Abstract
We demonstrate diffraction limited multiphoton imaging in a massively parallel, fully addressable time-resolved multi-beam multiphoton microscope capable of producing fluorescence lifetime images with sub-50ps temporal resolution. This imaging platform offers a significant improvement in acquisition speed over single-beam laser scanning FLIM by a factor of 64 without compromising in either the temporal or spatial resolutions of the system. We demonstrate FLIM acquisition at 500 ms with live cells expressing green fluorescent protein. The applicability of the technique to imaging protein-protein interactions in live cells is exemplified by observation of time-dependent FRET between the epidermal growth factor receptor (EGFR) and the adapter protein Grb2 following stimulation with the receptor ligand. Furthermore, ligand-dependent association of HER2-HER3 receptor tyrosine kinases was observed on a similar timescale and involved the internalisation and accumulation or receptor heterodimers within endosomes. These data demonstrate the broad applicability of this novel FLIM technique to the spatio-temporal dynamics of protein-protein interaction.
OCIS codes: (180.0180) Microscopy, (170.2520) Fluorescence microscopy, (180.4315) Nonlinear microscopy, (040.1240) Arrays, (090.1970) Diffractive optics
1. Introduction
Fluorescence lifetime imaging microscopy (FLIM) is a powerful technique for high resolution imaging of functional spatio-temporal dynamics in situ. Förster resonance energy transfer (FRET) is, by far, the most extensively studied technique for observation of protein-protein homo- and hetero-dimer interactions in intact cells [1, 2]. For intermolecular FRET, a key benefit of performing donor FLIM (when compared to intensity based ratiometric techniques), is that fluorescence-lifetime measurements of donor emission are independent of acceptor concentration and is therefore suited to imaging studies in intact cells [3–5]. Multiphoton microscopy confers additional advantages in terms of inherent optical sectioning and enhanced depth penetration for in vivo imaging [6–9]. However, the data acquisition rate for FLIM is a significant limitation in current implementations of laser scanning microscopy.
There are a number of techniques currently available in multifocal multiphoton microscopy to generate multiple scanning points. These include the use of Nipkow spinning disks [10], cascaded beamsplitter arrays [11], microlens arrays [12] and diffractive patterned illumination approaches [13]. These approaches restrict flexibility in relation to light efficiency (number of beams) or the use of patterned illumination. One technique utilizing a holographic pattern projected onto a diffractive spatial light modulator (SLM), (developed originally for use in optical tweezing applications [14]) is gaining a wider acceptance in multifocal multiphoton beam scanning implementations [15].
For high precision multiphoton FLIM, time-correlated single photon counting (TCSPC) is unparalleled in its measurement accuracy [16, 17]. In terms of imaging speed, TCSPC is fundamentally limited with respect to photon counting rate, since the stochastic nature of the emission process requires that the detection rate is much less than one photon per excitation event to prevent inaccuracies in lifetime determination [18]. Consequently, acquisition times for laser scanning FLIM are in the order of minutes, whereas many dynamic biological events occur on significantly faster timescales [19]. In order to overcome this limitation, parallel signal acquisition using arrays of laser beams with either photomultiplier arrays [20–22] or time-gated camera detection systems [23, 24] have been employed. However, in implementations to date, accurate determination of fluorescence lifetime with large numbers of channels in parallel is limited either due to cross-talk in multi-anode PMT’s [25] or subject to systematic error due to measurement methodology in camera systems [21, 26]. For a single beam scanning system, count rate limitations can be abrogated by parallel detection in a multi-anode PMT [21]. Whilst, significantly increasing frame rates, this is at the cost of significant pulse pile-up [18], leading to count rate dependent errors in the observed fluorescence lifetime. Furthermore, the count rates reported for in vivo imaging with a low axial and lateral resolution are unobtainable in high NA lenses without excitation saturation and significant phototoxicity and photobleaching. Thus, the ability to quantitatively measure complex biological events with high temporal resolution remains a significant challenge.
Single-photon avalanche photodiodes (SPAD) array cameras developed for microscopy and spectroscopy techniques have been designed utilizing either custom or standard CMOS technologies [27, 28]. Whilst recent studies have shown the use of custom CMOS in a parallelized 8 × 1 SPAD array for use in single molecule fluorescence correlation spectroscopy [29], there are issues in scalability due to electrical power requirements (6 W for 8 SPADs) [30]. Standard CMOS SPAD arrays are better suited for creating large, power efficient (100mW for 1024 SPADs) and affordable SPAD array cameras while maintaining good (50ps) timing resolution. In previous studies, the 32 × 32 Megaframe SPAD array has been presented where each individual pixel contained separate timing circuitry [31]. Due to the relative size of the timing circuitry and associated logic, the active region of each pixel is ~1% of the total device area. Hence, when operated as a 32 × 32 pixel widefield camera, due to the low fill factor there is a low collection efficiency [32].
In this paper, we demonstrate a novel multifocal multiphoton FLIM microscope (MM-FLIM) which dramatically improves the acquisition rate of high resolution fluorescence lifetime imaging by parallelizing the excitation and detection process. The system consists of a two dimensional array of ultrafast beams (generated holographically via an SLM) which is optically conjugate and precisely aligned to a SPAD array camera. By reimaging the collected fluorescence beamlets directly onto the active region of the SPADs, we optically amplify the fill factor to 100%. With each individual SPAD operating in TCSPC mode, the microscope system effectively consists of 64 individual multiphoton FLIM microscopes operating in parallel to enable high data acquisition rates. The Megaframe SPAD array acquires time-resolved data from single-photon events on 32 × 32 pixels at up to 500 kcts/sec/pixel [31] leading to a frame rate which scales linearly with detector number. Such high count rates generate too much data to transfer via a USB2.0 interface (effective throughput up to 35 MB/s or 280 Mbit/s) and in this paper we optimize data transfer by limiting acquisition to 64 detectors and achieve a maximum data acquisition rate of 16 Mcts/sec. This is 20 times faster than a typical TCSPC acquisition speed with a single detector (assuming photon counting rates of 1% for an 80MHz laser repetition rate). The FLIM system presented here, represents a paradigm shift in acquisition of fluorescence lifetime data, whereby there is no longer a need to make a compromise between accuracy, speed and spatial resolution. We perform a number of live cell imaging experiments to highlight the capabilities of the system.
2. Methods
2.1 Optical set-up
The MM-FLIM microscopy system is shown schematically in Fig. 1 . For multifocal beam generation, a phase-domain spatial light modulator based, arbitrary pattern holographic projection system was implemented, using techniques originally developed for optical tweezing [14, 24] and single molecule imaging with fluorescent correlation spectroscopy [29].
Fig. 1.
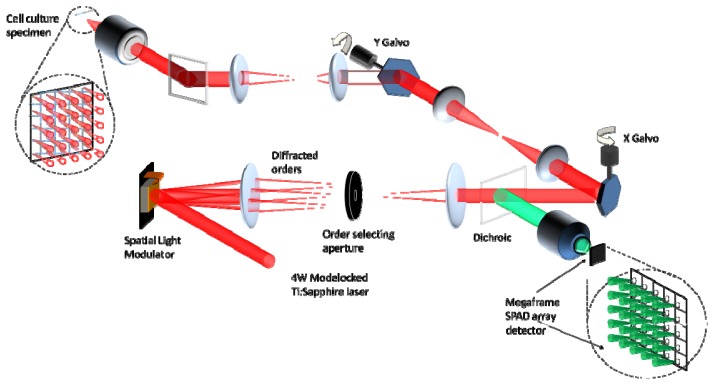
Operational schematic of the multifocal multiphoton system.
