Abstract
Although the biological importance of exosomes has recently gained an increasing amount of scientific and clinical attention, much is still unknown about their complex pathways, their bioavailability and their diverse functions in health and disease. Current work focuses on the presence and the behavior of exosomes (in vitro as well as in vivo) in the context of different human disorders, especially in the fields of oncology, gynecology and cardiology.
Unfortunately, neither a consensus regarding a gold standard for exosome isolation exists, nor is there an agreement on such a method for their quantitative analysis. As there are many methods for the purification of exosomes and also many possibilities for their quantitative and qualitative analysis, it is difficult to determine a combination of methods for the ideal approach.
Here, we demonstrate nanoparticle tracking analysis (NTA), a semi-automated method for the characterization of exosomes after isolation from human plasma by ultracentrifugation. The presented results show that this approach for isolation, as well as the determination of the average number and size of exosomes, delivers reproducible and valid data, as confirmed by other methods, such as scanning electron microscopy (SEM).
Keywords: Biochemistry, Issue 95, Exosome, exosome separation, microvesicle, nanoparticle-tracking analysis, nanoparticle counting, particle size analysis, ultracentrifugation, exosome isolation, blood
Introduction
The exact function of circulating exosomes remained unknown for a long period of time. Even now the full path mechanism of exosomes is not fully understood. Since exosomes carry antigens, proteins and RNA (mRNA and miRNA) that relates them to their parental cell of origin, their function as cell-cell signaling transmitters has mainly been given priority.
Many different methods have been described in the literature for the isolation and quantitative detection of exosomes 1,2. However, no consensus on a ‘gold standard’ has been reached. Meanwhile the majority of scientists active in the field of exosome research agree that a consistent method of isolation is highly warranted to achieve a higher degree of comparability between different reports and studies.
Fluorescence activated cell sorting (FACS) is the most common and prevalent tool for exosome analysis 3. FACS has the benefit that, via fluorescence labeling, cells from different origins can be compared in one step. The major disadvantages of FACS are that this method is not sensitive enough to identify particles smaller than 0.5 µm 4, whereas exosomes are generally between 30-120 nm in diameter 5, even less to measure their size.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) are other tools for the analysis of particle size and morphology of exosomes. However, both SEM and TEM have the disadvantage that the preparation of samples is time-consuming, both methods involve labor-intensive steps and each has some risk of artifact generation. Neither method is suitable for high sample throughput and characterization of several thousands of single particles of one sample. Moreover, a quantitative analysis for clinical daily routine where samples have often to be analyzed simultaneously or at least in a very short period is difficult to perform. New generation techniques now allow us to analyze exosomes without prior intensive preparatory work (e.g., environmental SEM). These modern techniques are still rather inconvenient for analyzing large volume suspensions containing exosomes to determine their average number and size distribution 6.
Another highly sensitive method for visualization and analysis of exosomes is nanoparticle-tracking analysis (NTA). This method takes advantage of two different principles of physics. First, particles are detected by the light scattered when they are irradiated with a laser beam. The second phenomenon is known as Brownian motion, according to which the diffusion of different particles in a liquid suspension is inversely proportional to their size. In the latter case the movement also depends on the temperature and the viscosity of the liquid. Yet, this rate is directly related to particle size and is used by NTA. Using software-based analysis, digital images of scattered light from single particles are recorded. Plots of scattered light spots and their speed of motion provide the data that facilitate the determination of total particle count and size distribution. This technique is particularly powerful for analyzing particles with a mean diameter of less than 100 nm.
