Abstract
Photoperiodic diapause is an important adaptation that allows individuals to escape harsh seasonal environments via a series of physiological changes, most notably developmental arrest and reduced metabolism. Global gene expression profiling via RNA-Seq can provide important insights into the transcriptional mechanisms of photoperiodic diapause. The Asian tiger mosquito, Aedes albopictus, is an outstanding organism for studying the transcriptional bases of diapause due to its ease of rearing, easily induced diapause, and the genomic resources available. This manuscript presents a general experimental workflow for identifying diapause-induced transcriptional differences in A. albopictus. Rearing techniques, conditions necessary to induce diapause and non-diapause development, methods to estimate percent diapause in a population, and RNA extraction and integrity assessment for mosquitoes are documented. A workflow to process RNA-Seq data from Illumina sequencers culminates in a list of differentially expressed genes. The representative results demonstrate that this protocol can be used to effectively identify genes differentially regulated at the transcriptional level in A. albopictus due to photoperiodic differences. With modest adjustments, this workflow can be readily adapted to study the transcriptional bases of diapause or other important life history traits in other mosquitoes.
Keywords: Genetics, Issue 93, Aedes albopictus Asian tiger mosquito, photoperiodic diapause, RNA-Seq de novo transcriptome assembly, mosquito husbandry
Introduction
Rapid advances in next-generation sequencing (NGS) technologies are providing exciting opportunities to probe the molecular underpinnings of a wide range of genetically complex ecological adaptations in a broad diversity of non-model organisms1–3. This approach is extremely powerful because it establishes a basis for population and functional genomics studies of organisms with an especially interesting and/or well-described ecology or evolutionary history, as well as organisms of practical concern, such as agricultural pests and disease vectors. Thus, NGS technologies are leading to rapid advances in the fields of ecology and have the potential to address problems such as understanding the mechanistic bases of biological responses to rapid contemporary climate change4, the spread of invasive species5, and host-pathogen interactions6,7.
The extraordinary potential of NGS technologies for addressing basic and applied questions in ecology and evolutionary biology is in part due to the fact that these approaches can be applied to any organism at a moderate cost that is feasible for most research laboratories. Furthermore, these approaches provide genome-wide information without the requirement of a priori genetic resources such as a microarray chip or complete genome sequence. Nevertheless, to maximize the productivity of NGS experiments requires careful consideration of experimental design including issues such as the developmental timing and tissue-specificity of RNA sampling. Furthermore, the technical skills required to analyze the massive amounts of data produced by these experiments, often up to several hundred million DNA sequence reads, has been a particular challenge and has limited the widespread implementation of NGS approaches.
Recent RNA-Seq studies on the transcriptional bases of diapause in the invasive and medically important mosquito Aedes albopictus provide a useful example of some of the experimental protocols that can be employed to successfully apply NGS technology to studying the molecular basis of a complex ecological adaptation in a non-model organism8–10. A. albopictus is a highly invasive species that is native to Asia but has recently invaded North America, South America, Europe, and Africa11,12. Like many temperate insects, temperate populations of A. albopictus survive through winter by entering a type of dormancy referred to as photoperiodic diapause. In A. albopictus, exposure of pupal and adult females to short (autumnal) day lengths leads to the production of diapause eggs in which embryological development is completed, but the pharate larva inside the chorion of the egg enters a developmental arrest that renders the egg refractory to hatching stimulus15–17. Diapause eggs are more desiccation resistant5,18 and contain more total lipids19 than non-diapause eggs. Photoperiodic diapause in A. albopictus is thus a maternally controlled, adaptive phenotypic plasticity that is essential for surviving the harsh conditions of winter in temperate environments. Despite the well-understood ecological significance of photoperiodic diapause in a wide range of insects20,21, the molecular basis of this crucial adaptation is not well characterized in any insect22. In organisms such as A. albopictus that undergo an embryonic diapause at the pharate larval stage, it remains a particularly compelling challenge to understand how the photoperiodic signal received by the mother is passed to the offspring and persists through the course of embryonic development to cause arrest at the pharate larval stage.
This protocol describes mosquito rearing, experimental design and bioinformatics analyses for NGS experiments (transcriptome sequencing) performed to elucidate transcriptional components of photoperiodic diapause in A. albopictus. This protocol can be used for additional studies of diapause in A. albopictus, can be adapted to investigate diapause in other closely related species such as other aedine mosquitoes that undergo egg diapause23, and is also more generally relevant to employing NGS approaches to study the transcriptional bases of any complex adaptation in any insect.
Protocol
1. Larval Rearing of Two A. albopictus Groups to Adulthood
- Set two photoperiod cabinets with programmable lighting at 21 °C for optimal diapause expression16 and approximately 80% relative humidity.
- Program one cabinet for a 16L:8D light:dark cycle (a non-diapause inducing LD photoperiod). Set the second cabinet for an 8L:16D light:dark cycle (diapause inducing)13.
- Program ‘lights on’ at the same time in both cabinets to synchronize circadian time between photoperiods.
- Calculate the quantity of eggs needed to perform the experiment. Aim for 300-500 eggs per cage. At least three replicate diapause cages and three replicate non-diapause cages are needed for RNA generation. This totals to ca. 1,800-3,000 eggs per experiment.