Horizontally polarized laser light from a Ti:Sapphire Chameleon Ultra II laser was expanded and collimated to overfill a near-IR (NIR) optimized SLM (Boulder Non-linear Systems Inc.). Using a doubly weighted Gerchberg-Saxton (DWGS) iterative phase retrieval algorithm [33], the appropriate holographic phase pattern was calculated and projected onto an SLM, which was positioned in the Fourier plane. Coupled with the incident laser light, the required laser beamlets were generated experimentally at the focal plane. A blazed grating phase pattern was convolved with the beamlet phase pattern to efficiently project the beamlet pattern into the first order diffraction pattern. The patterned NIR light was then optically relayed through a set of x-y galvanometer scanners and transmitted through a long-pass dichroic filter onto the back aperture of a x40 1.3N.A. Plan Fluor oil immersion microscope objective [Nikon Instruments Ltd] such that the back focal plane was conjugate to the SLM. Due to issues associated with projecting a square pattern onto a circular aperture, the beam size was set so that it slightly under-filled the pupil plane of the objective. Therefore the multifocal microscope system functioned with an effective excitation N.A. of 1.2. Since the focal plane was conjugate with the image plane of the SLM, the 2D beamlet array was re-created at the focal plane and was raster scanned across the sample in beam scanning mode (using the galvanometer scanners). Two photon excited fluorescence was then collected from the focal plane by the objective, de-scanned by the galvanometer scanners and re-imaged via a dichroic mirror onto the back aperture of a x10 0.3N.A. Plan Fluor objective [Nikon] and focused onto the Megaframe SPAD array.
The SLM based beamlet generation technique offers significant flexibility in terms of near-field resolution as well as having complete programmable control of the number and positioning of each beamlet. The excitation beamlet array generated fluorescence in the microscope focal plane via a two photon absorption process and was back-projected through the system to the detection plane. For parallelized TCSPC detection, each fluorescence image of the beamlet array was focused onto a single SPAD of a detector array (Fig. 2 ).
Fig. 2.
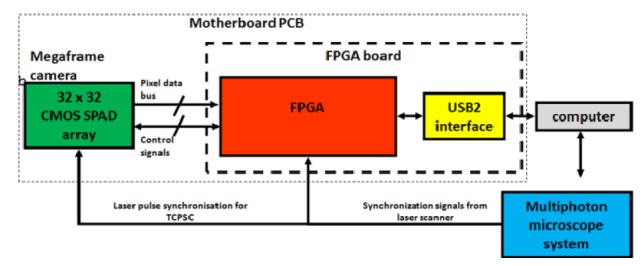
A flow chart detailing the Megaframe camera internal architecture, consisting of the motherboard PCB holding the Megaframe camera and programmable logic (FPGA). The FPGA controls the Megaframe camera readout and forwards the data to PC via USB2.0 link.
To ensure optimal efficiency of detection, the fluorescent beamlet projected onto the detector aperture was significantly smaller (1.8 µm FWHM) than the active area of the SPAD (6 µm dia.). The field of view of the microscope was determined by the magnification between object and detection planes, with beamlets separated in the detection plane by the detector spacing, 50 µm. With a 40x objective (Nikon Plan Fluor Oil Immersion 1.3 NA) the field of view was 100 μm for an 8 × 8 beamlet array. Precise alignment and matching of the beamlet spacing and angular orientation onto the detector array was performed to ensure an effective fill factor of 100%. The DWGS algorithm [33] was implemented (incorporating direct feedback from the detector array) to compensate for poor intensity fidelity between beamlets [34] leading to significant improvement in uniformity of the beamlet intensity variation.
2.2 Time-resolved detection and megaframe architecture
Based on the gated ring oscillator method [35, 36], the Megaframe SPAD array camera device is composed of a low power 32 × 32 10-bit time-to-digital converter (TDC) array (with an average temporal bin size of 52.5 +/− 0.7ps for a particular 8 × 8 array of SPADs) with integrated low dark-count SPAD, having a photon detection probability of 28% at 500 nm. Each pixel is 50 µm × 50 µm in size with a SPAD active region of 6 µm diameter [31].
In the current architecture, up to 0.5x106 time-stamp/s are delivered to the field programmable gate array (FPGA) for each pixel. Each timestamp is 10 bits long resulting in 56 ns dynamic range covering the needs for most fluorophores and light sources commonly used in microscopy. The FPGA circuit board (Opal Kelly XEM3050) contains the Spartan 3 FPGA from Xilinx (XC3S4000) and provides a USB2 interface to the host PC. The test software included with the FPGA circuit (pipetest.exe, FrontPanel 4.2.5, Opal Kelly) board can perform ~32MB/s streaming to the host PC. The firmware was written in Verilog using Xilinx’s ISE Design Tools, System Edition, version 13.2. FPGA board and Megaframe camera are interfaced via a custom motherboard printed circuit board (PCB). The motherboard PCB provides the inputs to laser pulse and laser scanner synchronization. Laser pulse synchronization signal is fed directly into the Megaframe camera for reverse start-stop TCSPC timing. Laser scanner synchronization signals are interfaced to the FPGA.
The Megaframe was operated in either time-correlated (lifetime) or time-uncorrelated modes (intensity). On-pixel TDCs generated raw time-correlated data, which was post-processed to generate an image. Due to the small size of the SPAD active area (6 µm diameter, ~28 µm2), the focused beamlet array required careful alignment for which a protocol was developed [37].
2.3 Image acquisition and reconstruction
All aspects of the microscope system were controlled via custom developed software written in the LabVIEW graphical programming environment. The appropriate region of interest for each sample was first selected whilst operating the microscope in wide-field fluorescent mode. Depending on the sample, the most suitable acquisition time and beamlet pattern laser power were chosen. The MM-FLIM system functioned in either single acquire, time-lapse or z-stack imaging modes. For each individual image acquisition, the system was set up to process 32 × 32 data points for 8 × 8 detectors producing 256 × 256 pixel images. The user sets the scan period and software generates the appropriate x-y galvanometer signals and pixel trigger clock. Once initialized, the buffered data reads were saved sequentially with the data containing an embedded x- address signal to enable sorting. Processing on the Megaframe camera was controlled through firmware loaded onto an on-chip FPGA.
Data lines from columns of the Megaframe camera were de-serialized and prepared for transfer over USB2. At each pixel clock cycle, one bit of a row was transferred from the top half (32 data lines) and bottom half (32 data lines). As each time-to-digital converter (TDC) was 10 bits, 10 pixel clock cycles were needed to transfer one top-half row and one bottom half row. The deserializer provided 640 lines (containing all data for two rows) to the module which checked if the two rows currently transferred contained pixels of interest. The data rate between the Megaframe camera and the FPGA is 5 Gbits/s (64 data lines at 80 MHz). This translates to a frame rate of 500 kHz, where each frame is 1024 pixels (32 × 32). As 5 Gbits/s data rate cannot be sustained to the PC and the FPGA did not include sufficient RAM for buffering full frame transfers, our approach was to select 64 pixels and transfer these at fastest possible rate over USB2.0 bus. The TDC timestamps for selected pixels were placed in the first-in-first-out (FIFO) buffer. Each timestamp is 16 bits long to allow for 10 bits of time data, 5 bits of addressing of 64 pixels and one spare bit. Our USB2 link allowed for up to 32 MBytes/s transfer rate. In practice, this was lower due to overheads in firmware and also lower photon budget. The interface between the microscope and detector system (Megaframe, FPGA circuit board and the interface board) was via TTL triggers whereby the scanner triggered the detector on each pixel thus ensuring that histograms were calculated for the correct pixels of interest.