The size and concentration measurements are performed with the ZetaView Brownian and Electrophoresis Motion Video Analysis Microscope. This is a semi-automated desk top nanoparticle analysis instrument for liquid samples (hereafter referred to as the particle tracking instrument). It consists of the particle tracking analyzer as well as a laptop with the software used for the data analysis. Heterogeneous biological samples are as suitable for this method as more homogeneous suspensions of inorganic particles. A laser scattering microscope with a video camera is used for the detection of particles and for the observation of their movement. While the microscope axis is horizontal and focused into the cell channel filled with a suspension containing exosomes, the laser beam is oriented vertically. The particles irradiated by the laser scatter the light, which is recorded under 90° by a digital video camera via the microscope (Figure 1). The intensity of the scattered light allows observation of particles larger 60 nm diameter. In such a setting the brightness of a particle is not the only indication of particle size. When no electrical field is applied, particle movement only follows Brownian motion and may serve as an indicator for calculating particle size. However, the instrument is also capable of applying an electrical field across the cell channel. When subjected to this field, the potential, polarity and level of ionic-charge of the suspended exosomes become further determinants of the direction of their movement. Velocity and direction result in an electrophoretic mobility histogram.
While finding an optimal method to analyze isolated exosomes is one problem, another one lies in the effective isolation of exosomes from different media, such as blood, ascites, urine, milk, amniotic fluid or cell media. Different methods have been described thus far, which are based on ultracentrifugation 1, industrial separation reagents (such as Exoquick) 7, magnetic beads for antigen employing separation 8 or ultrafiltration steps 9.
In this protocol we demonstrate the entire process of exosome isolation via ultracentrifugation and show how to analyze the resulting exosome containing suspension via the particle tracking instrument. Specific considerations for the analysis of human plasma or cell culture medium derived exosomes are provided.
Protocol
NOTE: The experiments presented in this work have been approved by the institutional ethical board of the University of Dusseldorf.
1. Exosome Preparation
Collect whole blood in 3 citrate tubes via venipuncture (total of 9 ml). Pour the blood into a 15 ml Falcon tube.
Centrifuge the sample at 1,500 x g for 20 min at 4 °C to initiate separation of cells from plasma. Transfer the supernatant to a new 15 ml falcon tube.
Centrifuge the sample at 2,800 x g for 20 min at 4 °C to remove all cells from plasma (cell-free plasma; CFP). Transfer the CFP to ultracentrifugation tubes, 1 ml per tube.
Centrifuge at 100,000 x g for 90 min at 4 °C to deplete exosomes. Remove 900 µl of supernatant. Re-suspend the pellet in the remaining 100 µl in the ultracentrifugation tube. Add 900 µl PBS.
Centrifuge again at 100,000 x g for 30 min at 4 °C. Remove 900 µl of supernatant. Re-suspend the pellet with the remaining 100 µl.
Transfer 5 to 20 µl of re-suspension in 40 ml distilled water. Filter the suspension through a 450 nm filter to separate exosomes from larger particles. Use this final suspension for particle measurement.
2. Startup Procedure of the Particle Tracking Instrument
Start the program by double clicking on the software-icon. Click on the various software tabs (“Cell Check”, “Measurement“, “Analysis”) to switch between them throughout the protocol.
Follow the instructions on the screen for an automated implementation of the startup procedure. Select the boxes for both cell quality check and auto alignment. NOTE: Either of these steps can be repeated separately if necessary by pressing button (A) or (B) on the “cell check” tab.
Open the inlet and outlet port of the NTA instrument and inject 10 ml distilled water by syringe into the cell channel through the inlet port. Close the outlet port as the last amount of water is being injected. Place a beaker under the outlet port to collect waste solution. Ensure that the measurement cell is free of air bubbles and do not syringe air bubbles into the system. Close the inlet port immediately.
Perform the cell quality check by clicking on “OK”. The software will display the cell quality results after a few seconds.
If any particles are still visualized in the live-view screen of the software or if the result of the quality check is only good or poor, repeat step 2.3 until the remaining particles are removed from the measurement cell. Also repeat step 2.3 after each measurement to avoid accumulation of particles. If repeating the distilled water injection and cell check does not produce a “good” outcome, proceed to 2.9.
Prepare a control suspension containing uniform 200 nm sized polystyrene particles to be used to align the foci of the laser and microscope. Add one drop of the suspension concentrate, provided by the instrument manufacturer, to 500 ml of distilled water to obtain the required concentration such that 600 ± 100 particles are displayed per screen in the live-view.