- Hatch the eggs by submerging egg papers into ca. 500 ml of deionized H2O.
- Add ca. 1 ml food slurry consisting of ground dog food and brine shrimp as previously described24. Cover container with mesh, and keep the mesh in place with a rubber band.
- Place in the LD photoperiod cabinet for ca. 24 hr.
- Transfer hatched larvae to 10 x 10 x 2 cm Petri dishes filled with ca. 90 ml deionized H2O.
- Maintain ca. 30 larvae per dish. Transfer larvae to clean dishes every 48–72 hr, for example every Monday, Wednesday, and Friday (M-W-F)24.
- Feed ca. 1 ml food slurry consisting of ground dog food and brine shrimp in deionized water every M-W-F as previously described24.
- Set up three to four adult cages for each photoperiod treatment, where each cage comprises a biological replicate.
- From opposite sides of 9.5 L buckets, cut out one 10-by-14 cm hole, and another hole with 15 cm diameter. Cover the first with mesh. Cut approximately one foot length of an orthopedic stocking, and glue one end around the inside of the other hole.
- Cut the foot end of the stocking off and knot shut — open only when access to the interior of the cage is needed. For the cage lid, cut out all of the interior, leaving only the rim, and replace the interior plastic with mesh24.
- Note the photoperiod, replicate number, cage start date, and other information relevant to the experiment with permanent marker on the side of the cage.
Line the bottom of the adult cages with wet filter paper. Dampen the filter paper with enough deionized H2O to increase local humidity in the cage, but avoid standing water, which can stimulate oviposition on the filter paper24. Check the filter paper daily for drying, re-wet when necessary.
To produce sufficient eggs for an RNA library, include at least 100 females/9.5 L cage, with no more than 500 mosquitoes per cage.
Collect pupae M-W-F and place in a small cup of clean H2O at a density of no more than 50 pupae per 25 ml H2O. Transfer the pupae cup to an adult cage. Place cages in the respective photoperiod cabinet — A. albopictus pupae are photosensitive15.
Ensure daily that H2O in cups is clean and clear, and remove dead pupae, because build-up of dead pupae can cause mass mortality. Remove H2O cups after all pupae emerge.
Place organic raisins on the top mesh of the cage to provide sugar for emerged adults. Monitor raisins, and change them every 3-5 days to prevent mold accumulation.
2. Maintenance of Adults to Allow Mating and Egg Production
Maintain cages at high humidity (approx. 80%) by lining the cage bottom with a moist filter paper, and provide access to non-moldy raisins, as described above (Sections 1.5 and 1.9).
3. Blood-feeding
Prepare to blood-feed females between two to six days after eclosion to ensure that females have been exposed to at least eight unambiguous short days before oviposition commences for nearly 100% diapause eggs14.
Prepare the Hemotek Membrane Feeding system. Plug the feeding units into the power supply. Adjust the temperature of each unit to 37 °C using the adjustment screw. Use an electronic thermometer and probe to measure the temperature of the feeding unit during calibration.
- Prepare the meal reservoir. Stretch a square of collagen feeding membrane over the aperture of the meal reservoir and secure it with an O-ring. Carefully pull the corners to remove wrinkles; trim the excess membrane with scissors. NOTE: Collagen membrane may not work well for all mosquito species, and it may be necessary to try several types to find the optimal membrane if working with a species other than A. albopictus. Parafilm works well with Culex pipiens.
- If blood is stored frozen, thaw at room temperature for at least 1 hr before using.
- Hold the reservoir so the membrane is facing down, unsupported, and the filling ports are facing up. Use a transfer pipette or syringe to fill the reservoir with approximately 3 ml of whole blood from chickens that has sodium citrate as an anti-coagulant. Seal filling ports with plastic plugs.
Attach the prepared reservoir to the feeder by screwing it onto the stud on the heat transfer plate on the bottom of the feeder. Invert the feeder and place it on top of the cage, membrane side down, so that mosquitoes can feed through the mesh of the cage. Keep the feeder on the cage for approximately 45 min to maximize feeding.
4. Stimulate Oviposition
Four to five days post blood meal, equip each cage with a dark colored 50 ml cup lined with unbleached seed germination paper (egg paper) or textured non-bleached paper towel and fill halfway with deionized water9. If more than 250 mosquitoes are in a cage, use two cups. NOTE: For small cages or single-female vials, “hay infusion” in oviposition containers may increase oviposition due to the odor of the microbial flora25.
5. Collect and Store Eggs
Commence egg collection within 4-5 days of blood feeding because egg production typically peaks approximately five days after blood feeding, and then subsides over the next week.
Vary egg collection frequency on necessities of the experiment. For general purposes, collect egg papers on an M-W-F schedule. Remove egg papers from each cage and replace with fresh paper. Place recently removed papers in Petri dishes and store in SD photoperiod cabinet to avoid confounding effects of egg storage.
Allow egg papers to remain wet for 2 days post-oviposition to allow serosal cuticle formation, which increases egg desiccation resistance26.
Approximately 48 hr post collection, dry eggs in open air. Dry the paper such that it is limp and slightly damp to the touch, but not so wet that the paper is dark from H2O or stimulates hatching of eggs. NOTE: A 6.5” x 4” paper may take approximately 3.5 hr to dry. Be cautious not to over-dry egg papers, as this will result in egg desiccation27.
