Abstract
Rigid tumor tissues have been strongly implicated in regulating cancer cell migration and invasion. Invasive migration through cross-linked tissues is facilitated by actin-rich protrusions called invadopodia that proteolytically degrade the extracellular matrix (ECM). Invadopodia activity has been shown to be dependent on ECM rigidity and cancer cell contractile forces suggesting that rigidity signals can regulate these subcellular structures through actomyosin contractility. Invasive and contractile properties of cancer cells can be correlated in vitro using invadopodia and traction force assays based on polyacrylamide gels (PAAs) of different rigidities. Invasive and contractile properties of cancer cells can be correlated in vitro using invadopodia and traction force assays based on polyacrylamide gels (PAAs) of different rigidities. While some variations between the two assays exist, the protocol presented here provides a method for creating PAAs that can be used in both assays and are easily adaptable to the user’s specific biological and technical needs.
Keywords: Medicine, Issue 95, invadopodia, traction force, degradation, contractility, rigidity, polyacrylamide
Introduction
The rigidity of the tumor-associated ECM has been identified as a significant factor in driving malignant behavior by increasing actomyosin contractility1-3. While this effect has primarily been demonstrated with breast cancer cells, matrix rigidity has been found to alter invasive properties of cells derived from a variety of cancers4-8 suggesting that tumor rigidity may play a role in other type of cancers. To penetrate cross-linked tissues during invasive migration, cancer cells utilize actin-rich adhesive protrusions known as invadopodia that localize proteinases to focally degrade the ECM9. Invadopodia are considered a hallmark of invasive cells and have been implicated in tumor cell invasion and metastasis10,11. Previous work has shown that matrix rigidity can regulate invadopodia numbers and associated ECM degradation4,12 through myosin II activity and mechanosensitive proteins12. Given the correlation between tumor density and cancer aggressiveness13,14, these results suggest a mechanism by which cancer cells may respond to rigid tumor tissues to drive invasion and metastasis through actomyosin contractility.
In vitro ECM rigidity and in vivo tissue density have been shown to regulate invasive behavior of cancer cells1,15-17. While actomyosin contractility appears to be important in this process, current studies conflict as to whether metastatic capacity is correlated to increased or decreased contractile forces6,18-20. Furthermore, it remains unknown whether these forces directly mediate invadopodia activity21. We recently found that cancer cell contractile forces were dependent on matrix rigidity and were predictive of ECM degradation by invadopodia5. These results suggest that cellular forces may play an important role in cancer progression by mediating invadopodia activity in response to the mechanical properties of the tumor microenvironment.
In order to correlate invasive and contractile properties of cancer cells5, we modified a protocol for creating PAAs with different rigidities that was previously used to investigate rigidity-dependent invadopodia activity4,12,22. By chemically crosslinking human plasma fibronectin throughout the PAAs, these modified hydrogels can be used as the basis for both invadopodia and traction force assays to ensure that cells experienced the same rigidities in both experiments5. In the invadopodia assays, the fibronectin provides a natural binding domain for gelatin to link the overlaid ECM to the PAAs to detect matrix degradation. In the traction force assays, the fibronectin provides a ligand for direct cellular adhesion to detect microsphere displacements used to calculate cellular traction forces. This method results in what we have called soft, hard, and rigid PAAs that are bound to glass bottom dishes and have elastic moduli, E, of 1,023, 7,307, and 22,692 Pa5 which span the range of mechanical properties reported for normal and cancerous tissues23.
Protocol
1. Preparation of Glass Coverslips for PAAs
Clean 12 mm coverslips with low lint wipes.
Flame the 12 mm coverslips and the 14 mm coverslip in the microwell of each 35 mm glass bottom dish by passing them through a Bunsen burner flame using tweezers.
Treat the microwells with 200 µl of 0.1 N NaOH for 5 min at room temperature.
Aspirate and air dry the microwells for 30 min.
Treat the microwells with 50-100 µl of 3-aminopropyltrimethoxysilane for 10 min at room temperature in the fume hood. This chemical reacts with plastic; therefore, use glass pipettes and do not fill the microwells completely to avoid contact with the dish plastic.
Wash the microwells with ultrapure water for approximately 10 min until the 3-aminopropyltrimethoxysilane becomes clear.
Rinse the microwells with ultrapure water twice using a squeeze bottle.
