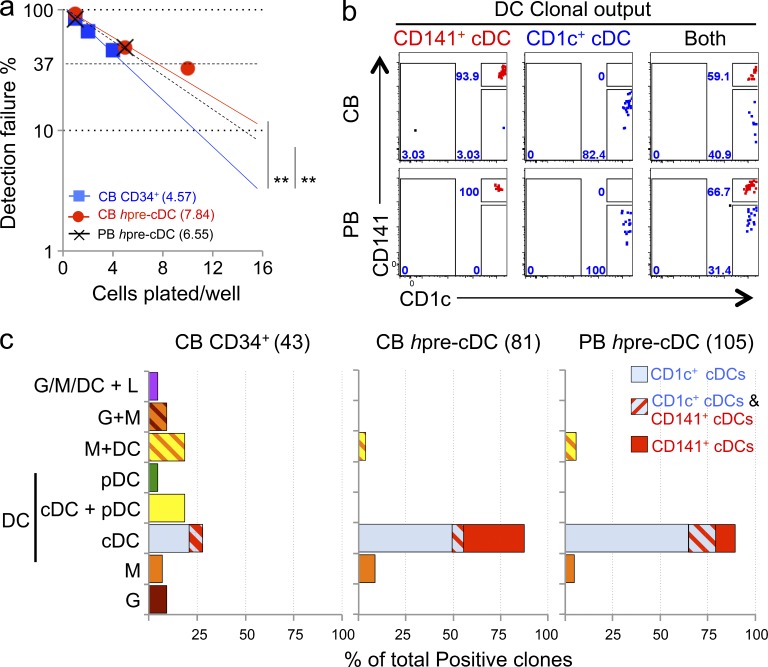
To determine the clonal efficiency of hpre-CDCs, we compared them to CD34+ HSPCs and progenitors in limiting dilution assays. Whereas 1 in 4.57 CD34+ cells produced CD45+ cells (i.e., granulocytes, monocytes, DCs, or lymphoid cells), 1 in 7.84 and 1 in 6.55 hpre-CDCs from cord blood or peripheral blood, respectively, produced CD45+ cells during the culture period (Fig. 5 a). This is in line with our observation that some hpre-CDCs remained undifferentiated in our cultures (Fig. 3 b). Although CD34+ HSPCs had higher clonal potential, only 12 out of 43 (28%) were able to produce cDCs (Fig. 5 c and Fig. S1), and both cord blood and peripheral blood hpre-CDCs generated mainly cDCs (Fig. 5 c, Fig. S2, and Fig. S3). Individual hpre-CDCs produced either CD1c+ cDCs or CD141+ cDCs or both (Fig. 5 b). Of 81 cells assayed, ∼88% of all productive clones of cord blood hpre-CDCs produced cDCs; 32% yielded only CD141+ cDCs, 50% only to CD1c+ cDCs, and 6% produced both CD1c+ and CD141+ cDCs (Fig. 5 c and Fig. S2). Of 105 cells assayed, 90% of hpre-CDCs from the PBMC produced only cDCs; 11% gave rise to CD141+ cDCs only, 65% only to CD1c+ cDCs, and 14% produced both CD1c+ and CD141+ cDCs (Fig. 5 c and Fig. S3). This demonstrates that hpre-CDCs purified from peripheral or cord blood have clonal efficiencies and differentiation potential similar to CD1c+ and CD141+ cDCs (Fig. 5 c). Only a very small fraction of hpre-CDCs produced monocytes alone or monocytes and DCs (Fig. 5 c; 11.5 and 10.5% from cord blood and peripheral blood, respectively). Thus, the hpre-CDC population in peripheral and cord blood contains single cells with potential to differentiate into both major subsets of human cDCs.
Figure 5.
Clonal efficiency and potential of hpre-CDC. (a) Limiting dilution outgrowth assay show clonal efficiency of CD34+ HSPCs (n = 3) or hpre-CDCs from peripheral (PB, n = 4) and cord blood (CB, n = 3) in MS5+FSG cultures for 5 d (for hpre-CDCs) or 14 d (for CD34+ HSPCs; Materials and methods). **, P < 0.05, pairwise test by ELDA. (b) Representative flow cytometry plots of gated CD45+ cells derived from single hpre-CDC clones indicated in (a) show output of CD1c+ cDCs (blue) and CD141+ cDCs (red). (c) Graphs summarize the lineage output of single clones from CD34+ HSPCS, and hpre-CDCs from cord (CB) or peripheral blood (PB). G, granulocyte; M, monocyte; L, lymphocytes.
Progenitors of hpre-CDCs
We have identified a common DC progenitor in BM and cord blood that produces pDCs and cDCs (hCDP; Lee et al., 2015). To determine whether there exists a progenitor–progeny relationship between the hCDP and hpre-CDC (Fig. 6 a), we purified hCDPs from cord blood (Fig. 6 b) and tested whether they differentiate into hpre-CDCs in MS5+FSG culture. hCDPs down-regulated CD34 as early as 1 d after culture and diverged into two populations: one group of cells up-regulated CD303 and assumed a pDC phenotype, the other group was CD303− but expressed CD45RA, CD117, and CD116 resembling hpre-CDCs. The number of hpre-CDCs peaked at day 2, and this was followed by increase of CD1c+ cDCs and CD141+ cDCs (Fig. 6 c). We conclude that hCDPs lose CD34 expression and give rise to hpre-CDCs and pDCs in culture.