Reserve additional eggs from both LD and SD photoperiods to assess diapause incidence and interpret the photoperiodic effect (see Measuring Diapause, Section 6).
For long-term storage, keep egg papers at 21 °C and approximately 80% humidity in Petri dishes. Keep Petri dishes in a Tupperware storage container with a flask of water to maintain local humidity as embryonic development takes four to five days at 21 °C.
6. Measure Diapause Incidence
Use additional reserved embryos (see Stimulate Oviposition, Section 4) that are 7–20 days old to quantify the diapause response.
Record the number of eggs present on each egg paper.
Stimulate eggs to hatch by completely submerging individual egg papers in a 90 ml Petri dish with approximately 80 ml deionized H2O. Add approximately 0.25 ml food slurry.
After 24 hr tally the number of hatched first instar larvae. Place the Petri dish on a black surface to visualize larvae and place a light source on one side of dish. Larvae will move away from the light source, allowing for a clear tally of individual larvae. Remove individual larvae with a pipette while counting to prevent recounting individual larvae.
Place egg papers in a new Petri dish and re-dry. Re-hatch eggs after ~1 week and again tally eggs hatched using the above method.
Place egg papers with the remaining un-hatched eggs in new 90 ml Petri dishes with approximately 80 ml bleaching solution28. Ensure that the egg papers are completely submerged in the bleaching solution and leave under a fume hood overnight to avoid the odor of bleach. NOTE: Bleaching solution can be stored for ~1 week at 4 °C, but should otherwise be made fresh.
Inspect eggs using a light microscope as the bleaching will clear the chorion and allow visualization of embryonated, un-hatched eggs. If the egg is embryonated, the egg will have an off-white color with eyes appearing as two small black dots opposite each other on the dorsal side. Tally the number of un-hatched, embryonated eggs13.
Determine diapause incidence with the following formula: % diapause = no. embryonated un-hatched eggs / (no. hatched eggs + no. embryonated un-hatched eggs) x 100 13.
7. RNA Extraction from Eggs/pharate Larvae
NOTE: Use Trizol in a laminar flow hood.
- Brush mosquito eggs containing developing embryos or pharate larvae at distinct development time points from egg papers to glass grinders using a camel-hair brush. Grind the eggs in Trizol (1 ml per 50–100 mg of tissue) until completely pulverized. Use at least 400 eggs per library to yield sufficient RNA.
- Alternatively, snap freeze eggs in liquid nitrogen and stored at -80 °C in microcentrifuge tubes for up to a month before grinding in Trizol.
Perform RNA extraction in Trizol followed by isopropanol precipitation according to manufacturer’s instructions.
- Treat the bench with RNase decontamination solution or other agents to remove any residual nucleases to avoid RNA degradation.
- Treat the extracted RNA with DNase. According to manufacturer’s instructions, incubate the RNA samples with DNase for 30 min at 37 °C. Use 1 µl DNase for up to 10 µg of RNA in a 50 µl reaction. Increase the amount of DNase if there are more than 10 µg of RNA in one reaction.
- Inactivate DNase by adding 5 µl suspended DNase inactivation reagent. Incubate 5 min at room temperature, mixing three times during incubation period (gentle vortexing).
- Centrifuge at 10,000 x g for 1.5 min. Transfer the supernatant containing the treated RNA samples to fresh tubes for subsequent steps.
- Assess the quality of the total RNA samples by fluorometry. Send the samples to a specialty facility with proper instrument for this task. The facility will perform on-chip gel electrophoresis to determine the sizes of RNA species in the sample, visualized by fluorescent dye instilled in the chip. The results will be returned as an electropherogram.
- Determine the integrity of total RNA samples by the presence or absence of degradation products, as evidenced by the presence of peaks between the 18S and 5S ribosomal RNA peaks on the resulting electropherogram (Figure 1B).
8. RNA Sequencing
Send total RNA samples with sufficiently high quality (Figure 1A) and quantity (usually >3 μg per library) to a commercial sequencing center for construction of enriched paired-end mRNA libraries and mRNA sequencing, following standard protocols.
If more than one lane is being used for sequencing a single experiment, split individual libraries into two lanes for sequencing to account for technical variation among lanes during sequencing.
9. lllumina Read Cleaning
NOTE:Figure 2 summarizes the bioinformatics portion of this protocol. For a full list of all programs and resources used in the bioinformatics section of this protocol, refer to Table 1. In addition, Supplemental File 1 contains command line examples for each of the following bioinformatics protocol steps.
Use ssaha229 (Table 1) to identify matches of 95% identity or higher to the NCBI UniVec Core database (Table 1), A. albopictus rRNA sequence (GenBank #L22060.1), and sequencing adapters (detailed command-line examples provided in Supplemental File 1). Remove read pairs with matches using Perl or a similar scripting tool, for example by adapting the provided Perl script (Supplemental File 2).
- Clean remaining reads with the SolexaQA package30 (Table 1; Supplemental File 1): trim regions with a phred score equivalent of less than 20 using the default settings of DynamicTrim.pl.
- Remove reads shorter than 25 bp with LengthSort.pl on both forward and reverse reads simultaneously. Evaluate the quality of the cleaned fastq files with FastQC (Table 1) — in particular, verify that the per-base sequence quality and the per-sequence quality scores are above 20.