Wash the microwells with 2 ml of ultrapure water at room temperature on a rocker set at a medium speed (~1 Hz) for 10 min.
Aspirate and air dry the microwells for 30 min.
Treat the microwells with 2 ml of 0.5% glutaraldehyde solution at room temperature on a rocker set at a medium speed (~1 Hz) for 30 min.
Wash the microwells with 2 ml of ultrapure water at room temperature on a rocker set at medium speed (~1 Hz) for 10 min. Repeat two more times for a total of 30 min.
Dry microwells at a steep angle (60º or greater) for 30 min. Note: Dishes can be stored for 2 months in a dessicator.
2. Preparation of PAAs for Invadopodia Assays
For a 1 ml solution of the soft PAA (8% acrylamide and 0.05% BIS), combine 200 µl of 40% acrylamide, 25 µl of 2% BIS, and 574 µl of ultrapure water (Table 1).
For a 1 ml solution of the hard PAA (8% acrylamide and 0.35% BIS), combine 200 µl of 40% acrylamide, 175 µl of 2% BIS, and 409 µl of ultrapure water (Table 1).
For a 1 ml solution of the rigid PAA (12% acrylamide and 0.60% BIS), combine 300 µl of 40% acrylamide, 300 µl of 2% BIS, and 169 µl of ultrapure water (Table 1).
Degas the solutions for 15 min. Add 200, 215, and 230 µl of 1 mg/ml human plasma fibronectin in ultrapure water to the soft, hard, and rigid PAA solutions, respectively. For all solutions, add 1 µl of 10 mg/ml acrylic acid N-hydroxysuccinimide (NHS) ester, 5 µl of a 100 mg/ml ammonium persulfate, and 2 µl of N,N,N′,N′-tetramethylethylenediamine (TEMED; Table 1). Mix with gentle pipetting and avoid creating bubbles in solutions.
| PAA (1 ml) | 40% AA (μl) | 2% BIS (μl) | Ultrapure Water (μl) | 1 mg/ml FN (μl) | 10 mg/ml NHS Ester (μl) | 100 mg/ml APS (μl) | TEMED (μl) | Elastic Modulus (Pa) |
| Soft | 200 | 25 | 574 | 200 | 1 | 5 | 2 | 1023 |
| Hard | 200 | 175 | 409 | 215 | 1 | 5 | 2 | 7307 |
| Rigid | 300 | 300 | 169 | 230 | 1 | 5 | 2 | 22692 |
Table 1: Ingredient volumes and resulting elastic moduli for soft, hard, and rigid PAAs used in the invadopodia assays. AA = acrylamide, FN = fibronectin, APS = ammonium persulfate.
Pipette 8.48 µl of the PAA solution on to the center of each microwell. Note: This volume theoretically yields a PAA of 75 µm in thickness.
Gently lower the flamed side of the 12 mm coverslip on to the droplet in the microwell using tweezers.
Allow the sandwiched PAA solution to polymerize for 15-30 min. Verify polymerization by checking the leftover solution.
Add 2 ml of 1x PBS to each dish and remove the 12 mm coverslips with tweezers. Note: 10x PBS is made in the laboratory and composed of 0.011 M KH2PO4, 1.54 M NaCl, and 0.056 M Na2HPO4.
Wash the microwells with 2 ml of 1x PBS at room temperature for 5 min. Repeat two more times for a total of 15 min.
3. Preparation of PAAs for Traction Force Assays
For a 1 ml solution of the soft PAA (8% acrylamide and 0.05% BIS), combine 200 µl of 40% acrylamide, 25 µl of 2% BIS, and 566 µl of ultrapure water (Table 2).
For a 1 ml solution of the hard PAA (8% acrylamide and 0.35% BIS), combine 200 µl of 40% acrylamide, 175 µl of 2% BIS, and 401 µl of ultrapure water (Table 2).
For a 1 ml solution of the rigid PAA (12% acrylamide and 0.60% BIS), combine 300 µl of 40% acrylamide, 300 µl of 2% BIS, and 161 µl of ultrapure water (Table 2).