Before analyzing the data, it must be pre-processed to compensate for non-linearities of the time-to-digital conversion (TDC) [38]. These manifest as:
Differential Non-Linearity (DNL) – cyclic intensity variations in time due to phases generated by the four delay-element oscillator (see Fig. 3 ). This was corrected individually for each pixel. The four delay-element oscillator in the pixel has 8 phases (essentially four positive transitions round the ring followed by four negative transitions). This results in a histogram with a repeating pattern of 8. By reducing the bin size to 8 for a measurement taken with a temporally uniform illumination source, one can determine an appropriate correction factor to scale each bin number.
Fig. 3.
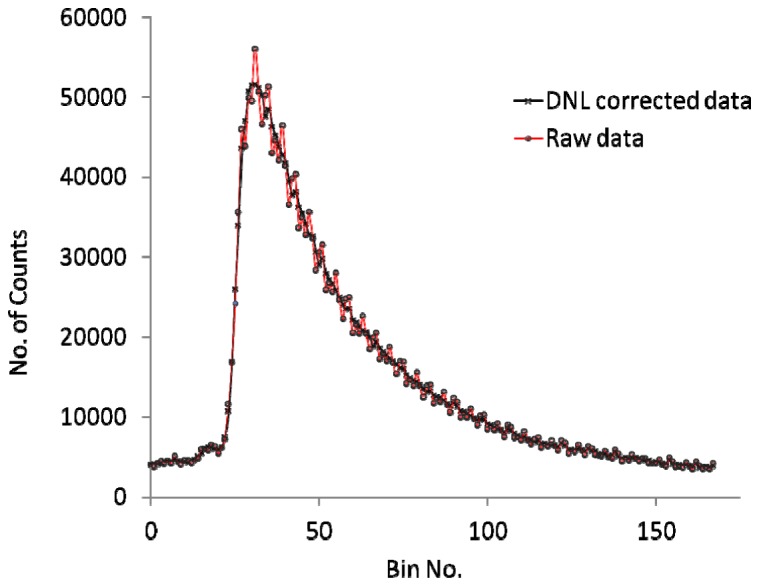
Comparing raw data with a DNL corrected histogram for a 7x7 detector array. Correction of the DNL gives a marked reduction of the intensity variation of the transient.
Integral Non-Linearity (INL) - Variations in the temporal bin size between detectors as a result of the CMOS fabrication process (see Fig. 4 ). This was compensated using a rational resampling technique. This technique incorporated first interpolation and decimation to change the sampling rate by a rational factor, thereby rescaling the temporal data taken from each detector into the same time bin size.
Fig. 4.
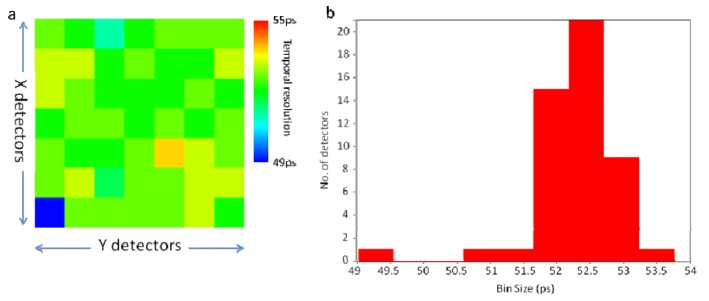
Presents data on the 7 × 7 detector array used for imaging (a) the temporal bin size of each detector in the 7 × 7 array. Average temporal resolution of the 7 × 7 array is 52.5 +/− 0.7ps. Variation in the temporal bin sizes are due to minor discrepancies between individual SPADs caused in the fabrication process of the chip. (b) Presents a histogram of the distribution of bin sizes of the 7 × 7 detector array.
Data was saved and subsequently analyzed using TRI2 lifetime analysis software [39].
2.4 Sample preparation of plasmids, cell lines, and cell culture conditions
The plasmids encoding human EGFR and human ErbB2 tagged at the c-terminus with enhanced green fluorescent protein (EGFP) were constructed by inserting the cDNA for the respective receptor into a modified version of the pEGFP-N3 vector (Clontech) in which the dimerisation-defficient Ala206 to Lys mutation had been incorporated into the EGFP sequence. The plasmid encoding human ErbB3 tagged at the c-terminus with mRFP1 was constructed by inserting the ErbB3 cDNA sequence upstream of mRFP1 within a modified pcDNA3.1 vector. The plasmid encoding Grb2-mCherry was a gift from Dr Michela Perani and Prof Peter Parker (King’s College London, UK).
MCF7 breast carcinoma cells were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS), penicillin G (100 U/ml)/streptomycin (100 mg/ml) and 1% L-glutamine, and cultured in an atmosphere containing 5% CO2/95% air (v/v). For imaging, cells were plated into 3.5 cm diameter glass-bottom culture dishes (PAA Laboratories) 24 hours prior to transfection. For EGFR-EGFP/Grb2-mCherry and HER2-EGFP/HER3-mCherry co-expression studies cells were co-transfected using Fugene 6 (Promega, Madison,WI, USA) with 1.5 µg and 3 µg of donor and acceptor plasmids respectively. Following transfection, cells were maintained at 37°C in a humidified 5% CO2 atmosphere for 24h to allow expression of exogenous proteins. For live cell imaging, cell culture media was replaced with phenol red-free OptiMEM (Gibco), supplemented with 10% FBS and 25 mM HEPES. The microscope body was equipped with an environment chamber (Solent Scientific Ltd, UK) enabling cell cultures to be stably maintained at 37°C for the duration of imaging experiments. Human recombinant EGF and Neuregulin were purchased from PeproTech, New Jersey, USA.
3. Experimental results
3.1 Resolution measurements
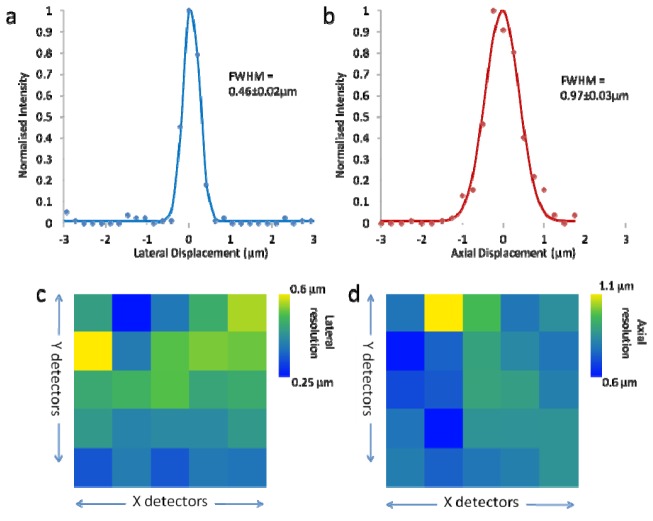
To demonstrate the diffraction limited imaging performance of the microscope across all beamlets, sub-resolution (100 nm dia.) fluorescent beads were imaged. Diffraction limited performance in lateral and axial resolutions of 0.44 ± 0.01 µm and 1.03 ± 0.01 µm respectively [N = 55 PSF’s distributed over 25 SPADs] was demonstrated with an effective 1.2 N.A. (due to under-illumination of the back pupil of the 1.3 N.A. objective). In Fig. 5(a) and Fig. 5(b) lateral and axial PSFs (used to calculate the resolution) are displayed respectively for a single bead. In Fig. 5(c) and Fig. 5(d) the average resolutions for a 25 SPADs are displayed on a colour chart.