Figure 6.
hpre-CDCs descend from hCDPs. (a) Flow cytometry plots show comparison of hCDP (purple) and hpre-CDC (orange) in cord blood. (b) Flow cytometry plots of gated CD45+Lin(CD3/19/56/14)−CD34+ show phenotype and purity of magnetically enriched cord blood (CB) CD34+ cells (presorting, upper) and sorted cells (post-sorting, bottom). hCDPs were gated as CD34+CD38hiCD45RA+CD10−CD123hi. (c) Differentiation kinetics of hCDPs purified from CB in MS5+FSG cultures for 1, 2, or 4 d. FACs plots show culture output of live human CD45+ cells, including CD34+ cells (purple), pDC (green), CD1c+ cDC (blue), CD141+ cDCs (red), and hpre-CDCs (orange). Representative of four independent experiments are shown. Graphs summarize composition of indicated populations among total hCD45+ cells. Bars are mean values from four independent experiments, and error bars are SEM.
Flt3L injection induces expansion of hpre-CDCs in human blood
Flt3L stimulates DC hematopoiesis (Karsunky et al., 2003) and is an acute phase reactant that can be elevated in infection in both mice and humans, accounting for DC expansion in patients with malaria infection and in volunteers injected with Flt3L (Maraskovsky et al., 2000; Pulendran et al., 2000; Guermonprez et al., 2013). In humans, Flt3L injection is associated with a rapid increase in circulating DCs and in CD11c−CD123+ cells that produce large amounts of IFN-α when cultured with influenza virus (Pulendran et al., 2000). The latter were initially thought to be DC progenitors but were subsequently shown to be pDCs (Siegal et al., 1999; Liu, 2005).
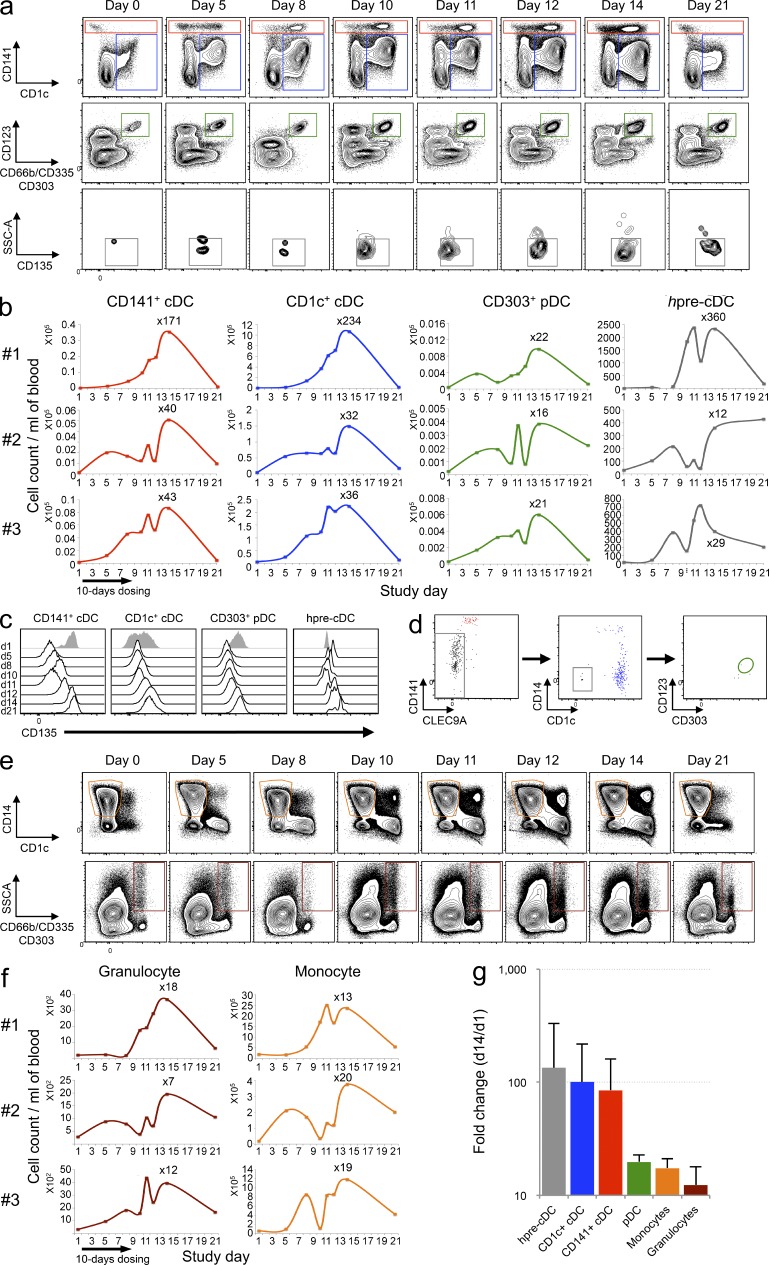
To determine whether Flt3L mobilizes DC progenitors into the blood, we made use of samples from three volunteers subcutaneously injected with 25 µg/kg of Flt3L for 10 consecutive days. 14-color flow cytometry was used to assess the number of cDCs, pDCs, and hpre-CDCs (Fig. S4). The number of circulating cDCs increased on day 5 and peaked on day 14 after the initial injection (Fig. 7, a and b). The degree of cDC expansion varied with a 32–234-fold increase in CD1c+ cDCs, a 40–171-fold increase in CD141+ cDCs, and 16–22-fold increase in pDCs in the blood (Fig. 7 b). Similar to CD141+ cDCs obtained from MS5+FSG culture (Lee et al., 2015), Flt3L-induced CD141+ cDCs express variable levels of CD1c, and all DC subtypes in Flt3L-injected individuals down-regulated expression of the Flt3L receptor, CD135 (Fig. 7 c; Waskow et al., 2008). Thus, as others have found, both subtypes of cDCs and pDCs increase in the blood in response to Flt3L injection (Pulendran et al., 2000).