Inject the alignment suspension into the NTA instrument as described in step 2.3. Press “OK” to start auto alignment, an automated routine through which the system will automatically find the optimum position of the two foci.
When a prompt appears stating the system is now ready for experiments, click on OK to start the measurement.
Occasionally, clean the cell channel manually between experiments by flushing the cell with a 30% solution of ethanol. Clean the channel whenever the software displays an automatic error report.
3. Measurement of Sample
Flush the channel with distilled water prior to each sample measurement (as described in step 2.3).
Inject the exosome suspension (prepared in section 1) in to the channel, as described in step 2.3.
- Adjust the following main parameters in the software as needed:
- Sensitivity. In order to find the optimal sensitivity range, click on the button “Number of Particles vs. Sensitivity” to display a curve for measured particles per screen for different sensitivity levels. Chose a sensitivity level before the maximum slope of the curve. A higher sensitivity level allows visualization of more small particles but also increases issues related to background noise.
- Minimum Brightness. Chose a starting brightness of 20 for exosome measurement to use this function as a filter. Regulate this parameter up to blank out strongly light-scattering particles. Regulate it down to amplify weakly scattering artifacts. Vary brightness as needed throughout the experiment. NOTE: Particles that light-scatter too strongly overlap and can be excluded from the analysis by mistake.
- Min and Max Size. Set a digital filter to eliminate pixel noise and unwanted scatter from oversized particles by regulating the minimum and maximum size. Use a range of 10 to 500 pixels for measurement of exosomes.
- Shutter. Adjust the period of time that the camera is open to 1/300 sec.
- Live read out parameters:
- Switch between a digital and an analog view modus at any point by clicking on the “Live-image” button.
- Throughout the experiment, monitor the scattering Intensity bar, which displays the saturation status of the sample as a color coded indicator. Do not analyze exosomes if the scattering bar is red. In such a case further dilute the sample to avoid this phenomenon. If the experimental condition prohibits a manipulation of the sample suspension, down-regulate the sensitivity or up-regulate the brightness in the software-settings to improve the measurement results. NOTE: When samples with an extremely high concentration of particles are analyzed the scatter will fuse individual particles and they are counted as one single particle.
- Note the number of particles counted in the field of vision from the display.
Click on the “Measurement” tab.
- Choose settings in the “run options” array.
- Before each acquisition, select “check movement’ for a repeated check particle drift test. Perform the test at least once before starting the whole analysis. If the drift is higher than 20 µm/sec, wait before continuing the measurement so that the sample stops flowing.
- Select the number of experiments (5) and the time delay between them (0).
- Perform a “sample check’ if necessary. This test is part of the auto alignment in the start up procedure and can be repeated afterwards as needed.
- Finally click on the “run video acquisition” button. In the new window, define the number of cycles (15) and the number of measurement positions (11). NOTE: The laser head and the microscope can be moved to analyze the particle number at 11 different positions. Depending on the experimental demand, decide whether the experiment should be performed on one single position or on different positions.
- Confirm the minimal time duration a particle has to be tracked to be counted as one individual particle by setting the video resolution (low). A lower resolution results in a shorter tracking duration.
- Select a file-name and click on “OK” to start measurement.
4. Interpretation of the Results
- View the following results and parameters displayed after measurement on the “results” tab: (1) total number of particles traced, (2) average number of particles per position, and (3) particle concentration.
- View the result average table for the numerical ratio of particle size (diameter, surface and volume) to the percentage of particle numbers, as well as values of mean and standard differentiation.
- Use the peak analysis table if more than one peak exists in the graphical analysis. This table shows the distribution of particles that are smaller than the first or respectively second peak.
View the graph calculated after the measurement to see a distribution of particles by size. Use the icons in the display mode and above the graph to change the settings.
Representative Results
The sample used for this demonstration shows an optimum setting for measurement with a sensitivity of 85%, before the maximum slope of the sensitivity curve (Figure 2). The brightness, the min/max values were chosen as recommended in the protocol. A concentration of 5.3 particles/ml x 106 was measured, while the mean size of the particles was 0.149 µm, most of them were 0.137 µm.