10. Digital Normalization
Perform one round of digital normalization on the cleaned reads using the khmer tool31 (Table 1; Supplemental File 1), specifically normalize-by-median.py (using k-mer size 20, a coverage cut-off of 20, and x = 1e10).
Alternatively, if a machine with high RAM is available (hundreds of GBs), use Trinity's normalize_by_kmer_coverage.pl script (Table 1).
11. De Novo Transcriptome Assembly
Obtain access to a computer or computer cluster with up to 256 Gb of RAM and 24 cpus, depending on the size of the assembly.
Use Trinity32 (Table 1; Supplemental File 1) to assemble the digitally normalized read set into contigs. To reduce memory usage, use --min_kmer_cov 2.
12. Assembly Evaluation
Run assemblathon_stats.pl from the Assemblathon2 project33 on the Trinity contig output. This script performs basic calculations relevant to evaluating assembly quality, such as number of scaffolds, N50, assembly composition, and more (Table 1; Supplemental File 1).
13. Annotation of the Assembled Transcriptome
Perform Blastx (Table 1) of the assembly against a reference protein set; for mosquitoes, Drosophila melanogaster, Anopheles gambiae, Culex pipiens, and Aedes aegypti are suitable references. Specifically, format the reference protein fasta file for blast, followed by blastx (Supplemental File 1).
14. Map Reads to the Assembly Using RSEM34 (Table 1)
Create a 'transcript-to-gene-map' file, in which the first column contains the reference gene IDs, and the second column the contig IDs. In a spreadsheet editor, swap the first and second columns from the Blastx output, and write these columns to a .txt file. Use the LineBreak program to convert line breaks in the resulting .txt file to Unix format.
Create a reference dataset from the transcriptome fasta file using the rsem-prepare-reference script, provided in the RSEM package (Supplemental File 1).
- Calculate the expression values separately for each library using the rsem-calculate-expression command, provided in the RSEM package (Supplemental File 1). As reads, use the paired fastq files resulting from the read-cleaning step (step 9.2).
- If RNA from a biological replicate was split into two lanes for sequencing, include both fastq files in the expression calculation to generate a single file.
Convert the expression results from each library to a matrix easily processed by other programs using the provided script rsem-generate-data-matrix, provided in the RSEM package (Supplemental File 1).
15. Differential Expression Analysis
Install R and EdgeR (Table 1).
Use read.delim to load the RSEM results from step 14.3 (Supplemental File 1). If necessary, round the counts to the nearest integer. NOTE: The EdgeR guide recommends limiting the dataset to genes with high enough expression to detect significance.
To format the data for EdgeR, generate a DGEList object from the loaded data file (Supplemental File 1). Then, normalize the data using TMM normalization (Supplemental File 1). Estimate the common and tagwise dispersions of the data (Supplemental File 1).
Identify differentially expressed genes with a Benjamini-Hochberg corrected p-value <0.05 (Supplemental File 1). Plot the distribution of log-fold-change vs. abundance (Supplemental File 1).
Representative Results
Fluorometry of two representative RNA samples showed two bands at approximately 2,000 nt (Figure 1A, B). The insect 28S ribosomal RNA is comprised of two polynucleotide chains held together by hydrogen bonds, which are easily disrupted by brief heating or agents that disrupt hydrogen bonds35. The resulting two components are approximately the same size as the 18S ribosomal RNA. The second RNA sample showed high levels of degradation (Figure 1B).
Photoperiodic treatment of a representative group of A. albopictus mosquitoes resulted in high diapause incidence in short-day-reared mosquitoes, and low diapause incidence in long-day-reared mosquitoes, although there was some variation among replicates (Table 2). For example, replicate SD2 shows lower (80%) diapause incidence than the remaining replicates (87.18% – 97.67%). This replicate also had the smallest sample size, so it is recommended to set aside a sufficient number of eggs (>150) for the diapause measurement in order to obtain an accurate result.
Post-sequencing read cleaning on one representative library from adult A. albopictus females removed a substantial number of reads (from 83,853,322 to 52,736,065 total reads for one representative library). Digital normalization further reduced the number of total reads to 41,435,934. A Trinity assembly of these reads generated 76,377 contigs, with an N50 of 1,879, mean contig length of 1,023.1, and a maximum contig length of 20,892 (Figure 3). Differential expression analyses from a similar workflow of embryos reared under diapause-inducing conditions at 11 and 21 days post-oviposition revealed 3,128 differentially expressed genes between these two time periods (Figure 4).