Figure 7.
Flt3L administration increases circulating DC subsets and hpre-CDCs in humans. PBMCs from Flt3L-treated volunteers (n = 3; 25 µg/kg for 10 consecutive days) were analyzed by flow cytometry over a 21-d period to assess the expansion of DC subsets (CD1c+ cDCs [blue], CD141+ cDCs [red], and pDCs [green]; hpre-CDCs (gray); monocytes (orange); and granulocytes (brown). (a) Representative flow cytometry dot plots show DC subsets and hpre-CDCs in blood (gating strategy in Fig. S4). (b) Graphs show the kinetics of cell number of cDC subsets, pDCs, and hpre-CDCs over 21 d in 3 Flt3L-treated volunteers. The absolute numbers per milliliter of blood were obtained by multiplying the number of cells (obtained by flow cytometry) by the total number of PBMCs per milliliter of blood. (c) Representative histograms show CD135 expression on CD141+ cDCs, CD1c+ cDCs, pDCs and hpre-CDCs over 21 d in one Flt3L-treated volunteer. (d) Differentiation potential of purified hpre-CDCs from blood of Flt3L-injected volunteers in MS5+FSG cultures after 7 d. Flow cytometry plots of gated CD45+ cells from culture show output of CD141+ cDCs (red), CD1c+ cDCs (blue), and lack of pDCs (green gate). (e) Representative flow cytometry dot plots show CD14+ monocytes and CD66b+ granulocytes in blood (gating strategy in Fig. S4). (f) Graphs show the kinetics of cell number of monocytes and granulocytes over 21 d in 3 Flt3L-treated volunteers. The absolute numbers per milliliter of blood were obtained by multiplying the number of cells (obtained by flow cytometry) by the total number of PBMCs per milliliter of blood. (g) Graph showing the mean fold change increase from the three patients (d1 vs. d14) of hpre-CDCs (gray), CD1c+ cDCs (blue), CD141+ cDCs (red), pDCs (green), monocytes (orange), and granulocytes (brown) per milliliter of blood. Error bars are SDs.
Like the more differentiated DCs, hpre-CDC also increased in the blood of the same patients, starting on day 5 but peaking earlier, on days 11–12 after injection (Fig. 7, a and b). Flt3L-expanded hpre-CDCs were similar to their uninduced counterparts and cord blood derived hpre-CDCs in producing CD1c+ and CD141+ cDCs in MS5+FSG culture (Fig. 7 d). In contrast, the immediate precursors of hpre-CDCs, hCDPs, and more distant progenitors such as hGMDPs or hMDPs were undetectable in the peripheral blood of individuals injected with Flt3L at any time point examined (Fig. S4). We conclude that Flt3L expands human DCs and their progenitors and recruits hpre-CDCs, but not earlier DC progenitors, into the circulation.
Other myeloid populations, such as monocytes and granulocytes also expand in response to Flt3L, starting on day 5 and peaking on day 14 after the initial injection (Fig. 7, e and f). Interestingly, comparison of the fold change at day 14 indicates that Flt3L preferentially increases the number of human cDCs and their progenitors rather than the other myeloid subsets, i.e., pDCs, monocytes, and granulocytes in blood (Fig. 7 g).
DISCUSSION
DCs turn over in tissues and must be replaced continuously to maintain homoeostasis (Liu et al., 2007; Liu and Nussenzweig, 2010). In the mouse, lymphoid and nonlymphoid tissue resident DCs are continually replenished by pre-CDCs, which are produced in the BM, enter the circulation and then emigrate into tissues to differentiate into both major subsets of cDCs (Naik et al., 2006; Bogunovic et al., 2009; Ginhoux et al., 2009; Liu et al., 2009; Varol et al., 2009). Defining the mouse pre-CDC was essential to establishing that DCs represent a unique cell lineage because pre-CDCs do not produce monocytes, or pDCs; their differentiation potential is restricted to cDCs.
Human DC development has been difficult to delineate, in part because of limited access to human tissues and limited tissue culture methods. Earlier human trials showed increased numbers of DCs in the blood after Flt3L injection, and suggested that a CD11c−CD123+ IFN-α producing cell might be a DC progenitor (Pulendran et al., 2000). However, progenitor potential was never tested directly and it is likely that those cells represent pDCs, which are CD123+ and induced by Flt3L injection (Fig. 7 a). More recently, circulating cDCs were shown to be less mature than their tissue-derived counterparts and it has been proposed that these less mature cells are the precursors of tissue cDCs (Collin et al., 2011; Segura et al., 2012; Ginhoux and Schlitzer, 2014). Our experiments identify a human pre-CDC that resides in the BM, peripheral blood, and peripheral lymphoid organs that is the direct progenitor of both major subsets of human cDCs. Whether the hpre-CDC differentiates into cDCs in the blood or tissues or both has yet to be determined.