The values received after measurement can be saved in a report with a file format of .pdf or as a .txt for exporting into a database. The graph can be adjusted as preferred as described in the protocol (section 4). The video sequence is also saved and can be used for later off-line re-analysis. However, in such an off-line analysis, the pre-acquisition settings of the camera cannot be changed retrospectively.
In order to find the optimum parameter settings for a measurement, we here describe the optimization of the instrument settings on the example of a 100 nm polystyrene size standard. The influence of two parameters, sensitivity and min/max size on video image and particle size distribution is discussed in detail. All other parameters are summarized in Table 1.
A visual impression of the impact of sensitivity (ranging from 50 to 94) on analog and digital images is visualized in Figure 3. The quantitative information derived from the images is presented in Figure 4, based on the settings in Table 1, Min Size = 5 and Max Size = 200. A typical relationship of the number of detected particles vs. sensitivity is shown in Figure 4A. Between 50 and 90, the number of detected particles increased with sensitivity and rose dramatically for sensitivities >90. An optimum range of sensitivity was found between 66 and 86 (A). Particle size distributions obtained with different sensitivity settings are shown in Figure 4B. The particle size distributions represent the mean of three individual measurements. For too low a sensitivity (sensitivity = 62, red curve) only a few particles were analyzed resulting in rather poor statistics. The number of analyzed particles increased with sensitivity and reached an optimum between 70 (yellow curve) and 86 (tan curve). Further increasing the sensitivity lead to a deterioration of the particle size distribution with the number of particles dropping and size distribution shifting towards smaller sizes (sensitivity = 94, blue curve). Figure 4C shows the trend of the number based x50 diameter (50% of particles are smaller than this diameter) as a function of sensitivity. In the beige interval, the RSD of particle size was less than 8% and corresponds to the optimal interval in A. The red regions indicate RSD > 8% as a result of poor statistics (sensitivity too low) or broad distributions with shift to smaller sizes (sensitivity too high).
The settings of Min Size and Max Size are filters applied to digital images in order to remove particles with spot sizes smaller than Min Size and larger than Max Size. Due to the ability to scatter light, a particle creates a spot of a certain size on a digital image. The size of the spot is measured as a number of pixels (px). When a particle scatters light very well (e.g., particles > 200 nm or aggregates), the spot size is quite large, e.g., > 500 px. The spot size is quite small (e.g., < 10 px) for small particles (e.g., < 20 nm) depending on the particle material. Spot size (px) may not be interchanged with particle size (nm) as they are not identical and there is no direct relationship between these two variables. An optimization of Min and Max Size allows the user to filter out unwanted objects such as agglomerates (Max Size) or small objects such as background noise (Min Size). The influence of Min/Max Size on the particle size distribution of a 100 nm size standard is shown in Figure 4D (sensitivity = 82). When the interval is set to small spot sizes (e.g., min = 1, max = 52; orange curve), the number of analyzed particles is reduced and the number based x50 diameter is slightly shifted towards smaller sizes. A setting for larger spots (min = 40, max = 1,000; red curve) results in a broad particle size distribution shifted towards larger sizes. In order to obtain equal total numbers of particles, the interval boundaries of both orange and red distributions were adjusted to match 80 particles. The distribution with optimal settings (min = 5, max = 200; tan curve) consists of 360 particles.
A series of successful experiments were performed with exosomes isolated by ultracentrifugation and measured by NTA using the presented system. The resulting data were highly consistent and confirmed a high level of reproducibility. Other isolation methods should show similar results. However, the dilution step was identified as a particularly critical step and its impact on the calculated total numbers of particles has to be re-evaluated.