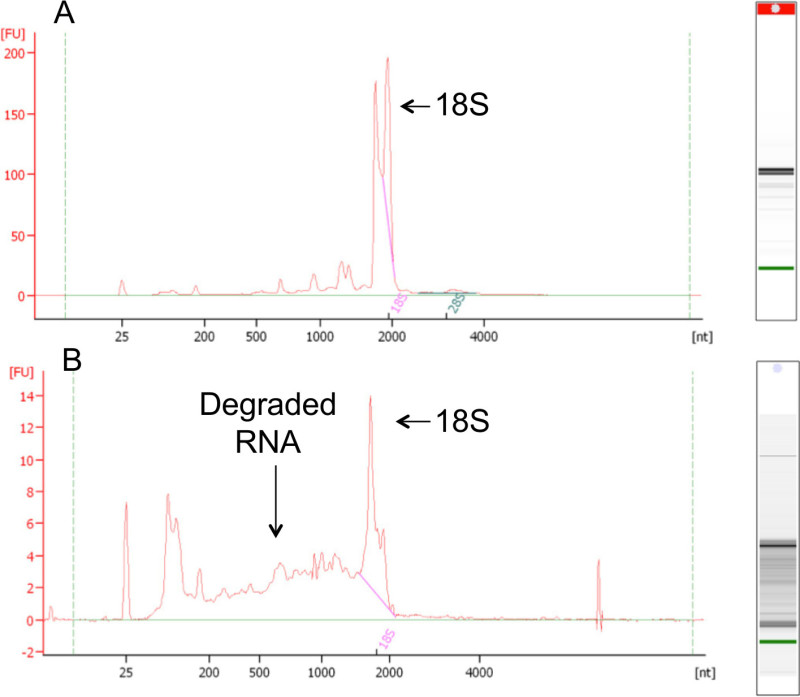
Figure 1: Fluorometry profiles of example high-quality (A) and low-quality (B) RNA extractions from A. albopictus. The x-axis represents the sizes of the nucleotide fragments, and the y-axis represents the fluorescent readings. Note the difference in the y-axis scale between panels (A) and (B). Arrows mark the positions of the different ribosomal RNAs. The apparent bands close to the green marker band indicate degradation.
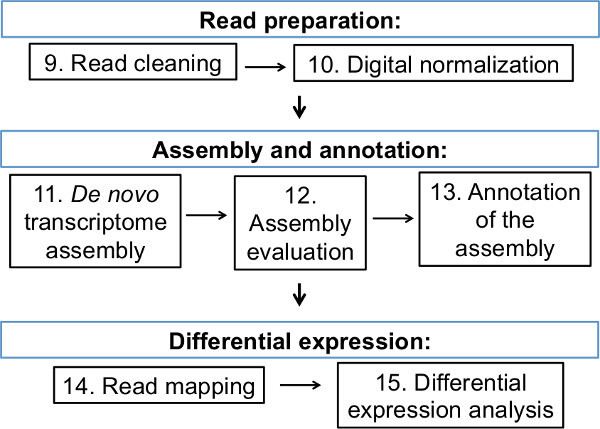
Figure 2: Summary of the bioinformatics workflow from read preparation to differential expression. Each box represents a step in the bioinformatics section of this protocol, accompanied by the corresponding number of each protocol step.
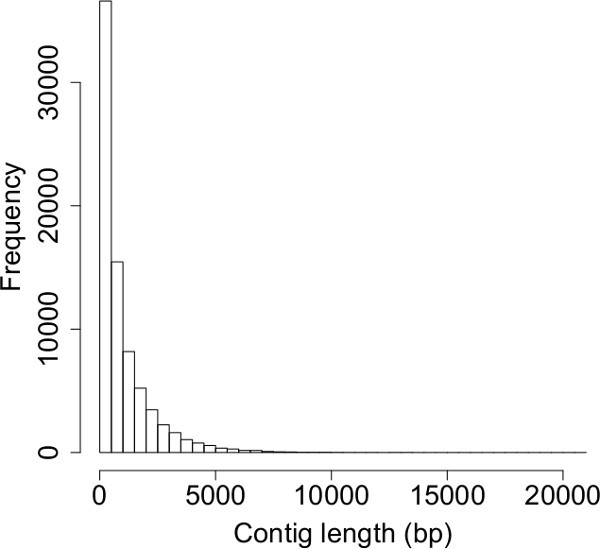 Figure 3: Histogram of contig lengths from a Trinity de novo transcriptome assembly. The average contig length is 1,023.1. Note that the distribution of contig lengths is heavily skewed towards shorter contigs; this is typical of de novo transcriptome assembly.
Figure 3: Histogram of contig lengths from a Trinity de novo transcriptome assembly. The average contig length is 1,023.1. Note that the distribution of contig lengths is heavily skewed towards shorter contigs; this is typical of de novo transcriptome assembly.
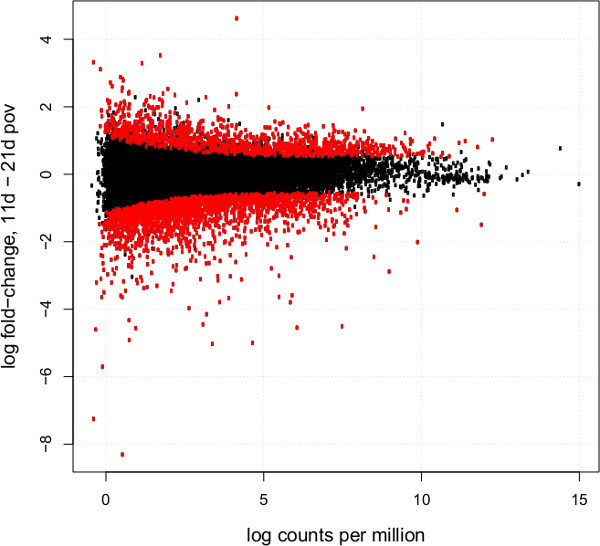 Figure 4: Log2-fold-change vs. abundance of TMM-normalized gene expression of diapause pharate larvae at 11 days vs. 21 days post-oviposition. Each point designates a unigene; differentially expressed unigenes are in red. Unigenes with higher expression at 11 days post-oviposition have positive fold-change values, whereas unigenes with higher expression 21 days post-oviposition have negative fold-change values.