The cell surface markers used to define mouse DCs and their progenitors are not entirely conserved by their human counterparts. For example, lack of cell surface MHCII expression, one of the key features that distinguishes mouse pre-CDCs from fully differentiated DCs, cannot be used to define human DC progenitors because MHCII is expressed on the surface of most early human hematopoietic cells. Similarly, CD11c used as marker to identify pre-CDCs in the mouse is expressed at low levels on hpre-CDCs (Fig. 2 f) and should not be used to identify this cell in humans. However, both human and mouse pre-CDCs express CD135. This receptor marks the entire DC pathway in both species from the earliest BM precursor to the fully differentiated cDCs in tissues (Chicha et al., 2004; Fogg et al., 2006; Naik et al., 2007; Onai et al., 2007; Waskow et al., 2008; Doulatov et al., 2010; Lee et al., 2015).
In the mouse, DC progenitors can be distinguished from other cells based on increasingly restricted expression of cytokine receptors (Karsunky et al., 2003; Onai et al., 2007; Waskow et al., 2008; Liu et al., 2009). We applied this principle to identify the hpre-CDC and made use of differential expression of CD116 (GM-CSF receptor), CD135 (Flt3L receptor), CD115 (M-CSF receptor), CD117 (SCF receptor), and CD45RA (marker for DC progenitors) on monocytes, cDCs, and pDCs to uncover the hpre-CDC, which is Lin−CD34−CD117+CD135+CD115−CD116+CD45RA+. In contrast, otherwise identical cells that are CD115+ appear to be the immediate precursors of monocytes, resembling the mouse common monocyte progenitor (cMOP), in its differentiation potential as well as in its absence in the circulation (Hettinger et al., 2013).
hpre-CDCs enter the circulation and emigrate into tissues as incompletely differentiated progenitors. In the mouse, this feature of the cDC lineage enables pre-CDC differentiation into specialized subsets in different tissues. We speculate that similar tissue specific heterogeneity in cDC subtypes will also be found in human. In contrast, the closely related human monocyte progenitor is restricted to the BM (Fig. 3). Similar to monocytes, pDCs enter the circulation as fully differentiated effector cells.
Like all cDCs and their early progenitors, hpre-CDCs express CD135, the receptor for Flt3L. Our experiments show that like more mature cDCs (Maraskovsky et al., 1997; Pulendran et al., 2000), hpre-CDCs are mobilized into the blood by Flt3L injection. However, the immediate precursor of hpre-CDCs, hCDPs, are not. Flt3L levels in serum increase in response to DC depletion in mice and are also increased in humans that lack cDCs and other myeloid cells due to GATA2 mutation (Dickinson et al., 2014). In addition, levels of this hematopoietin are also increased in the serum of humans infected with Plasmodium falciparum, and in mice infected with Plasmodium chaubodii or Plasmodium yoelli or cytomegalovirus (Eidenschenk et al., 2010; Guermonprez et al., 2013). Moreover, in humans infected with P. falciparum, increased serum Flt3L is associated with an increase in the number of circulating cDCs. Thus, cDC levels in humans are regulated in part by hpre-CDC recruitment from the BM in response to Flt3L.
In both humans and mice, pre-CDCs constitute a very small population of cells in the peripheral blood (0.001 and 0.02%, respectively). Although present in only small numbers, the flux of pre-CDCs through the blood stream is remarkably high due to their short residence time in circulation. In the mouse, 65% of pre-CDCs are cleared from the circulation within 1 min after entering the blood, indicating a t1/2 < 1 min (Liu et al., 2007, 2009). Thus, a cell with a half-life in circulation of 24 h would have to be present in circulation at a 1,440-fold higher concentration than the pre-CDC to achieve the same overall flux. Similar behavior has been demonstrated for HSCs in the circulation (Wright et al., 2001). Like their mouse counterparts, hpre-CDCs have limited expansion potential (Fig. 4 a; Liu et al., 2009), but their immediate progeny, CD1c+ and CD141+ cDCs, can proliferate to further expand the DC pool in the periphery (Fig. 4 c; Kabashima et al., 2005; Liu et al., 2009; KC et al., 2014). We speculate that this highly dynamic pool of specialized antigen-presenting cells allows rapid adaptation to acute antigenic challenges.
MATERIALS AND METHODS
Cell samples.
Human umbilical cord blood and leukophoretic peripheral blood (buffy coat) were purchased from New York Blood Center. Human BM was obtained from total hip arthroplasty at Hospital for Special Surgery (New York, NY). Tonsils were obtained from routine tonsillectomies performed at the Babies and Children’s Hospital of Columbia-Presbyterian Medical Center (New York, NY). Informed consent was obtained from the patients and/or samples were exempt from informed consent being residual material after diagnosis and fully de-identified. All samples were collected according to protocols approved by the Institutional Review Board at Columbia University Medical Center and The Rockefeller University (New York, NY). The specimens were kept on ice immediately after surgical removal. Tonsil samples were minced, treated with 400 U/ml collagenase (Roche) at 37°C for 20 min, and put into cell isolation. BM samples were preserved in solution containing 1000 U/ml heparin (National Drug Code #63323-540-11) until digestion. The samples were then digested in RPMI containing 20 mg/ml collagenase IV (#C-5138; Sigma-Aldrich) for 15 min at 37°C. After density centrifugation using Ficoll-Hypaque (GE Healthcare), aliquots of mononuclear BM cells were frozen and stored in liquid nitrogen for future analysis.