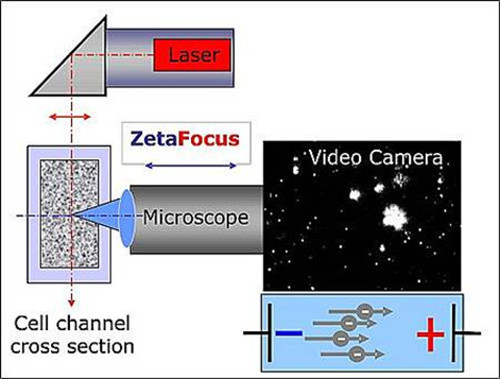 Figure 1. Schematic of the setup of the NTA. The microscope / video axis and laser beam are orientated orthogonally to each other, crossing at the cell channel cross section. Light scattered by the particles is displayed in the “live-view” window of the software. Please click here to view a larger version of this figure.
Figure 1. Schematic of the setup of the NTA. The microscope / video axis and laser beam are orientated orthogonally to each other, crossing at the cell channel cross section. Light scattered by the particles is displayed in the “live-view” window of the software. Please click here to view a larger version of this figure.
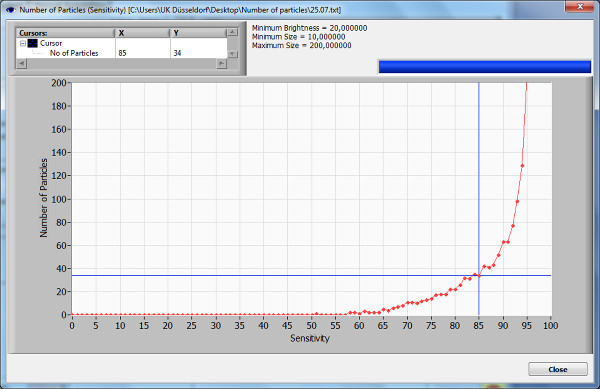 Figure 2. Number of particles vs. sensitivity curve. The number of particles vs. sensitivity curve displays the particles in one position at one moment during an automatic sensitivity scan. This graphical visualization is used to determine the best preferences for the initial measurements. A sensitivity value is chosen prior to the maximum slope of the graph. It is important to remember that artifacts are not eliminated in this test experiment and can affect the graph. Please click here to view a larger version of this figure.
Figure 2. Number of particles vs. sensitivity curve. The number of particles vs. sensitivity curve displays the particles in one position at one moment during an automatic sensitivity scan. This graphical visualization is used to determine the best preferences for the initial measurements. A sensitivity value is chosen prior to the maximum slope of the graph. It is important to remember that artifacts are not eliminated in this test experiment and can affect the graph. Please click here to view a larger version of this figure.
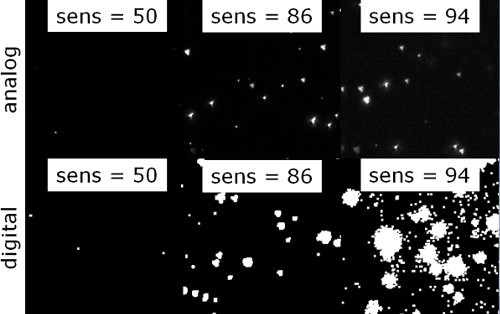 Figure 3. Impact of sensitivity on analog and digital image. Visualization of particles on the live view screen is displayed for sensitivities between 50 and 94 for both analog (top row) and digital (bottom row) views. When the sensitivity is too low, only a few particles are detected (left). At optimum sensitivity the particles appear as single dots well isolated from each other (middle). At a comparatively high sensitivity particles merge together leading to poor image quality (right). Please click here to view a larger version of this figure.
Figure 3. Impact of sensitivity on analog and digital image. Visualization of particles on the live view screen is displayed for sensitivities between 50 and 94 for both analog (top row) and digital (bottom row) views. When the sensitivity is too low, only a few particles are detected (left). At optimum sensitivity the particles appear as single dots well isolated from each other (middle). At a comparatively high sensitivity particles merge together leading to poor image quality (right). Please click here to view a larger version of this figure.