Fig. 5.
Presents resolution imaging performance of the microscope across all beamlets which were calculated by imaging sub-resolution (100 nm dia.) fluorescent beads. (a) A typical lateral PSF of a single bead. (b) A typical axial PSF of a single bead. (c) The average lateral resolutions per detector for 5 × 5 detector array (d) The average axial resolutions per detector for a 5 × 5 array.
3.2 Comparing lifetime analysis techniques for image acquisition of live cells at high speed
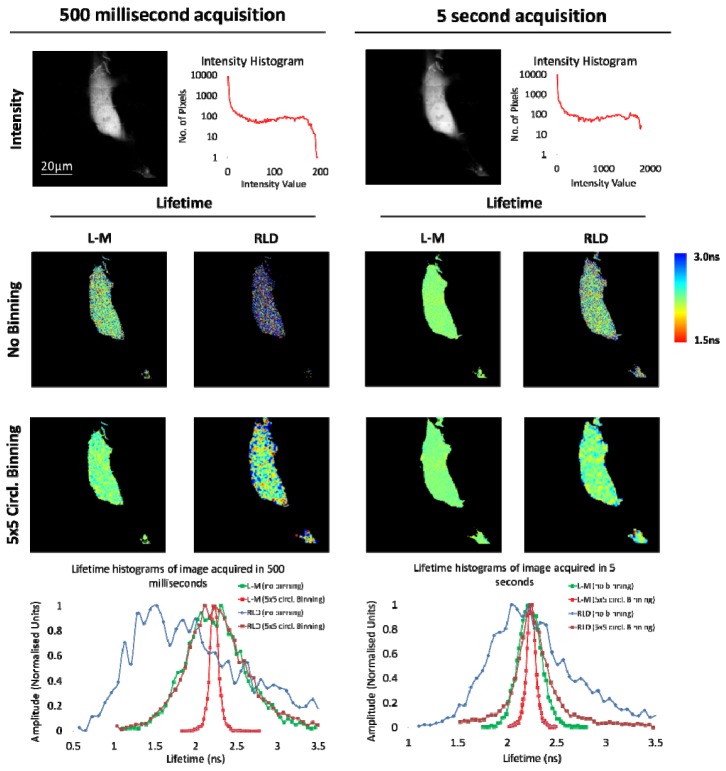
In order to compare the accuracies of lifetime determination techniques for image acquisition of live cells at high speed, data sets of MCF-7 human carcinoma cells transfected with EGFP were imaged for 500 milliseconds and 5 seconds per frame (Fig. 6 ).
Fig. 6.
Comparing lifetime analysis techniques for image acquisition of live cells at high speed. MCF-7 human breast carcinoma cells were transiently transfected with EGFP only and data sets were acquired for a 7 × 7 array for 500 milliseconds and 5 seconds. In a 256 × 256 data set, a maximum of 200 photons and 2000 photons were collected per pixel at 500 milliseconds and 5 second acquisitions respectively. In order to simulate the analysis of time gated camera to compare with Levenberg-Marquardt (L-M) fitting using Tri2 we used the 2 gated Rapid Lifetime Determination (RLD) approach for single exponential decays with 2 ns gate size. Lifetime images are displayed for both techniques with no binning and 5 × 5 circular binning for both 500 milliseconds and 5second acquisitions and their histograms compared. L-M clearly outperforms the RLD in all data analysis performed giving a much more accurate determination of the lifetime. The average calculated lifetimes for 500 ms acquisitions are: RLD no binning = 2.12 +/− 0.83 ns, RLD 5 × 5 circ. binning = 2.30 ± 0.46 ns, L-M no binning = 2.25 ± 0.39 ns and L-M 5 × 5 circ. binning = 2.23 ± 0.07 ns. The average calculated lifetimes for 5 s acquisitions are: RLD no binning = 2.31 ± 0.49 ns, RLD 5 × 5 circ. binning = 2.28 ± 0.29 ns, L-M no binning = 2.23 ± 0.12 ns and L-M 5 × 5 circ. binning = 2.24 ± 0.06 ns.
Levenberg-Marquardt (L-M) lifetime fitting was compared with Rapid Lifetime Determination (RLD) [40], commonly used for analysis in gated image intensifier (GOI) based FLIM systems. These data demonstrate that L-M fitting with TCSPC data, even at low photon numbers gives a much more accurate determination of the lifetime per pixel than RLD. It should be noted that this comparison is at Poisson limited signal/noise which is superior to GOI performance. Our data also show that frame rates in excess of 2 Hz are readily achievable with the system, without the limitations imposed by GOI based systems [41–44].
3.3 Imaging protein-protein interactions in live cells
To demonstrate the dynamic imaging capability of the system we chose to image two critical steps involved in the signalling of receptor tyrosine kinases, namely, (1) receptor dimerization and (2) receptor recruitment of adaptor protein. In both cases we chose to image signalling components of the Epidermal Growth Factor Receptor (EGFR) family of receptor tyrosine kinases and all experiments were conducted in live human breast carcinoma cells.
3.4 HER2-EGFP and HER3-mRFP1
MM-FLIM was applied to the analysis of ligand-dependent receptor heterodimerisation. We examined the interaction between the EGFR family members HER2 and HER3, given the clinical significance of this heterodimer pair [45]. HER3, which lacks intrinsic tyrosine kinase activity, can form competent signalling complexes through heterodimerisation with other EGFR family members following activation by its ligand neuregulin. As a potent driver of the PI3K/AKT-dependent cell survival pathway this novel kinase-deficient EGFR family member is gaining greater attention as a potential therapeutic target in cancer [45]. In particular, the role that the HER2/HER3 heterodimer plays in tumourigenesis and treatment resistance in breast cancer has become the focus of intense study. In HER2-amplified breast cancers, it is the HER2/HER3 heterodimer which is largely responsible for driving tumour growth. Poor response to HER2-targeted therapies (for instance trastuzumab) in these patients is believed, in part, to be associated with increased expression of HER3 [46].