Degas the solutions for 15 min. Add 200, 215, and 230 µl of 1 mg/ml human plasma fibronectin to the soft, hard, and rigid PAA solutions, respectively. Sonicate 8 µl of 200 nm red fluorescent microspheres (excitation/emission of 580/605 nm) for 30 sec. For each solution, add 8 µl of 200 nm red fluorescent microspheres, 1 µl of 10 mg/ml acrylic acid NHS ester, 5 µl of a 100 mg/ml ammonium persulfate, and 2 µl of TEMED (Table 2). Mix with gentle pipetting and avoid creating bubbles in solutions.
| PAA (1 ml) | 40% AA (μl) | 2% BIS (μl) | Ultrapure Water (μl) | 1 mg/ml FN (μl) | Micro-spheres (μl) | 10 mg/ml NHS Ester (μl) | 100 mg/ml APS (μl) | TEMED (μl) | Elastic Modulus (Pa) |
| Soft | 200 | 25 | 566 | 200 | 8 | 1 | 5 | 2 | 1023 |
| Hard | 200 | 175 | 401 | 215 | 8 | 1 | 5 | 2 | 7307 |
| Rigid | 300 | 300 | 161 | 230 | 8 | 1 | 5 | 2 | 22692 |
Table 2: Ingredient volumes and resulting elastic moduli for soft, hard, and rigid PAAs used in the traction force assays. AA = acrylamide, FN = fibronectin, APS = ammonium persulfate.
Pipette 8.48 µl of the PAA solution on to the center of each microwell. Note: This volume theoretically yields a PAA of 75 µm in thickness.
Gently lower the flamed side of the 12 mm coverslip on to the droplet in the microwell using tweezers.
Allow the sandwiched PAA solution to polymerize for 15-30 min. Verify polymerization by checking leftover solution.
Add 2 ml of 1x PBS to each dish and remove the 12 mm coverslips with tweezers.
Wash the microwells with 2 ml of 1x PBS at room temperature for 5 min. Repeat two more times for a total of 15 min.
4. Preparation of ECM for Invadopodia Assays
Heat the gelatin solution (1% gelatin and 1% sucrose) to 37 °C.
Treat the PAAs with 150 µl of the gelatin solution for 1 min then carefully aspirate from the bottom of the microwells when holding the glass bottom dishes at a 45° angle.
Dry the remaining thin layer of gelatin on the PAAs in the microwells at a steep angle (60° or greater) to optimize drying for 60 min.
Treat the microwells with 2 ml of a chilled 0.5% glutaraldehyde solution on ice for 15 min followed by room temperature for 30 min.
Wash the microwells with 2 ml of a 1x PBS for 5 min. Repeat two more times for a total of 15 min.
Treat the microwells with 2 ml of a sodium borohydride solution for 1 min. Gently tap dishes on bench to remove bubbles that form on the surface of the gelatin.
Aspirate and wash the microwells with 2 ml of a 1x PBS for 5 min. Repeat two more times for a total of 15 min.
- Dilute a FITC-labeled human plasma fibronectin solution to 50 µg/ml with a 1x PBS and centrifuge at 175,000 x g at 4 °C for 15 min. Either purchase commercially available labeled fibronectin or label it as follows:
- Dissolve 5 mg of fibronectin in 10 ml of borate buffer (170 mM sodium metaborate tetrahydrate and 40 mM NaCl) and place in dialysis tubing.
- Label the fibronectin by placing the tubing in 200 ml of borate buffer containing 6 mg of dissolved FITC at room temperature on a magnetic stirrer set at the lowest setting for 1.5 hr in the dark.
- Dialyze with 500 ml of a 1x PBS on a magnetic stirrer set at the lowest setting for 3 days in a dark cold room (4 °C) with two volume changes a day.
- Calculate FITC-labeled fibronectin concentration based on the optical density of diluted aliquots (1:200) at 280 nm and 493 nm as [OD280 – (0.36 x OD493)] / 1.4 times the dilution factor of 200 times 2 (which accounts for the glycerol concentrating the fibronectin in the next step) resulting in mg/ml units.
- Dialyze overnight in a 50% glycerol solution on a magnetic stirrer set at the lowest setting overnight in a dark cold room at 4 °C.)
Treat the microwells with 150 µl of the FITC-labeled fibronectin solution at room temperature for 60 min in the dark.