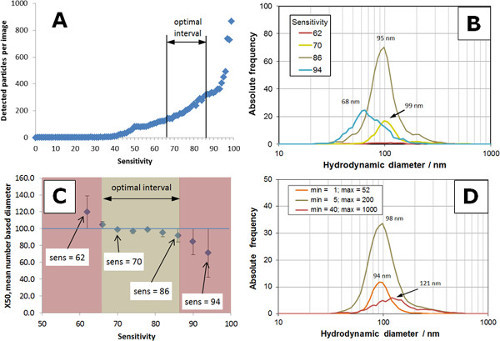 Figure 4. Influence of sensitivity min and max size settings for a control 100 nm polystyrene particle sample. (A) Plot of sensitivity versus detected number of particles; the optimal interval is from 66 to 86, prior to the maximum slope of the curve. (B) Particle size distributions obtained with several sensitivity settings (62 to 94); graphs for too low (62) or too high (94) sensitivity do not capture the particle size distribution for the 100 nm polystyrene control sample. (C) Number based x50 diameter versus sensitivity; the error of x50 in the beige interval is less than 8%, optimal interval is from 66 to 86. (D) Impact of Min and Max Size on the particle size distribution; optimal parameters (min = 5 and max = 200) capture the correct distribution for the control sample. Please click here to view a larger version of this figure.
Figure 4. Influence of sensitivity min and max size settings for a control 100 nm polystyrene particle sample. (A) Plot of sensitivity versus detected number of particles; the optimal interval is from 66 to 86, prior to the maximum slope of the curve. (B) Particle size distributions obtained with several sensitivity settings (62 to 94); graphs for too low (62) or too high (94) sensitivity do not capture the particle size distribution for the 100 nm polystyrene control sample. (C) Number based x50 diameter versus sensitivity; the error of x50 in the beige interval is less than 8%, optimal interval is from 66 to 86. (D) Impact of Min and Max Size on the particle size distribution; optimal parameters (min = 5 and max = 200) capture the correct distribution for the control sample. Please click here to view a larger version of this figure.
| Pre-acquisition parameters | |
| Sensitivity | variable |
| Shutter | 40 |
| Frame rate | 30 fps |
| Resolution | High |
| Cycles | 10 |
| Multiple Acquisitions | 3 |
| Position | 1 |
| Post-acquisition parameters | |
| Min Brightness | 30 |
| Max Size | variable |
| Min Size | variable |
Table 1. Summary of pre- and post-acquisition parameters for setting of the particle tracking instrument.
Discussion
We demonstrate a detailed protocol for exosome isolation from blood and present nanoparticle tracking as a novel and innovative method to measure exosome size and concentration in a biological fluid. In the presented experiments human peripheral blood was used as the origin of the exosomes. However, other origins, such as urine, sputum, cell culture supernatant, etc., may also be used as test material.
Based on the biological variability of exosome concentration in humans, assays from different individuals may feature a remarkably varying concentration of exosomes. However, the concentration of particles in the biological test probe may have an impact on the results. Therefore, a reliable and standardized approach for the dilution of probes is needed. In the presented method exosome containing plasma was generated from 9 ml of peripheral whole blood. Using differential centrifugation steps an exosome pellet was isolated from 1 ml plasma and re-suspended in 5 ml distilled water to result in the working sample of suspended exosomes. This pre-defined setting has provided us with a proper concentration of particles, i.e., appropriate scatter intensity during NTA. Beside the volume and dilution aspect, the composition of the solutions that are used is also of importance. We have used distilled water for the final re-suspension of exosomes. Of course, different media for the final dilution step may also be used, according to the requirements of the experimental set-up. However, in the case of zeta potential measurements of ionic-solutions, particularly when testing samples with a buffering capacity, a prudent dilution in turbulence free conditions is useful. For the direct comparison of multiple samples we recommend keeping the same dilution level for all samples. All settings except the sensitivity and the video resolution can be changed afterwards by using the ZetaView Analysis software.