Cell isolation and flow cytometry.
Fresh mononuclear cells were isolated by density centrifugation using Ficoll-Hypaque (GE Healthcare). Samples from cord blood, peripheral blood, BM, and tonsil were incubated with fluorescent-labeled antibodies for direct analysis on the BDLSR II flow cytometers (BD) or further purification by fluorescence-activated cell sorting on the BD FACSAria or Influx, both using HeNe and argon lasers.
For isolation of rare hpre-CDCs from cord blood and peripheral blood, an enrichment step was performed before sorting. In brief, mononuclear cells were incubated with antibodies against CD135 (4G8; PE; BD) and CD117 (A3C6E2; Biotin; BioLegend) for 40 min at 4°C. After washing, antibody against PE (PE001; Biotin; BioLegend) was added and incubated for another 10 min at 4°C. After washing, CD117+ and CD135+ cells were positively selected using anti-biotin MicroBeads and LS MACS magnetic columns (Miltenyi Biotec). For sorting hpre-CDCs, enriched cells (from CB or PB) or total mononuclear cells (from CB, PB, BM, or tonsils) were stained for CD14 (TuK4; Qdot-655; Invitrogen), CD3 (OKT3; Brilliant Violet [BV] 650; BioLegend), CD19 (HIB19; BV650; BioLegend), CD56 (HCD56; BV650; BioLegend), CD66b (G10F5; PerCP-Cy5.5; BioLegend), CD303 (201A; PerCP-Cy5.5; BioLegend), CD1c (L161; APC-Cy7; BioLegend), CD141 (M80; PE-Cy7; BioLegend), CD34 (581; Alexa Fluor 700; BioLegend), CD117 (104D2; BV421; BioLegend), CD135 (4G8; PE; BD), CD45RA (HI100; BV510; BioLegend), CD116 (4H1; FITC; BioLegend), and CD115 (9-4D2-1E4; APC; BioLegend) for 40 min on ice. hpre-CDCs were isolated as Lin(CD3/19/56/14)−Granulocyte(CD66b)−pDC(CD303)−cDC(CD1c/CD141)−CD34−CD117+CD135+SSClo CD116+CD115−CD45RA+.
In addition to these antibodies, for surface marker phenotype characterization of fully differentiated cells, CD45 (HI30; Alexa Fluor 700; BioLegend), CD34 (581; APC-Cy7; BioLegend), CD3 (UCHT1; Biotin; BD), CD19 (HIB19; Biotin; BD), CD56 (B159; Biotin; BD), CD115 (12-3A3-1B10; PE; eBioscience), CD123 (9F5; PE; BD) and CD45RA (12–0458-42; eBioscience; PE) were used alternatively, followed by a secondary stain with streptavidin-conjugated PerCP-Cy5.5 for 40 min on ice.
Cultured cells were harvested and stained with LIVE/DEAD (Life Technologies), CD45 (AF700), CD56 (B159; Pacific Blue; BD), CD66b (G10F5; PerCP-Cy5.5; BioLegend), CD19 (HIB19; APC-Cy7; BioLegend), CLEC9a (8F9; PE; BioLegend), CD14 (Qdot-655), CD1c (L161; PE-Cy7; BioLegend), CD303 (201A; FITC; BioLegend), CD123 (6H6; BV510; BioLegend), CD141 (AD5-14H12; APC; Miltenyi Biotec) for 40 min on ice and analyzed for lineage potential by flow cytometry. Absolute cell counts have been calculated relative to a well containing a known number of CD45+ cells (i.e., 10,000 cells), which were added on the day of harvest.
For CFSE assays, cultured cells were stained with LIVE/DEAD (Life Technologies), CD45 (AF700), CD34 (APC-Cy7), CLEC9a (PE), CD14 (Qdot-655), CD1c (PE-Cy7), and CD141 (APC) for 40 min on ice.
For progenitor–progeny relationship experiments, CD34+ cells were first enriched from cord blood using CD34 MicroBead kit and LS MACS magnetic columns (Miltenyi Biotec). Enriched CD34+ cells (40–95% purity) were then incubated with Lin (CD3/19/56/14; BV650), CD34 (AF700), CD38 (HIT2; BV421; BioLegend), CD10 (HI10a; PE-Cy7; BioLegend), CD45RA (BV510), CD123 (PE), CD116 (FITC), and CD115 (APC). hCDPs were sorted as Lin−CD34+CD38hiCD10−CD45RA+CD123hiCD115−. Cultured cells were harvested at specific time points and stained with LIVE/DEAD, CD45 (AF700), CD14 (Qdot-655), CD3 (BV650), CD19 (BV650), CD56 (BV650), CD303 (FITC), CD1c (PE-Cy7), CD141 (APC), CD34 (APC-Cy7), CD117 (BV421), CD123 (PE), CD45RA (BV510), and CD116 (FITC) for 40 min on ice.