Carefully aspirate the FITC-labeled fibronectin solution from the bottom of the microwells when holding the glass bottom dishes at a 45° angle and then fill the glass bottom dishes with 70% ethanol solution at room temperature for 10 min in a dark cell culture hood for sterilization. Also fill glass bottom dish lids or wipe clean with low lint wipes soaked in 70% ethanol.
Wash glass bottom dishes by filling them with 1x PBS for 5 min. Repeat two more times with 2 ml of 1x PBS for a total of 15 min.
5. Preparation and Imaging of Invadopodia Assays
Add 25,000 cancer cells in 2 ml of invadopodia medium (1:1 DMEM and RPMI 1640 with 5% low-protein serum, 10% fetal bovine serum, and 20 ng/ml of freshly added epidermal growth factor) to the glass bottom dishes and incubate overnight.
Aspirate and treat the microwells with 2 ml of a 3.7% paraformaldehyde solution at room temperature in the dark for 20 min to fix the cells.
After two quick washes with 2 ml of 1x PBS, treat the microwells with 2 ml of a 0.1% Triton X-100 solution at room temperature in the dark for 5 min to permeabilize the cells.
After one quick wash with 2 ml of 1x PBS, treat the microwells with 3% bovine serum albumin blocking solution at room temperature in the dark for 60 min to block the cells.
Incubate the microwells with 150 µl of mouse anti-cortactin antibody (1:750) in blocking solution at room temperature in the dark for 60 min.
Wash microwells with 2 ml of 1x PBS for 5 min. Repeat two more times for a total of 15 min.
Incubate the microwells with 150 µl of goat anti-mouse 633 antibody (1:500) and 546 phalloidin (1:750) in blocking solution at room temperature in the dark for 60 min.
Wash microwells with 2 ml of 1x PBS for 5 min. Repeat two more times for a total of 15 min.
Add six drops of mounting medium to fill each microwell and cover with 22 x 22 coverslips.
Image cortactin and actin to identify invadopodia using excitation and emission filters of 632/647 nm and 556/570 nm, respectively, using a high NA, 40X oil immersion lens on a wide-field inverted microscope. Similarly, image fibronectin to identify ECM degradation using an excitation and emission filter of 492/518 nm.
6. Preparation and Imaging of Traction Force Assays
Add 15,000 cancer cells in 2 ml of invadopodia medium to the glass bottom dishes and incubate overnight.
Aspirate medium in the glass bottom dishes and replace with 2 ml of L-15 medium, with the same supplements (5% low-protein serum, 10% fetal bovine serum, and 20 ng/ml of freshly added epidermal growth factor), pre-warmed to 37 °C.
Incubate the glass bottom dishes in an environmental chamber on an inverted microscope pre-equilibrated to 37 °C with high humidity for 1 hr.
- Take four sets of images using a high NA, 40X lens on a wide-field inverted microscope with an automated stage to mark cellular positions:
- Image cells on top of the PAAs using phase contrast (“phase” image).
- Image fluorescent microspheres using excitation and emission filters of 560/645 nm by focusing slightly under the cells until the first layer of microspheres at the top of the PAAs comes into focus (“stressed” image).
- In addition, focus to the bottom of the PAAs (using the microspheres as markers) and take an image (“bottom” image). Note: The purpose of this image is to record the z-position to get the actual PAA thickness at each position for traction force calculations as described in Representative Results.
- After these images have been acquired at multiple positions, gently mix in 220 µl of 10% Triton-X solution to remove cells. Image microspheres at the top of the PAAs at each position as previously described (“null” image).
Representative Results
In the invadopodia assay, invadopodia are typically identified by colocalization of markers like actin and cortactin at punctate structures within the cell body (Figure 1). Both actively degrading and non-degrading invadopodia can be counted and are differentiated by whether these structures are colocalized with black areas lacking fluorescent signal in the FITC-labeled fibronectin (Figure 1). Invadopodia are manually counted, and ECM degradation per cell is determined by manually thresholding these black areas within outlines of the cells.