Figure 4: Log2-fold-change vs. abundance of TMM-normalized gene expression of diapause pharate larvae at 11 days vs. 21 days post-oviposition. Each point designates a unigene; differentially expressed unigenes are in red. Unigenes with higher expression at 11 days post-oviposition have positive fold-change values, whereas unigenes with higher expression 21 days post-oviposition have negative fold-change values.
| Program/Resource | Website URL (accessed 1/13/2014) |
| perl | http://www.perl.org/get.html |
| python | http://www.python.org/download/ |
| ssaha2 | http://www.sanger.ac.uk/resources/software/ssaha2/ |
| NCBI UniVec Core | ftp://ftp.ncbi.nih.gov/pub/UniVec/UniVec_Core |
| SolexaQA | http://solexaqa.sourceforge.net/ |
| FastQC | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| khmer | https://github.com/ged-lab/khmer |
| Trinity | http://trinityrnaseq.sourceforge.net/ |
| Trinity normalization script | http://trinityrnaseq.sourceforge.net/trinity_insilico_normalization.html |
| Assemblathon 2 evaluation scripts | https://github.com/ucdavis-bioinformatics/assemblathon2-analysis |
| BLAST+(which includes makeblastdb and blastx) | ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ |
| linebreak | https://code.google.com/p/linebreak/ |
| RSEM | http://deweylab.biostat.wisc.edu/rsem/ |
| EdgeR User's Guide | http://www.bioconductor.org/packages/2.13/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf |
| R | http://www.r-project.org/ |
| EdgeR | http://www.bioconductor.org/packages/2.13/bioc/html/edgeR.html |
Table 1: Programs and resources used for the bioinformatics procedures in this protocol. URLs are listed to easily access each of the resources needed in this protocol.
| Treatment | Replicate | No. of eggs from 1st hatch | No. of eggs from 2nd hatch | No. embryonated, unhatched eggs | % Diapause |
| SD | 1 | 10 | 0 | 68 | 87.18 |
| SD | 2 | 3 | 0 | 12 | 80.00 |
| SD | 3 | 1 | 0 | 42 | 97.67 |
| SD | 4 | 5 | 0 | 46 | 90.20 |
| SD | 5 | 6 | 0 | 79 | 92.94 |
| LD | 1 | 79 | 4 | 9 | 9.78 |
| LD | 2 | 28 | 0 | 4 | 12.50 |
| LD | 3 | 92 | 1 | 6 | 6.06 |
| LD | 4 | 30 | 0 | 5 | 14.29 |
| LD | 5 | 43 | 0 | 3 | 6.52 |
Table 2: Diapause incidence calculations. Results from five replicates per photoperiod of diapause incidence calculations. Numbers of hatched larvae from two separate hatchings are included, as are the number of un-hatched, embryonated eggs, all of which are necessary to calculate diapause incidence.
Discussion
This protocol presents methods to discover differentially expressed genes due to photoperiodically induced diapause in A. albopictus. The protocol is significant in that it uniquely combines mosquito rearing and bioinformatics techniques to make all experimental aspects of a molecular physiology program accessible to novice users — in particular for those focusing on the photoperiodic diapause response. Existing methods, to our knowledge, do not provide as much detail in the rearing protocol — which is often necessary to identify rearing mistakes — nor do they provide insight on experimental design during the rearing stage that will enable successful bioinformatic analysis downstream. The methods presented here have been optimized for A. albopictus, especially the rearing methods, which generally take six weeks from one laboratory generation to the next. However, in future applications this method could be adapted with modest adjustments to other mosquito species that exhibit photoperiodic diapause23. Furthermore, the general experimental design and bioinformatics workflow are applicable to the study of other polyphenisms.
Several points not detailed in the protocol should be considered when rearing A. albopictus larvae. First, A. albopictus can be found in a wide variety of natural and artificial container habitats as described in previous papers36, 37. Used tire lots are a common source of larvae for establishing laboratory colonies. Populations collected above 32N latitude in North America can be expected to exhibit a strong diapause response13. The A. albopictus strain used in this protocol was collected from Manassas, VA, and was reared in a laboratory setting for more than eight generations prior to experimental manipulation. Second, lighting in the photoperiod cabinets should be chosen with care. Bulbs in cabinets with built-in lighting functions can cause temperature spikes within the cabinet when the lighting turns on or off. Anecdotal observation suggests these temperature spikes can disrupt the diapause response. To prevent this, built-in lighting functions should be disabled and cabinets should be equipped with a 4-watt cool-fluorescent bulb. Third, larvae are sensitive to H2O quality and food abundance. Therefore, over-feeding may lead to bacterial accumulation and larval mortality. Fourth, there are alternative methods to blood-feed adult female mosquitoes. Glass membranes are an alternative artificial membrane system38, 39, although the HemoTek system performs better in the authors’ experience. Live animals (usually chicken or rodent) can also be used38 — in this case, it is essential to first obtain appropriate certification from your Institutional Animal Care and Use Committee (IACUC). Fifth, although there is no clear published evidence that eggs are photosensitive15, anecdotal observations suggest that eggs from an SD photoperiod treatment exhibit slightly reduced diapause incidence when exposed to an LD photoperiod within 10 days of oviposition. Thus, store both SD and LD eggs under SD conditions to produce a maximal diapause response in the SD eggs and avoid any confounding effect of photoperiod (SD vs. LD) during egg storage.