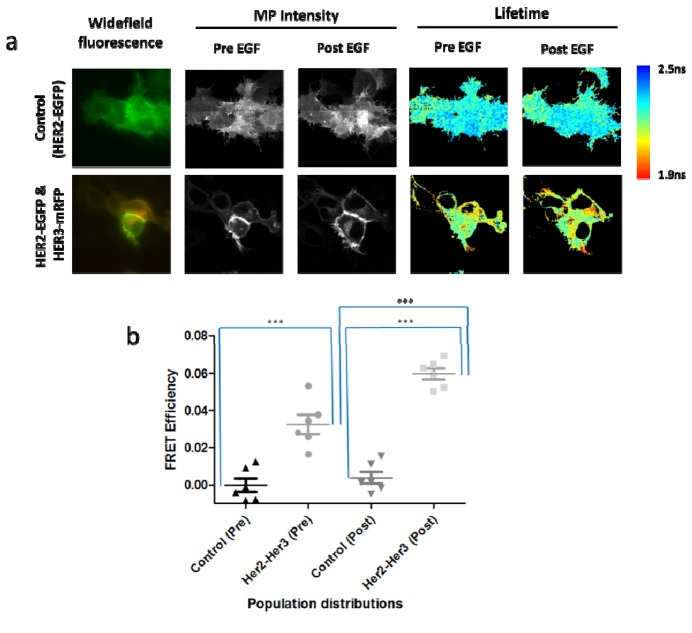
For live cell imaging experiments, MCF-7 cells were transiently transfected with expression constructs encoding EGFP-tagged HER2 (HER2-EGFP) and mRFP1-tagged HER3 (HER3-mRFP1) and compared with control cells transfected with EGFP-tagged HER2 only, before and after treatment with neuregulin ligand (50 ng/ml). Cells were imaged every 15s for 40 minutes with the addition of neuregulin occurring at the 0 seconds time point. A 7 × 7 beamlet pattern was applied which gave sufficient field of view (87.5 × 87.5 µm) to illuminate small clusters of cells over the course of time-lapse experiments. The lifetimes of 6 control and test samples were measured and the FRET efficiencies calculated (Fig. 7 ). In Fig. 6(a) the wide-field fluorescence, MP intensity and fluorescence lifetimes of representative control and test cells are presented. Statistical analysis comparing FRET efficiencies is shown in Fig. 6(b). The donor lifetime of control cells expressing HER2-EGFP alone (2.28 ± 0.02ns) remained unchanged throughout the course of time-lapse experiments. A basal constitutive association between HER2 and HER3 was observed within the perinuclear compartment with an average FRET efficiency of 3.3 ± 1.2%. On addition of neuregulin ligand, a significant increase in this FRET interaction occurred, with FRET efficiency rising to 6.1 ± 0.9%.
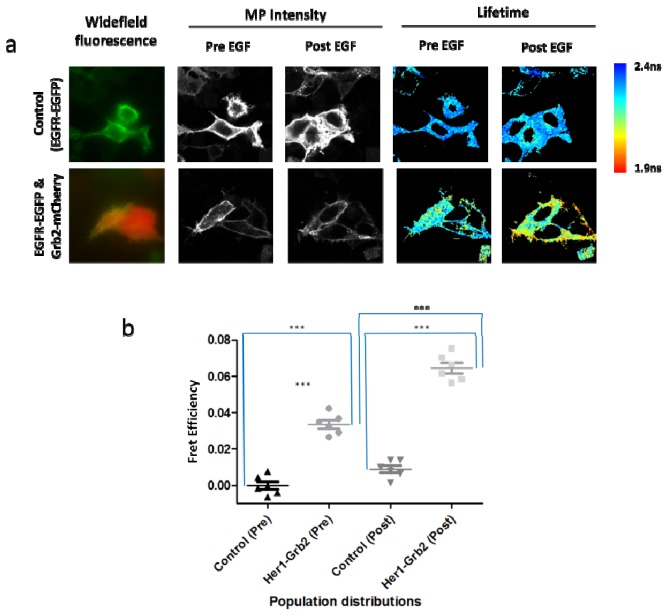
Fig. 7.
(a) Comparing widefield fluorescence, MP intensity and fluorescence lifetime data sets of test MCF-7 cells expressing EGFR-EGFP (control vs EGFR-EGFP & Grb2-mCherry, pre and post treated with EGF ligand. Acquisition time per frame is 15 seconds and all images have an 87.5µm x 87.5µm field of view. (b) Statistical analysis comparing FRET efficiencies of control MCF-7 cells expressing HER2-EGFP with MCF-7 cells coexpressing HER2-EGFP & HER3-mRFP1, both pre and post addition of neuregulin ligand. In order to compare two different populations (control vs. test) for statistical significance, two-tailed unpaired Student t-test was used. For the same populations (pre vs. post treatment) paired t-test was used. *** between populations denotes a highly significant difference in lifetime values (p<0.0001).
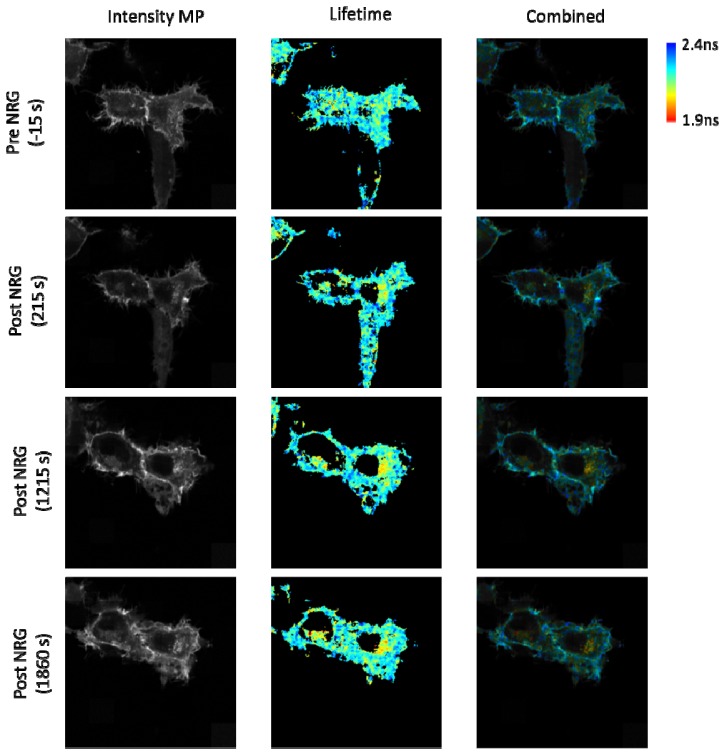
Time-lapse imaging data of one cell, representative of HER2-HER3 interaction, is presented in Fig. 8 . Following expression, HER2-EGFP and HER3-mRFP1 both exhibited a distinct pattern of spatio-temporal localization within MCF-7, which would be extremely challenging to reveal using convention lifetime imaging techniques. These proteins accumulated at the cell periphery and within a perinuclear compartment, the latter of which was far more pronounced in the case of HER3-mRFP1 (Fig. 8(a) and 8(b)). Following the addition of neuregulin, time-lapse imaging revealed a gradual fall in the lifetime of the HER2-EGFP donor specifically within the perinuclear compartment (from 2.21 +/− 0.02ns to 2.15 +/− 0.02 ns) due to accumulation of vesicles with short lifetimes whilst donor lifetime at the cell periphery remained relatively constant (Fig. 8(c)). These data constituted a FRET efficiency increase from 3.1 ± 1.3% to 5.7 ± 0.9% after neuregulin addition. The fall in donor lifetime, indicating an accumulation of HER2/HER3 heterodimer within a perinuclear compartment, was detectable within the first 100s of ligand addition and stabilised after approximately 7 minutes, remaining low for the remainder of the observation period. Interestingly, the lack of any detectable change in donor lifetime at the cell periphery suggests that newly formed HER2/HER3 heterodimers are rapidly internalised and targeted to subcellular compartments following neuregulin stimulation and therefore active receptor complexes are predominantly intracellular.