In the traction force assay, four images are captured at each cellular location marked by stage position (Figure 2). First, “phase” and “stressed” images are taken of all of the cells of interest. A “bottom” image of the PAA is also taken at each position in order to calculate hydrogel thickness. After removing the cells, a “null” image is taken at each marked stage position. Deformations in the PAAs are calculated based on the change in microsphere positions between the “stressed” and “null” images. These displacements and the mechanical properties of the PAAs (E and Poisson’s ratio assumed as 0.5) are then used to calculate traction forces (Figure 2). Several different methods exist for calculating traction forces24 based on some formulation of the Boussinesq solution for an infinite elastic half space25. While the details of these analyses are beyond the scope of this text, we have licensed LIBTRC software from Micah Dembo at Boston University which uses a previously described method for calculating traction forces26. This particular computational method compensates for finite thicknesses in its calculations which requires the thickness of the PAAs at each position which is calculated based on the difference in the z-position of the “stressed” and “bottom” images. Many research groups have similar computer packages available or choose to write their own programs. In addition, other methods exist for calculating traction forces based on different mathematical approaches24.
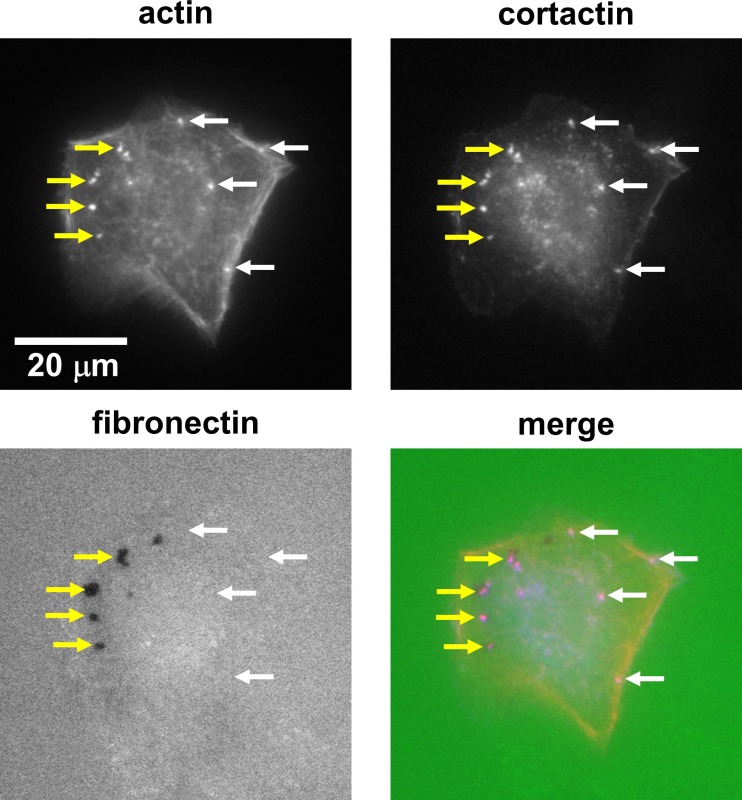 Figure 1: The invadopodia assay can be used to identify invadopodia and associated ECM degradation. Example wide-field fluorescence images of invadopodia in a SCC-61 (head and neck squamous cell carcinoma) cell on a hard PAA with 1% gelatin and FITC-labeled fibronectin. Invadopodia are typically identified by colocalization of two markers such as actin and cortactin (red and blue in the overlay image, respectively). Invadopodia can be quantitated as both actively degrading (colocalized with black areas lacking FITC signal as denoted by yellow arrows) and non-degrading (denoted by white arrows). Please click here to view a larger version of this figure.
Figure 1: The invadopodia assay can be used to identify invadopodia and associated ECM degradation. Example wide-field fluorescence images of invadopodia in a SCC-61 (head and neck squamous cell carcinoma) cell on a hard PAA with 1% gelatin and FITC-labeled fibronectin. Invadopodia are typically identified by colocalization of two markers such as actin and cortactin (red and blue in the overlay image, respectively). Invadopodia can be quantitated as both actively degrading (colocalized with black areas lacking FITC signal as denoted by yellow arrows) and non-degrading (denoted by white arrows). Please click here to view a larger version of this figure.