Morphological analysis.
Purified hpreDCs were analyzed by Giemsa staining of cytospin preparations. As few as 5 × 104 cells were cytospun for 5 min at 800 rpm on a glass slide and then stained with the Hemacolor stain kit as recommended by the manufacturer (HARLECO). Slides were then imaged on an Axioplan 2 microscope at 100× magnification (Carl Zeiss).
Cell culture.
MS5 BM stromal cells were maintained and passed in complete α-MEM medium (Invitrogen) with 10% FCS and penicillin/streptomycin (Invitrogen). 24 h before hpre-CDC culture, MS5 stromal cells were treated with 10 µg/ml of mitomycin C (Sigma-Aldrich) for 3 h, washed, and reseeded at 3.75 × 104 MS5 cells per well in 96-well plates. Purified populations were seeded in medium containing 100 ng of Flt3L/ml (Celldex), 20 ng/ml SCF (PeproTech), and 10 ng/ml GM-CSF (PeproTech). Cells were harvested between day 1 and 14 for flow cytometry analysis.
To determine cellular divisions in culture, input populations were labeled for 15 min with 5 µM CFSE (Molecular Probes) at 37°C.
Limiting dilution assay.
For limiting dilution experiments, cells were sorted directly into 96-well plates containing MS5+FSG at 1, 2, 4, 8, or 16 cells per well for CD34+ HSPCs, and at 1, 5 or 10 cells per well for hpre-CDCs. The percentage of detection failure for cDCs was calculated as 100% × (1-k/n), where k is the number of positive wells and n the total number of wells combined from three or four independent experiments (CB and PB, respectively). We classified each well as positive if more than 5 cells were detected for cDCs or other lineages among live human CD45+ cells. Then, for every positive clone, we scored positive for cDCs or other lineages based on their respective gates to determine the clonal lineage potential. The frequency of DC-producing precursors was determined by “Loi de Poisson” using the Extreme Limiting Dilution Analysis software provided by the Walter and Eliza Hall Institute of Medical Research Bioinformatics (Parkville, Victoria, Australia.
Flt3L injection in volunteers.
3 healthy volunteers received a 25 µg/kg/day subcutaneous injection of CDX-301 (a clinical formulation of recombinant human Flt3L; Celldex) for 10 d. Blood samples were collected before and at 5, 8, 10, 11, 12, 14, and 21 d after initial administration. PBMCs were isolated from heparinized blood using Ficoll-Hypaque and stored in liquid nitrogen for further analysis.
PBMCs were stained with CD116 (4H1; FITC; BioLegend), CD135 (4G8; PE; BD), CD66b (G10F5; PerCP-Cy5.5; BioLegend), CD335 (9E2, PerCP-Cy5.5, BioLegend), CD303 (201A; PerCP Cy5.5; BioLegend), CD141 (M80; PE-Cy7; BioLegend), CD117 (104D2; BV421; BioLegend), CD123 (6H6; BV510; BioLegend), CD20 (2H7; BV605; BioLegend), CD3 (OKT3; BV605; BioLegend), CD45RA (MEM-56; Qdot 655; Life Technologies), LIVE/DEAD Fixable Blue Dead Cell Stain (Life Technologies), CD14 (Tük4; Qdot 800; Life Technologies), CD115 APC (9-4D2-1E4; APC; BioLegend), CD34 (581; Alexa Fluor 700; BioLegend), CD1c (L161; APC-Cy7; BioLegend). For surface staining, titrated antibodies were added to 2 million cells in 50 µl PBS for 20 min at 4°C.
Washed cells were fixed in 2% formaldehyde and stored at 4°C until analysis, which was performed using an LSR II flow cytometer (BD). The whole sample was acquired and analysis was performed using Flow Jo 9.1 software (Tree Star). hpre-CDCs were isolated as Lin(CD3/20/335/14)− Granulocyte(CD66b)−DC(CD303−CD1c/CD141)−CD34−CD117+CD135+CD116+ CD115−CD45RA+CD123+.
Online supplemental material.
Fig. S1 shows clonal output of cord blood CD34+ HSPCs. Fig. S2 shows clonal output of cord blood hpre-CDCs. Fig. S3 shows clonal output of peripheral blood hpre-CDCs. Fig. S4 shows 14-color gating strategy for identification of human hpre-CDCs in blood and that hCDPs, hMDPs, and hGMDPs are undetectable in the blood before and after Flt3L administration in healthy volunteers. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20141441/DC1.
Supplementary Material
Acknowledgments
This work was inspired by Dr. Ralph M. Steinman. We thank Klara Velinzon (Flow Cytometry Core Facility, Laboratory of Molecular Immunology, The Rockefeller University) for technical support with polychromatic flow cytometry sorting. We thank Heidi Schreiber for help with human BM protocol (Laboratory of Molecular Immunology, The Rockefeller University). We acknowledge the assistance of Arlene Hurley and the clinical team at the Rockefeller University Hospital. We also thank James Pring, Popi Sarma and Christine Trumpfheller for the management of the Flt3L patient samples (Laboratory of Cellular Physiology and Immunology, The Rockefeller University).