Fig. 8.
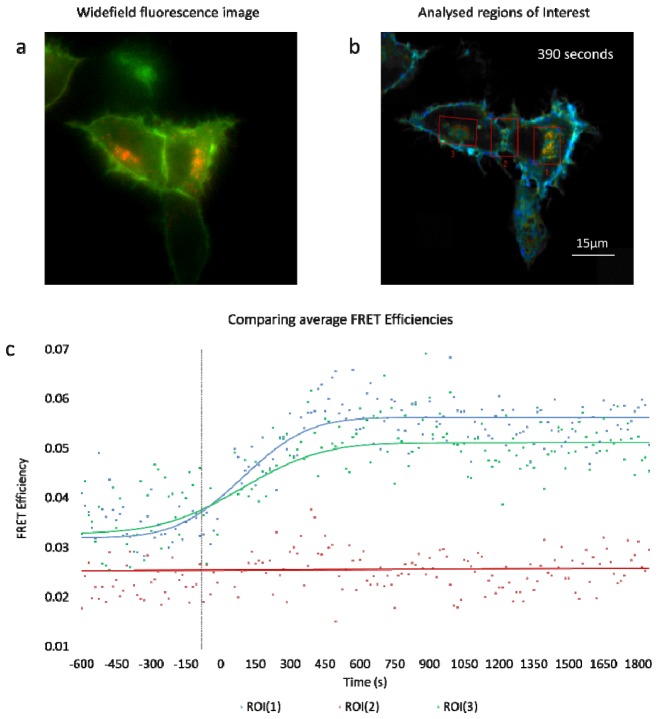
(a) Fluorescence widefield image highlighting the relative abundance of donor (green) and acceptor (red) regions in each cell. (b) A composite of Intensity and lifetime images are presented highlighting the ROIs chosen for the cells which were examined before and after neuregulin ligand was added. (c) Average FRET efficiencies values of the ROIs are presented in the accompanying graph with 0 seconds indicating the moment that neuregulin addition occurs. Cells were imaged every 15s for 40 minutes.
The diffraction limited spatial resolution afforded by the MM-FLIM system enabled lifetime changes within these discrete subcellular compartments to be monitored over the course of imaging experiments and regions of interest were selected such that lifetime changes within these compartments could be assessed (Fig. 9 , Media 1 (30.3MB, AVI) ).
Fig. 9.
Intensity, lifetime and composite images for a four frames in the time lapse ( Media1 (30.3MB, AVI) ).
3.5 EGFR-EGFP and Grb2-mCherry
Epidermal Growth Factor (EGF)-dependent activation of EGFR represents the archetypal mitogenic signalling pathway for receptor tyrosine kinases, coupling extracellular growth factors to Ras/MAPK activation [47]. In addition to its role in cell proliferation, EGFR activation can promote cell survival and differentiation and also drive the profound remodelling of the cell cytoskeleton, inducing migratory and invasive phenotypes in many cell types. The adaptor protein Grb2 is likely to represent a key node coupling EGFR activation to this diverse range of biological outputs as it provides a molecular bridge between active receptor and an array of downstream signalling protein classes including exchange factors, phosphatases, ubiquitin ligases and cytoskeletal remodelling factors [48–51]. Grb2 is recruited to activated receptor via its Src Homology 2 (SH2) domain which targets specific peptide consensuses flanking a critical central phosphotyrosine residue within the receptor’s intracellular domain. Grb2 serves to establish receptor-associated signalling scaffolds through which a downstream array of enzymatic pathways are rapidly activated: indeed Ras activation, which occurs following the Grb2-dependent recruitment of the exchange factor SOS to ligand-activated EGFR, is maximal within two minutes of addition of EGF to cultured cells [52]. Visualisation of EGF-induced EGFR/Grb2 association (which cannot be performed in a conventional signal beam scanning FLIM due to slow acquisition rates) represents an interesting challenge for testing the capabilities of the MM-FLIM system.
For live cell experiments, MCF-7 human breast carcinoma cells, transiently transfected with an EGFP fusion of EGFR (EGFR-EGFP) and a mCherry fusion of Grb2 (Grb2-mCherry) were subjected to fluorescence lifetime time-lapse imaging to assess the effects of EGF stimulation on EGFR-EGFP/Grb2- mCherry interaction. Cell imaging data was acquired for 30 minutes at 10 second acquisitions. The lifetimes of 6 control and test samples were measured and the FRET efficiencies calculated (Fig. 10 ). In Fig. 10(a) the widefield fluorescence, MP intensity and fluorescence lifetimes of representative control and test cells are presented. Statistical analysis comparing FRET efficiencies is shown in Fig. 10(b).
Fig. 10.
(a) Comparing widefield fluorescence, MP intensity and fluorescence lifetime data sets of control MCF-7 cells expressing EGFR-EGFP vs test MCF-7 cells expressing EGFR-EGFP & Grb2-mCherry, pre and post treated with EGF ligand. Acquisition time per frame is 10 seconds and all images have an 87.5µm x 87.5µm field of view.(b) Statistical analysis comparing control MCF-7 cells expressing EGFR-EGFP with MCF-7 cells coexpressing EGFR-EGFP & Grb2-mCherry, both pre and post addition of EGF ligand. In order to compare two different populations (control vs. test) for significance two-tailed unpaired Student t-test was used. For the same populations (pre vs. post treatment) paired t-test was used. *** between populations denotes a highly significant difference in lifetime values (p<0.0001).
No reduction in lifetime was observed for control cells expressing EGFR-EGFP donor alone, or for cells expressing both donor and acceptor molecules where the expression of acceptor was poor indicating reduced stoichiometry for the interaction (Fig. 10). The basal donor lifetime of EGFR-EGFP/Grb2-mCherry expressing cells (2.21 ± 0.01 ns) was lower than that of control cells (2.28 ± 0.01 ns), indicating the existence of a basal constitutive association between receptor and Grb2 and a FRET efficiency of 3.1 ± 0.4%. On addition of EGF ligand, a significant increase in this FRET interaction occurred, with FRET efficiency rising to 6.1 ± 0.9%.