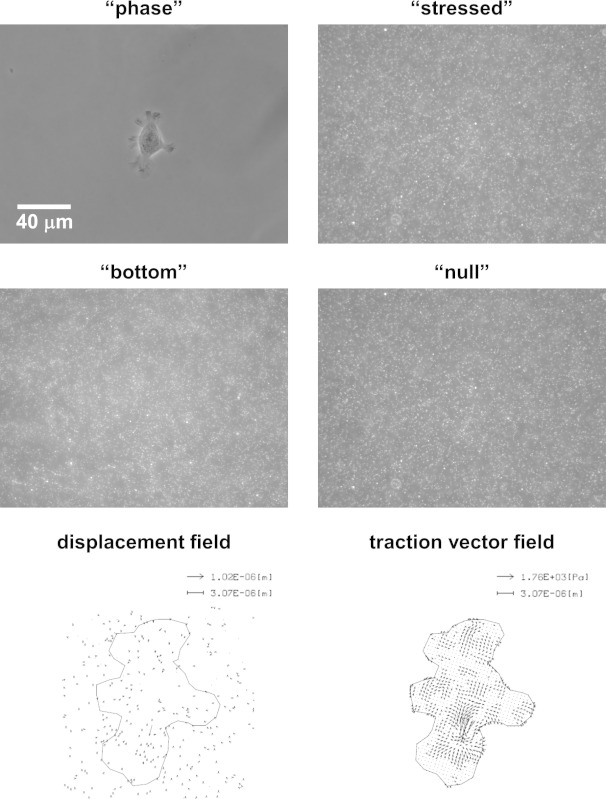 Figure 2: The traction force assay can be used to determine cellular traction forces generated by the actin cytoskeleton by tracking the displacement of embedded microspheres in the PAAs. Example wide-field phase and fluorescence images of a SCC-61 cell on a hard PAA (“phase”), and the microspheres directly under the cell at the top surface of the PAA (“stressed”). An image of the bottom of the PAA can also be taken to later calculate its local thickness (“bottom”). After the cell is removed, an image is once again taken of the microspheres at the top surface of the PAA (“null”). Microsphere positions between the “stressed” and “null” images can be tracked to yield a “displacement field.” Microsphere displacements and PAA mechanical properties can then be used to calculate a “traction vector field.” Please click here to view a larger version of this figure.
Figure 2: The traction force assay can be used to determine cellular traction forces generated by the actin cytoskeleton by tracking the displacement of embedded microspheres in the PAAs. Example wide-field phase and fluorescence images of a SCC-61 cell on a hard PAA (“phase”), and the microspheres directly under the cell at the top surface of the PAA (“stressed”). An image of the bottom of the PAA can also be taken to later calculate its local thickness (“bottom”). After the cell is removed, an image is once again taken of the microspheres at the top surface of the PAA (“null”). Microsphere positions between the “stressed” and “null” images can be tracked to yield a “displacement field.” Microsphere displacements and PAA mechanical properties can then be used to calculate a “traction vector field.” Please click here to view a larger version of this figure.
Discussion
We present a method for fabricating PAAs that can be used as the basis for invadopodia and traction force assays to correlate invasive and contractile cellular behaviors. While PAAs have long been used to look at rigidity effects on cells and calculate traction forces18,24,27, this protocol is the first to develop parallel assays based on PAAs with the same rigidities to correlate invasive and contractile cellular behaviors in response to matrix mechanical properties. Properly activating the coverslips of the glass bottom dishes ensures that the PAAs will bind to them and not come off. While we and others have relied on reagents such as sulfo-SANPAH and acrylic acid NHS ester to bind gelatin to the surface of the PAAs in the past4,12,28, we found that these methods did not produce reliably uniform surface layers of fibronectin. Therefore, fibronectin was embedded and crosslinked throughout the PAAs using acrylic acid NHS ester as performed by other groups29,30. However, increasing concentrations of fibronectin were required in order to yield the same ligand density at the surfaces of the soft, hard, and rigid PAAs5. Additionally, incorporation of fibronectin (but not the microspheres) reduced the mechanical properties of the soft, hard, and rigid PAAs from 1,071, 9,299, and 28,283 Pa4 to 1,023, 7,307, and 22,692 Pa5, respectively. However, these reduced elastic moduli values still encompassed the range of normal and cancerous tissues23. Storage moduli of the PAAs can easily be measured by rheometry and converted to elastic moduli1,4.