High RNA quality is essential for generating high quality RNA-Seq data. Abundant care should be taken during the RNA extraction to avoid any nuclease contamination. Low quality RNA samples, such as that shown in Figure 1B, are not appropriate for sequencing. Assessing the RNA quality before sending the samples for sequencing is imperative. Characteristic bands of RNA molecules might be visible for different types of insect tissue used for RNA extraction, such as the four bands smaller than 18S shown in the high quality RNA electropherogram in Figure 1A. Consistent patterns of RNA bands other than the two bands at 18S across samples under distinct biological treatments can strongly indicate that these bands do not result from degradation, but represent biological composition of the RNA molecules in the specific tissue types chosen in the experimental design.
The bioinformatics workflow outlined here allows a user with some command-line and scripting skills to obtain a list of differentially expressed genes from Illumina sequencing data generated from replicated RNA libraries from two contrasting experimental conditions. While this example concerns genes differentially expressed due to photoperiod, this workflow can be applied to any experimental design with two or more treatments, in any organism. There are many other ways to arrive at a list of differentially expressed genes; however, this protocol is likely to be the most straightforward approach for the novice user. More experienced bioinformaticians may want to take extra measures to improve the contiguity and redundancy of their assembly. Biologists with little to no bioinformatics experience may also complete at least part of this pipeline within the iPlant40 Discovery Environment, which is a free graphical-user-interface driven analysis environment. It is likely that iPlant’s functionality will grow larger in the future in order to accommodate full RNA-Seq pipelines from de novo transcriptome assemblies. Finally, note that the excellent User's Guide thoroughly discusses the many ways to use EdgeR41 (Table 1) for differential expression analysis.
In some cases, mis-assemblies can generate chimeric contigs. There are several methods that can help to identify these mis-assemblies, for example, Uchime42. However, from past experience, the number of detected chimeras is exceedingly low (< 0.1%); therefore, employing a chimera detection program may not be worth the extra effort.
Processing high-throughput, next-generation sequencing data requires the ability to 1) store large amounts of data (for a single project, >500 Gb); 2) manipulate large data files that cannot be opened in traditional word processors or spreadsheet programs; 3) perform analyses that require large amounts of RAM, e.g., for de novo assembly; and 4) analyze large datasets, either through programs driven by a command-line interface (which requires the ability to install these programs, which is often non-trivial), or through analysis suites with graphical user interfaces (e.g. Galaxy43 or iPlant40). Researchers with some proficiency in Unix command line and a scripting language will gain the most benefit from access to a local computing cluster - either University-owned, via a collaborator, or purchased for their own laboratory. For example, the above workflow was accomplished using a laboratory-owned Macintosh (12 cores, 64 GB RAM, 1 Tb hard drive), and a University-owned computer cluster for the Trinity assembly. If similar resources are not available, researchers can still turn to iPlant to perform large-scale analyses at no cost, and with relatively lower investment in training due to the graphical interface environment. However, those performing and interpreting the analyses still need to understand the assumptions of each program used.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by the National Institutes of Health grant 5R21AI081041-02 and Georgetown University.
References
- Bilyk KT, Cheng CHC. Model of gene expression in extreme cold - reference transcriptome for the high-Antarctic cryopelagic notothenioid fish Pagothenia borchgrevinki. BMC Genomics. 2013;14:634. doi: 10.1186/1471-2164-14-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Hiscock SJ, Filatov DA. Genomic divergence during speciation driven by adaptation to altitude. Mol. Biol. Evol. 2013;30:2553–2567. doi: 10.1093/molbev/mst168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, et al. Sympatric ecological speciation meets pyrosequencing: sampling the transcriptome of the apple maggot Rhagoletis pomonella. BMC Genomics. 2009;10:633. doi: 10.1186/1471-2164-10-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshis DJ, et al. Genomic basis for coral resilience to climate change. P. Natl Acad. Sci. USA. 2013;110:1387–1392. doi: 10.1073/pnas.1210224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski JM, Aruda A, Armbruster PA. A transcriptional element of the diapause program in the Asian tiger mosquito, Aedes albopictus, identified by suppressive subtractive hybridization. J. Insect Physiol. 2010;56:1147–1154. doi: 10.1016/j.jinsphys.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Huang YH, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat. Genet. 2013;45:776–783. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions OM, et al. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS Negl. Trop. Dis. 2013;7(3):2107. doi: 10.1371/journal.pntd.0002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. Deep sequencing reveals complex mechanisms of diapause preparation in the invasive mosquito, Aedes albopictus. P. R. Soc B. 2013;280 doi: 10.1098/rspb.2013.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. Transcriptome sequencing as a platform to elucidate molecular components of the diapause response in Aedes albopictus. Physiol. Entomol. 2013;38:173–181. doi: 10.1111/phen.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau MF, Reynolds JA, Denlinger DL, Elsik CG, Armbruster PA. A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation. BMC Genomics. 2011;12:619. doi: 10.1186/1471-2164-12-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict MQ, Levine RS, Hawley WA, Lounibos LP. Spread of the tiger: Global risk of invasion by the mosquito Aedes albopictus. Vector-Borne Zoonot. 2007;7:76–85. doi: 10.1089/vbz.2006.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounibos LP. Invasions by insect vectors of human disease. Annu. Rev. Entomol. 2002;47:233–266. doi: 10.1146/annurev.ento.47.091201.145206. [DOI] [PubMed] [Google Scholar]
- Urbanski JM, et al. Rapid adaptive evolution of photoperiodic response during invasion and range expansion across a climatic gradient. Am. Nat. 2012;179:490–500. doi: 10.1086/664709. [DOI] [PubMed] [Google Scholar]
- Lounibos LP, Escher RL, Lourenco-de-Oliveria R. Asymmetric evolution of photoperiodic diapause in temperate and tropical invasive populations of Aedes albopictus (Diptera Culicidae) Ann. Entomol. Soc. Am. 2003;96:512–518. [Google Scholar]
- Mori A, Oda T, Wada Y. Studies on the egg diapause and overwintering of Aedes albopictus in Nagasaki. Trop. Med. 1981;23:79–90. [Google Scholar]
- Pumpuni CB. Factors influencing photoperiodic control of egg diapause in Aedes albopictus [dissertation] 1989.