Although the authors believe that the herein introduced system currently represents the most suitable instrument, it is not the only available NTA system. A number of previous reports have employed other systems that apply the same NTA principles but provide another instrument design that goes along with some differing features 10. We believe that practical aspects concerning the sample handling and reproducibility of the results are of central importance in choosing the methodological sequence for exosome evaluation. We also believe that an ideal detection system should eliminate user-depending factors that may interfere with the results of the measurements. The exosome analyzing approach that is presented here fulfills the criterion of easy handling to a high degree. As a further advantage, the semi-automated analysis of the samples is performed in a fast process, generating results in a short time. An online visualization of exosomes aids the analyzer to gain an instant idea of the exosome characteristics, e.g., gross concentration.
A very simple but elegant addition to the measurement technique presented here is the use of antibody labeled exosomes and the capture of the scattered laser beam by using a preceding filter in front of the detector. In this way exosome subpopulations may be distinguished according to their surface antigens and other biologically relevant characteristics that are technically accessible to a selective fluorescent staining.
One disadvantage of the current version of our particle tracking instrument is the fact that at very high sensitivity levels artifacts such as background noise may become present, which is based on the cell channel wall. A technical solution and improvement to the current system may become available in the near future. By this method reflections of the laser scatter light on the channel wall will be reduced, thereby increasing the accuracy of the measured signals and obtained data and lowering the limit of detection.
Although the currently used protocol for exosome isolation is well established and the applied particle tracking instrument has a remarkably precise resolution with a lower size distribution of 30 nm, there is no guarantee that the detected particles are indeed entirely true exosomes. Other particles, such as dead cell fragments or larger protein complexes may also be present in the exosome suspension and lead to false positive signals. One reliable method by which such “pollution” can be excluded may be electron microscopy (EM), either transmission EM or scanning EM. Unfortunately, no general and specific marker for exosomes has been identified so far, although a number of previously proposed surface markers have gained an increasing amount of attention, including tetraspanin (Tspan), CD81, C63, CD9, etc. 11.
Irrespective of the future progress of research on exosome biology, particularly concerning exosome specific markers, the protocol that is presented here will provide a powerful method for the isolation and detection of exosomes. Concentration and size may be readily determined in a software-assisted straightforward fashion. The addition of selective fluorescent markers or fluorophore coupled antibodies will likely further enhance the possible applications of the presented approach of NTA.
Disclosures
This work was supported by the institutional funds of the Dept. of Cardiovascular Surgery, Medical Faculty, HHU. The publication costs of this study were provided by Particle MetrixGmbH.
Acknowledgments
The authors would like to thank Christina Ballázs, Hug Aubin and Jörn Hülsmann for the critical reading of the manuscript and excellent editorial assistance. Moreover, the authors thank Gisela Mueller for the technical assistance. The authors thank Particle Metrix GmbH for providing the funds covering the publication costs.
References
- Thery C, Amigorena S, Raposo G, Clayton A, et al. Bonifacino JS, editor. Chapter 3. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology / editorial board. 2006. [DOI] [PubMed]
- Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods in molecular biology. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012. p. e3037. [DOI] [PMC free article] [PubMed]
- Konokhova AI, et al. Light-scattering flow cytometry for identification and characterization of blood microparticles. Journal of biomedical. 2012;17:057006. doi: 10.1117/1.JBO.17.5.057006. [DOI] [PubMed] [Google Scholar]
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et biophysica acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Sokolova V, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids and surfaces. B, Biointerfaces. 2011;87:146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Exoquick, ExoQuick-TC and ExoQuick-LP. Polymer-based exosome preciption. System Biosciences. 2015. Available from: http://www.systembio.com/microrna-research/exoquick-exosomes/overview.
- Clayton A, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. Journal of immunological. 2001;247:163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- Cheruvanky A, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Amer. J. Phys. Renal Phys. 2007;292:F1657–1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr B, Warren J. Company profile: NanoSight: delivering practical solutions for biological nanotechnology. Nanomedicine. 2012;7:1129–1132. doi: 10.2217/nnm.12.43. [DOI] [PubMed] [Google Scholar]
- Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. International immunology. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]