While the protocol is straight forward, lowering and removing the 12 mm coverslips are the most difficult steps and require practice. The biggest challenge when lowering a coverslip is to ensure that the PAA solution spreads out evenly without any bubbles or splashing. Removing a coverslip requires a steady hand in order to not damage the 12 mm coverslip, the glass bottom 14 mm coverslip, or the PAA. We have tried hydrophobic coatings on the 12 mm coverslip to aid in removal but found that the PAAs are left with patterns in their surfaces. The use of fine tip and thin, paddle-style tweezers for lowering and removing the 12 mm coverslips, respectively, is highly recommended. In addition, the amount of light used to image the cells must be minimized since some cell types are sensitive to concentrated light on the microscope. Also, L-15 medium was used for the traction force assays since our environmental chamber on our microscope is not equipped for CO2. While the same supplements were used in the media for the invadopodia and traction force assays in an attempt to keep conditions the same, DMEM and RPMI 1640 could be used instead of L-15 if CO2 levels can be controlled.
While the volume of PAA solution was chosen to theoretically yield gels with a thickness of 75 µm, their thicknesses typically vary between 30-60 µm. However, this range is well above the value at which cells can sense the underlying rigidity of the coverslips of the glass bottom dishes31. In addition, the thickness of the ECM layer used to detect degradation (gelatin and FITC-labeled fibronectin overlaid on to the PAAs) has been previously reported as approximately 1 µm in thickness12; therefore, this thin layer does not shield the cells from the rigidities of the PAAs. However, solid debris, cracks, and other deformities in either the PAAs and/or ECM layer can affect individual cellular invasive and contractile properties. These problems can be caused by unclean 12 mm coverslips that introduce debris into the hydrogels, excessive drying of the gels and/or gelatin, and incomplete aspiration of sodium borohydride which can leave bubbles that deform the ECM layer. Therefore, care must be taken when selecting cells for imaging to ensure that they are not unduly influenced by local irregularities in the surfaces of the samples.
Relatively high concentrations of fibronectin in both the PAAs (mixed in at 200-230 µg/ml) and the ECM layer (overlaid on the gelatin at 50 µg/ml) have been used; however, the actual concentrations on either surface have not been directly measured. While each surface appears to be saturated with fibronectin, it is unclear whether cells experience the same exact ligand densities between the two assays. For the traction force assays, 200 nm microspheres at a dilution of 1:125 have proven optimal for imaging. However, it is quite common to find other groups using microspheres of different diameters or dilutions. In this system, smaller microspheres displayed Brownian motion within the PAAs, while larger microspheres caused cracks and deformities. These observations are most likely due to the physical properties of the PAAs (i.e., pore size, degree of crosslinking, etc.) that result from the specific amounts of acrylamide and BIS that were used. The microsphere dilution provides excellent resolution for tracking displacements in the PAAs, particularly since cancer cells exert relatively small forces. For improved optical resolution, particularly if using even smaller microspheres, high NA water or oil immersion objectives can be used.
Overall, many of these experimental factors can be adjusted based on the user’s requirements such as PAAs with different mechanical properties, cancer cells with varying invasive and contractile properties, and microscopy systems with other imaging capabilities. The ratio of acrylamide:BIS can be varied to produce PAAs with similar or different elastic moduli and crosslinking27. The sensitivity of the traction force assays can be adjusted to account for different cellular force levels by changing the rigidity and/or microsphere dilution. Different fluorophores can also be chosen for immunofluorescence and fibronectin labeling based on microscope specifications. While FITC does bleach, it is quite bright making ECM degradation easily identifiable. However, fluorescence imaging of invadopodia and small microspheres typically requires high NA objectives for optimal resolution. In addition, both assays can be performed on glass coverslips in well plates. However, glass bottom dishes are much easier to prepare and use throughout the experimental process. In the future, combining these assays into one would allow for direct visualization of both ECM degradation and cellular force generation. However, several technical challenges would arise including live cell imaging for invadopodia and microbead displacements, increased cellular exposure to light, bleaching of the FITC fibronectin, and whether traction forces are completely transduced through the fluorescently labeled and cross-linked ECM layer to the PAA surfaces.
Disclosures
This work was supported by the National Cancer Institute of the National Institutes of Health under Award Number K25CA143412 (Parekh). We would like to acknowledge that additional support was provided by the Department of Otolaryngology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgments
The authors have nothing to disclose.