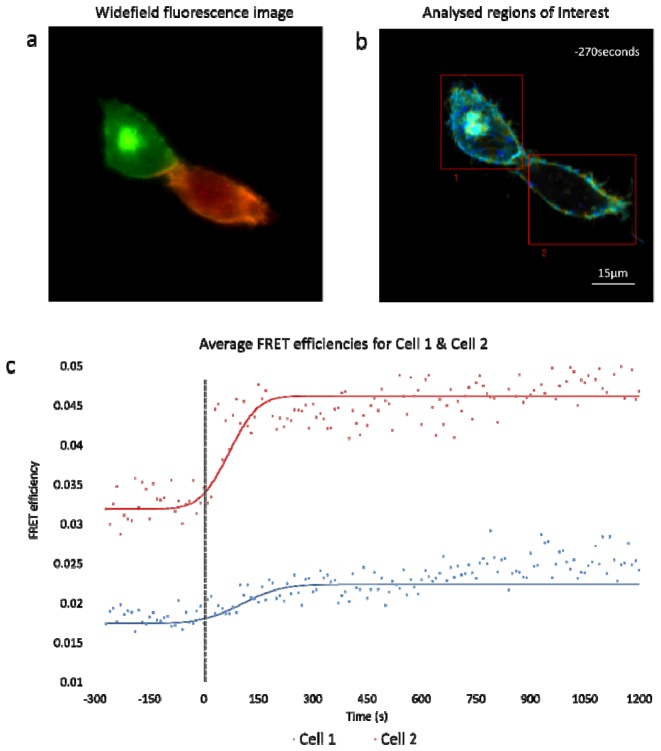
A continuous time lapse image data for one representative cell is presented in Fig. 11 . Cell imaging data was acquired continuously for 30 minutes (10 s/frame) ( Media 2 (16.6MB, AVI) ). Addition of EGF (100 ng/ml) occurred at the 0 seconds time point. Analysis of the time-lapse imaging revealed a rapid increase in FRET between EGFR-EGFP and Grb2-mCherry following stimulation as measured by a reduction in the lifetime of the EGFR-tagged EGFP donor fluorophore (Fig. 11(c)). Reduction in lifetime was detectable within 75 s of EGF stimulation and was sustained for the duration of the experiments, demonstrating rapid initiation of signalling downstream of the activated receptor.
Fig. 11.
(a) Fluorescence widefield image highlighting the relative abundance of donor (green) and acceptor (red) in each cell. (b) A composite of intensity and lifetime images are presented highlighting the ROIs chosen for the two cells which were examined before and after EGF ligand was added. (c) Average FRET efficiency values of the region of interest (ROI) are presented in the accompanying graph with 0 seconds indicating the moment that EGF addition occurs ( Media 2 (16.6MB, AVI) ).
The rapid association between EGFR and Grb2 observed in these live-cell imaging studies would be significantly more difficult to capture using conventional multiphoton excitation TCSPC techniques since association would be convolved with a long acquisition time or a loss of spatial resolution by integration of signal (c.f. increase in association shown by high speed multiphoton FLIM within 75s after adding ligand in Fig. 11). These experiments highlight the major advantage afforded by the increased temporal resolution of MM-FLIM.
In order to highlight the three dimensional sectioning capabilities of the system, a 40 image series of z-sections of the cells was taken before and after the time-lapse data (Fig. 12 , Media 3 (12.1MB, AVI) ). Each image stack was acquired in 400 s (10s/section). These data were therefore acquired on a time-scale comparable to that required to acquire a single z-section using conventional TCSPC.
Fig. 12.
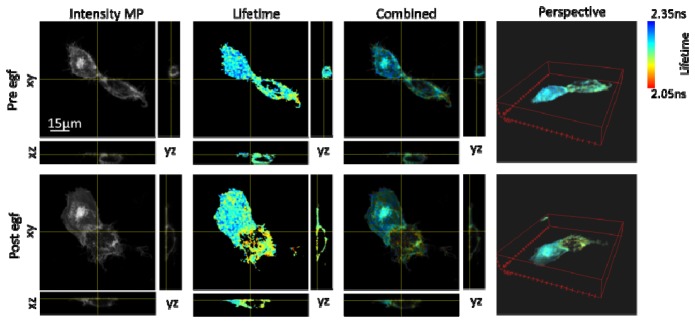
3D-image stack of a 40 z-sections acquired in 400 s (1 section/10s) of an MCF-7 cell, taken pre and post EGF addition ( Media 3 (12.1MB, AVI) ). Data taken before and after the time lapse data had been acquired.
Data from Fig. 11 were reanalysed by biexponential global analysis to determine the fraction of EGFR-EGFP interacting by FRET with Grb2-mCherry. The full data set was analysed simultaneously in a manner similar to that performed by Barber et al. [39]. The fluorescence lifetimes for the non-interacting and interacting fractions were determined as 2.28 ± 0.01 ns and 1.40 ± 0.01 ns by global fitting of the entire data set. The fractional contribution of each lifetime component was then determined for each time-point (Fig. 13 , Media 4 (17.2MB, AVI) ). These data show that the drop in average fluorescence lifetime for EGFR-EGFP in response to ligand stimulation in cell 2 is due to recruitment of Grb2 to ~35% of EGFR molecules. This interpretation of these FRET data is appropriate if we assume only two populations (interacting and non-interacting) are present in the data. More complex interpretations are beyond the scope of this paper and these data serve as an example for the significant advantage of acquiring TCSPC data for FLIM post analysis rather than assuming a monoexponential model.
Fig. 13.
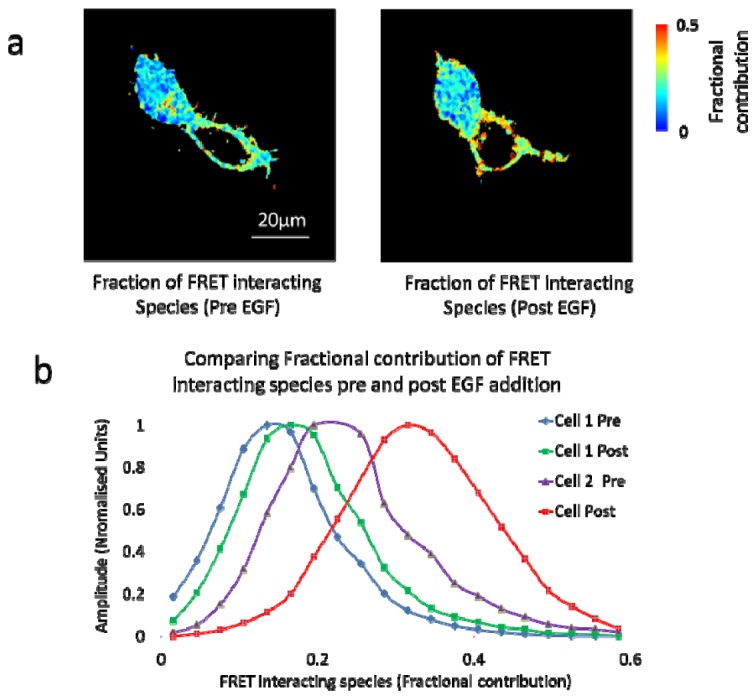
Data from Fig. 11 reanalysed by biexponential global analysis to determine the fraction of EGFR-EGFP interacting by FRET with Grb2-mCherry. (a) Presents the total fraction of FRET interacting species, pre and post EGF addition. (b) Histograms comparing fractional contributions of FRET interaction species for pre and post EGF ligand addition ( Media 4 (17.2MB, AVI) ).
4. Discussion and conclusions
This paper presents development of a multifocal multiphoton imaging system for high-frame rate FLIM imaging. We have demonstrated that multifocal multiphoton imaging with a 2D SPAD array is practical and offers significant speed advantages compared to single beam laser scanning or widefield FLIM microscopes. With this implementation of MM-FLIM, the minimum acquisition time for a homogeneous image scales as where N2 is the number of pixels in the image, Δt is the pixel dwell time to acquire the required 1000 photons for a satisfactory single exponential fit [53] and n is the number of beams. At the maximum count rate per pixel for the current system (500 kHz) and 64 beams, the minimum frame time is 2.05 s for a homogeneous sample, which is a drastic improvement when compared with 131 s required for a single beam scanning system. Often in FLIM imaging spatial resolution is sacrificed for temporal resolution and applying a 3 x 3 spatial binning reduces this acquisition further to 228 ms in the MM-FLIM. One can also fit with a lower number of photons to obtain faster acquisition rates (500 ms acquisitions) although this will affect the accuracy of the lifetime determination (Fig. 6). We have demonstrated long-term observation of living biological specimens with diffraction limited imaging performance with no significant degradation in signal to noise. This is due to the lower photon flux of excitation in each beamlet compared to a single beam scanning system [21].