- Wang RL. Observations on the influence of photoperiod on egg diapause in Aedes albopictus Skuse. Acta Entomol. Sinica. 1966;15:75–77. [Google Scholar]
- Sota T, Mogi M. Survival-time and resistance to desiccation of diapause and non-diapause eggs of temperate Aedes (Stegomyia) mosquitoes. Entomol. Exp. Appl. 1992;63:155–161. [Google Scholar]
- Reynolds JA, Poelchau MF, Rahman Z, Armbruster PA, Denlinger DL. Transcript profiling reveals mechanisms for lipid conservation during diapause in the mosquito, Aedes albopictus. J. Insect Physiol. 2012;58:966–973. doi: 10.1016/j.jinsphys.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrewartha HG. Diapause in relation to the ecology of insects. Biol. Rev. 1952;27:50–107. [Google Scholar]
- Danks HV. Biological Survey of Canada (Terrestrial Arthropods) Ottowa, Canada: 1987. Insect Dormancy: An Ecological Perspective. [Google Scholar]
- Denlinger DL. Regulation of diapause. Ann. Rev. Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- Rev Entomol Ann. 2014;59:93–122. [Google Scholar]
- Armbruster PA, Conn JE. Geographic variation of larval growth in North American Aedes albopictus (Diptera) Culicidae). Ann. Entomol. Soc. Am. 2006;99:1234–1243. [Google Scholar]
- Reiter P, Amador MA, Colon N. Enhancement of the CDC ovitrap with hay infusions for daily monitoring of Aedes aegypti populations. J. Am. Mosquito Contr. Association. 1991;7(1):52. [PubMed] [Google Scholar]
- Rezende GL, et al. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle. BMC Dev. Biol. 2008;8:182. doi: 10.1186/1471-213X-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munstermann L. Care and maintenance of Aedes mosquito colonies. In: Crampton J, Beard C, C Louis, editors. The Molecular Biology of Insect Disease Vectors. Netherlands: Springer; 1997. pp. 13–20. [Google Scholar]
- Trpis M. A new bleaching and decalcifying method for general use in zoology. Can. J. Zoolog. 1970;48:892–893. [Google Scholar]
- Ning ZM, Cox AJ, Mullikin JC. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11(10):1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. A reference-free algorithm for computational normalization of shotgun sequencing data [Internet] 2012. [cited 2014 May 6]. Available at: http://arxiv.org/abs/1203.4802.
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BN, De Lucca FL. Preparation and analysis of RNA. In: Turner RB, editor. Analytical Biochemistry of Insects. Philadelphia, PA: Elsevier Scientific Publishing Company; 1977. [Google Scholar]
- Hawley WA. The biology of Aedes albopictus. J. Am. Mosq. Contr. Assoc. 1988;4:1–39. [PubMed] [Google Scholar]
- Dowling Z, Ladeau SL, Armbruster P, Biehler D, Leisnham PT. Socioeconomic status affect mosquito (Diptera:Culicidae) larval habitat type availability and infestation level. J. Med Entomol. 2013;50:764–772. doi: 10.1603/me12250. [DOI] [PubMed] [Google Scholar]
- Benedict MQ. Methods in Anopheles Research. Malaria Research and Reference Reagent Resource Center(MR4); 2010. Chapter 2,4,10. Bloodfeeding: Membrane apparatuses and animals. [Google Scholar]
- Das S, Garver S, Ramirez JR, Xi Z, Dimopolous G. Protocol for dengue infections in mosquitoes (A. aegypti) and infection phenotype determination. J. Vis. Exp. 2007;5(220) doi: 10.3791/220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, et al. The iPlant collaborative: cyberinfrastructure for plant biology. Plant Sci. 2011;2(34) doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2220. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]