References
- Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nature. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Weaver VM. The tension mounts: mechanics meets morphogenesis and malignancy. Journal of mammary gland biology and neoplasia. 2004;9:325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- Parekh A, et al. Sensing and modulation of invadopodia across a wide range of rigidities. Biophysical. 2011;100:573–582. doi: 10.1016/j.bpj.2010.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerrell RJ, Parekh A. Cellular traction stresses mediate extracellular matrix degradation by invadopodia. Acta biomaterialia. 2014;10:1886–1896. doi: 10.1016/j.actbio.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraning-Rush CM, Califano JP, Reinhart-King CA. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE. 2012;7:e32572. doi: 10.1371/journal.pone.0032572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haage A, Nam DH, Ge X, Schneider IC. Matrix metalloproteinase-14 is a mechanically regulated activator of secreted MMPs and invasion. Biochemical and biophysical research communications. 2014. [DOI] [PubMed]
- Haage A, Schneider IC. Cellular contractility and extracellular matrix stiffness regulate matrix metalloproteinase activity in pancreatic cancer cells. FASEB J. 2014. [DOI] [PubMed]
- Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clinical & experimental metastasis. 2006;23:97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- Weaver AM. Invadopodia. Curr Biol. 2008;18:R362–364. doi: 10.1016/j.cub.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Current opinion in cell biology. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander NR, et al. Extracellular matrix rigidity promotes invadopodia activity. Curr Biol. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow WE, et al. Prospective breast cancer risk prediction model for women undergoing screening mammography. Journal of the National Cancer Institute. 2006;98:1204–1214. doi: 10.1093/jnci/djj331. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. Projecting absolute invasive breast cancer risk in white women with a model that includes mammographic density. Journal of the National Cancer Institute. 2006;98:1215–1226. doi: 10.1093/jnci/djj332. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nature reviews. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MH, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munevar S, Wang Y, Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophysical journal. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel D, et al. Up-regulation of Rho/ROCK signaling in sarcoma cells drives invasion and increased generation of protrusive forces. Mol Cancer Res. 2008;6:1410–1420. doi: 10.1158/1541-7786.MCR-07-2174. [DOI] [PubMed] [Google Scholar]
- Indra I, et al. An in vitro correlation of mechanical forces and metastatic capacity. Phys Biol. 2011;8:015015. doi: 10.1088/1478-3975/8/1/015015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh A, Weaver AM. Regulation of cancer invasiveness by the physical extracellular matrix environment. Cell adhesion & migration. 2009;3:288–292. doi: 10.4161/cam.3.3.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Page JM, Guelcher SA, Parekh A. Coutts AS, editor. Methods in Molecular Biology in Adhesion Protein Protocols. 3rd Edition. 2013. pp. 171–189. [DOI] [PubMed]
- Samani A, Zubovits J, Plewes D. Elastic moduli of normal and pathological human breast tissues: an inversion-technique-based investigation of 169 samples) Physics in medicine and biology. 2007;52:1565–1576. doi: 10.1088/0031-9155/52/6/002. [DOI] [PubMed] [Google Scholar]
- Wang JH, Lin JS. Cell traction force and measurement methods. Biomechanics and modeling in mechanobiology. 2007;6:361–371. doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophysical journal. 1996;70:2008–2022. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophysical journal. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Rehfeldt F, Sen S, Discher DE. Microtissue elasticity: measurements by atomic force microscopy and its influence on cell differentiation. Methods Cell Biol. 2007;83:521–545. doi: 10.1016/S0091-679X(07)83022-6. [DOI] [PubMed] [Google Scholar]
- Kandow CE, Georges PC, Janmey PA, Beningo KA. Polyacrylamide hydrogels for cell mechanics: steps toward optimization and alternative uses. Methods in cell biology. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- Leach JB, Brown XQ, Jacot JG, Dimilla PA, Wong JY. Neurite outgrowth and branching of PC12 cells on very soft substrates sharply decreases below a threshold of substrate rigidity. J Neural Eng. 2007;4:26–34. doi: 10.1088/1741-2560/4/2/003. [DOI] [PubMed] [Google Scholar]
- Zhou J, Kim HY, Wang JH, Davidson LA. Macroscopic stiffening of embryonic tissues via microtubules, RhoGEF and the assembly of contractile bundles of actomyosin. Development. 2010;137:2785–2794. doi: 10.1242/dev.045997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Rajagopal K, Brown AE, Discher DE. How deeply cells feel: methods for thin gels. J Phys Condens Matter. 2010;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]