Research reported in this publication was supported by the Empire State Stem Cell Fund through New York State Department of Health Contract #C029562” (to K.Liu), Helmsley Foundation (to K. Liu), National Institute of Allergy and Infectious Diseases of National Institutes of Health under award numbers of AI101251 (to K. Liu) and U19AI111825 (to M.C. Nussenzweig), National Institute of Neurological Disorder and Stroke of NIH under award number of NS084776 (to S. Puhr), Iris and Junming Le Foundation (to G.Breton), the Dermatology Foundation (to N. Anandasabapathy), National Institute of Arthritis and Musculoskeletal and Skin Diseases AR063461-01A1 (to N. Anandasabapathy), and CTSA, RUCCTS grant no. UL1RR024143 from the National Center for Research Resources, NIH, and by NIH grant AI13013. The Rockefeller University Center for Clinical and Translational Science is supported, in part, by a Clinical and Translational Science Award (CTSA), and the National Center for Advancing Translational Sciences (NCATS), part of the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Empire State Stem Cell Board, the NY State Department of Health or the State of New York or any other applicable federal funding agency. M.C. Nussenzweig is an HHMI Investigator.
T. Keler works for Celldex Therapeutics. The authors declare no additional financial interests.
Footnotes
Abbreviation used:
- cDCs
- conventional DCs
- CDP
- common dendritic progenitor
- pre-CDC
- cDC precursor
- MS5+FSG
- MS5 stromal cells with Flt3L, SCF, and GM-CSF cytokines
References
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., and Steinman R.M.. 1998. Dendritic cells and the control of immunity. Nature. 392:245–252 10.1038/32588 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicha L., Jarrossay D., and Manz M.G.. 2004. Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J. Exp. Med. 200:1519–1524 10.1084/jem.20040809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn L., Chatterjee B., Esselborn F., Smed-Sörensen A., Nakamura N., Chalouni C., Lee B.C., Vandlen R., Keler T., Lauer P., et al. 2013. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J. Exp. Med. 210:1049–1063 10.1084/jem.20121251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M., Bigley V., Haniffa M., and Hambleton S.. 2011. Human dendritic cell deficiency: the missing ID? Nat. Rev. Immunol. 11:575–583 10.1038/nri3046 [DOI] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Contreras V., Feuillet V., Dutertre C.A., Ventre E., Vu Manh T.P., Baranek T., Storset A.K., Marvel J., et al. 2010. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 207:1283–1292 10.1084/jem.20100223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R.E., Milne P., Jardine L., Zandi S., Swierczek S.I., McGovern N., Cookson S., Ferozepurwalla Z., Langridge A., Pagan S., et al. 2014. The evolution of cellular deficiency in GATA2 mutation. Blood. 123:863–874 10.1182/blood-2013-07-517151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Eppert K., Nguyen L.T., Ohashi P.S., and Dick J.E.. 2010. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11:585–593 10.1038/ni.1889 [DOI] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Park C.G., et al. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science. 315:107–111 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Eidenschenk C., Crozat K., Krebs P., Arens R., Popkin D., Arnold C.N., Blasius A.L., Benedict C.A., Moresco E.M., Xia Y., and Beutler B.. 2010. Flt3 permits survival during infection by rendering dendritic cells competent to activate NK cells. Proc. Natl. Acad. Sci. USA. 107:9759–9764 10.1073/pnas.1005186107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., and Geissmann F.. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311:83–87 10.1126/science.1117729 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., and Schlitzer A.. 2014. CD11b+ DCs rediscovered: implications for vaccination. Expert Rev. Vaccines. 13:445–447 10.1586/14760584.2014.893196 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granelli-Piperno A., Shimeliovich I., Pack M., Trumpfheller C., and Steinman R.M.. 2006. HIV-1 selectively infects a subset of nonmaturing BDCA1-positive dendritic cells in human blood. J. Immunol. 176:991–998 10.4049/jimmunol.176.2.991 [DOI] [PubMed] [Google Scholar]
- Grouard G., Rissoan M.C., Filgueira L., Durand I., Banchereau J., and Liu Y.J.. 1997. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 185:1101–1111 10.1084/jem.185.6.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P., Helft J., Claser C., Deroubaix S., Karanje H., Gazumyan A., Darasse-Jèze G., Telerman S.B., Breton G., Schreiber H.A., et al. 2013. Inflammatory Flt3l is essential to mobilize dendritic cells and for T cell responses during Plasmodium infection. Nat. Med. 19:730–738 10.1038/nm.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.N., Malinarich F., Malleret B., et al. 2012. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 37:60–73 10.1016/j.immuni.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Collin M., and Ginhoux F.. 2013. Ontogeny and functional specialization of dendritic cells in human and mouse. Adv. Immunol. 120:1–49 10.1016/B978-0-12-417028-5.00001-6 [DOI] [PubMed] [Google Scholar]
- Hettinger J., Richards D.M., Hansson J., Barra M.M., Joschko A.C., Krijgsveld J., and Feuerer M.. 2013. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14:821–830 10.1038/ni.2638 [DOI] [PubMed] [Google Scholar]
- Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V., et al. 2010. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207:1247–1260 10.1084/jem.20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K., Banks T.A., Ansel K.M., Lu T.T., Ware C.F., and Cyster J.G.. 2005. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 22:439–450 10.1016/j.immuni.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Kamphorst A.O., Guermonprez P., Dudziak D., and Nussenzweig M.C.. 2010. Route of antigen uptake differentially impacts presentation by dendritic cells and activated monocytes. J. Immunol. 185:3426–3435 10.4049/jimmunol.1001205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky H., Merad M., Cozzio A., Weissman I.L., and Manz M.G.. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313 10.1084/jem.20030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KC W., Satpathy A.T., Rapaport A.S., Briseño C.G., Wu X., Albring J.C., Russler-Germain E.V., Kretzer N.M., Durai V., Persaud S.P., et al. 2014. L-Myc expression by dendritic cells is required for optimal T-cell priming. Nature. 507:243–247 10.1038/nature12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Breton G., Oliveira T.Y.K., Zhou Y.J., Aljoufi A., Puhr S., Cameron M.J., Sékaly R.-P., Nussenzweig M.C., and Liu K.. 2015. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 212:xxx–xxx 10.1084/jem.20141441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J.2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275–306 10.1146/annurev.immunol.23.021704.115633 [DOI] [PubMed] [Google Scholar]
- Liu K., and Nussenzweig M.C.. 2010. Development and homeostasis of dendritic cells. Eur. J. Immunol. 40:2099–2102 10.1002/eji.201040501 [DOI] [PubMed] [Google Scholar]
- Liu K., Waskow C., Liu X., Yao K., Hoh J., and Nussenzweig M.. 2007. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 8:578–583 10.1038/ni1462 [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., and Nussenzweig M.. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397 10.1126/science.1170540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E., Pulendran B., Brasel K., Teepe M., Roux E.R., Shortman K., Lyman S.D., and McKenna H.J.. 1997. Dramatic numerical increase of functionally mature dendritic cells in FLT3 ligand-treated mice. Adv. Exp. Med. Biol. 417:33–40 10.1007/978-1-4757-9966-8_6 [DOI] [PubMed] [Google Scholar]
- Maraskovsky E., Daro E., Roux E., Teepe M., Maliszewski C.R., Hoek J., Caron D., Lebsack M.E., and McKenna H.J.. 2000. In vivo generation of human dendritic cell subsets by Flt3 ligand. Blood. 96:878–884. [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497. [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., and Mortha A.. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M.2013. Transcriptional control of dendritic cell development. Adv. Immunol. 120:239–267 10.1016/B978-0-12-417028-5.00009-0 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Metcalf D., van Nieuwenhuijze A., Wicks I., Wu L., O’Keeffe M., and Shortman K.. 2006. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat. Immunol. 7:663–671 10.1038/ni1340 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Nizzoli G., Krietsch J., Weick A., Steinfelder S., Facciotti F., Gruarin P., Bianco A., Steckel B., Moro M., Crosti M., et al. 2013. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood. 122:932–942 10.1182/blood-2013-04-495424 [DOI] [PubMed] [Google Scholar]
- O’Doherty U., Peng M., Gezelter S., Swiggard W.J., Betjes M., Bhardwaj N., and Steinman R.M.. 1994. Human blood contains two subsets of dendritic cells, one immunologically mature and the other immature. Immunology. 82:487–493. [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., and Manz M.G.. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Patterson S., Donaghy H., Amjadi P., Gazzard B., Gotch F., and Kelleher P.. 2005. Human BDCA-1-positive blood dendritic cells differentiate into phenotypically distinct immature and mature populations in the absence of exogenous maturational stimuli: differentiation failure in HIV infection. J. Immunol. 174:8200–8209 10.4049/jimmunol.174.12.8200 [DOI] [PubMed] [Google Scholar]
- Poulin L.F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J.L., Keller A.M., Joffre O., Zelenay S., Nye E., et al. 2010. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 207:1261–1271 10.1084/jem.20092618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., Banchereau J., Burkeholder S., Kraus E., Guinet E., Chalouni C., Caron D., Maliszewski C., Davoust J., Fay J., and Palucka K.. 2000. Flt3-ligand and granulocyte colony-stimulating factor mobilize distinct human dendritic cell subsets in vivo. J. Immunol. 165:566–572 10.4049/jimmunol.165.1.566 [DOI] [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W., See P., Shin A., Wasan P.S., et al. 2013. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 38:970–983 10.1016/j.immuni.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Valladeau-Guilemond J., Donnadieu M.H., Sastre-Garau X., Soumelis V., and Amigorena S.. 2012. Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 209:653–660 10.1084/jem.20111457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., and Liu Y.J.. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science. 284:1835–1837 10.1126/science.284.5421.1835 [DOI] [PubMed] [Google Scholar]
- Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.D., Shakhar G., and Jung S.. 2009. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 31:502–512 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., and Nussenzweig M.. 2008. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9:676–683 10.1038/ni.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D.E., Wagers A.J., Gulati A.P., Johnson F.L., and Weissman I.L.. 2001. Physiological migration of hematopoietic stem and progenitor cells. Science. 294:1933–1936 10.1126/science.1064081 [DOI] [PubMed] [Google Scholar]
- Yu C.I., Becker C., Wang Y., Marches F., Helft J., Leboeuf M., Anguiano E., Pourpe S., Goller K., Pascual V., et al. 2013. Human CD1c+ dendritic cells drive the differentiation of CD103+ CD8+ mucosal effector T cells via the cytokine TGF-β. Immunity. 38:818–830 10.1016/j.immuni.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.