Whilst wide-field fluorescence lifetime imaging is possible at up to video frame rates with gated image intensifiers [54,55, 56], this is not practical in a biological setting due to sample limitations (i.e. excited state fluorophore saturation), significant imaging artefacts and excitation photon flux that may be damaging to cells [19, 26, 27,29]. Moreover, the MM-FLIM system described here, provides a platform for future improvements in speed and signal-to-noise by increasing the number of beams or improving the sensor spatial resolution with lower pitch, higher fill factor SPADs. To demonstrate the capabilities of the system we have performed quantitative imaging of protein-protein interactions of the EGFR family signalling network within living cells at unprecedented spatial and temporal resolutions for time-domain FLIM. We observe the gradual accumulation of these HER2/HER3 receptor complexes within a discrete perinuclear compartment highlighting the spatial resolution of the MM-FLIM system. We show that the Grb2 adaptor protein is recruited to EGFR receptor within 75 s of cell stimulation with EGF, an observation that would be challenging with conventional TCSPC systems. In addition, we are able to visualise the rapid heterodimerisation of HER2 and HER3 receptor tyrosine kinases in response to cell stimulation with the ligand neuregulin. We believe this technology has the potential to transform functional FLIM/FRET imaging and other FLIM applications, to enable dynamic spatio-temporal protein interactions to be studied quantitatively for the first time in situ.
Acknowledgments
The authors would like to thank: The Biotechnology and Biological Sciences Research Council UK ( BB/I022074/1 and BB/I022937/1); STMicroelectronics (Imaging Division, Edinburgh, UK); Cancer Research UK, Engineering and Physical Sciences Research Council (UK), Medical Research Council (UK) and Department of Health (UK), KCL/UCL Comprehensive Cancer Imaging Centre; Dimbleby Cancer Care endowment fund to King’s College London and The Royal Society. The Megaframe project has been supported by the European Community within the Sixth Framework Programme IST FET Open, http://www.megaframe.eu.
References and links
- 1.Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., Solomon E., “The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress,” Nature 462(7275), 886–890 (2009). 10.1038/nature08593 [DOI] [PubMed] [Google Scholar]
- 2.Peter M., Ameer-Beg S. M., Hughes M. K., Keppler M. D., Prag S., Marsh M., Vojnovic B., Ng T., “Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions,” Biophys. J. 88(2), 1224–1237 (2005). 10.1529/biophysj.104.050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastiaens P. I., Pepperkok R., “Observing proteins in their natural habitat: the living cell,” Trends Biochem. Sci. 25(12), 631–637 (2000). 10.1016/S0968-0004(00)01714-X [DOI] [PubMed] [Google Scholar]
- 4.Wouters F. S., Verveer P. J., Bastiaens P. I., “Imaging biochemistry inside cells,” Trends Cell Biol. 11(5), 203–211 (2001). 10.1016/S0962-8924(01)01982-1 [DOI] [PubMed] [Google Scholar]
- 5.Pelet S., So P. T., Previte M. J., “Comparing the quantification of Förster resonance energy transfer measurement accuracies based on intensity, spectral, and lifetime imaging,” J. Biomed. Opt. 11, 034017 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Zipfel W. R., Williams R. M., Webb W. W., “Nonlinear magic: multiphoton microscopy in the biosciences,” Nat. Biotechnol. 21(11), 1369–1377 (2003). 10.1038/nbt899 [DOI] [PubMed] [Google Scholar]
- 7.Brown E. B., Campbell R. B., Tsuzuki Y., Xu L., Carmeliet P., Fukumura D., Jain R. K., “In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy,” Nat. Med. 7(9), 1069 (2001). 10.1038/nm0901-1069 [DOI] [PubMed] [Google Scholar]
- 8.Denk W., Strickler J. H., Webb W. W., “Two-Photon Laser Scanning Fluorescence Microscopy,” Science 248(4951), 73–76 (1990). 10.1126/science.2321027 [DOI] [PubMed] [Google Scholar]
- 9.Helmchen F., Denk W., “Deep tissue two-photon microscopy,” Nat. Methods 2(12), 932–940 (2005). 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- 10.Bewersdorf J., Pick R., Hell S. W., “Multifocal multiphoton microscopy,” Opt. Lett. 23(9), 655–657 (1998). 10.1364/OL.23.000655 [DOI] [PubMed] [Google Scholar]
- 11.Nielsen T., Fricke M., Hellweg D., Andresen P., “High efficiency beam splitter for multifocal multiphoton microscopy,” J. Microsc. 201(3), 368–376 (2001). 10.1046/j.1365-2818.2001.00852.x [DOI] [PubMed] [Google Scholar]
- 12.Buist A., Müller M., Squier J., Brakenhoff G., “Real time two-photon absorption microscopy using multi point excitation,” J. Microsc. 192(2), 217–226 (1998). 10.1046/j.1365-2818.1998.00431.x [DOI] [Google Scholar]
- 13.Sacconi L., Froner E., Antolini R., Taghizadeh M. R., Choudhury A., Pavone F. S., “Multiphoton multifocal microscopy exploiting a diffractive optical element,” Opt. Lett. 28(20), 1918–1920 (2003). 10.1364/OL.28.001918 [DOI] [PubMed] [Google Scholar]
- 14.Leach J., Wulff K., Sinclair G., Jordan P., Courtial J., Thomson L., Gibson G., Karunwi K., Cooper J., Laczik Z. J., Padgett M., “Interactive approach to optical tweezers control,” Appl. Opt. 45(5), 897–903 (2006). 10.1364/AO.45.000897 [DOI] [PubMed] [Google Scholar]
- 15.Nikolenko V., Watson B. O., Araya R., Woodruff A., Peterka D. S., Yuste R., “SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators,” Frontiers in neural circuits 2 (2008). [DOI] [PMC free article] [PubMed]
- 16.Gratton E., Breusegem S., Sutin J., Ruan Q., Barry N., “Fluorescence lifetime imaging for the two-photon microscope: time-domain and frequency-domain methods,” J. Biomed. Opt. 8(3), 381–390 (2003). 10.1117/1.1586704 [DOI] [PubMed] [Google Scholar]
- 17.Esposito A., Gerritsen H. C., Wouters F. S., “Optimizing frequency-domain fluorescence lifetime sensing for high-throughput applications: photon economy and acquisition speed,” J. Opt. Soc. Am. A 24(10), 3261–3273 (2007). 10.1364/JOSAA.24.003261 [DOI] [PubMed] [Google Scholar]
- 18.Suhling K., McLoskey D., Birch D., “Multiplexed single‐photon counting. II. The statistical theory of time‐correlated measurements,” Rev. Sci. Instrum. 67(6), 2238–2246 (1996). 10.1063/1.1147042 [DOI] [Google Scholar]
- 19.Miyawaki A., Griesbeck O., Heim R., Tsien R. Y., “Dynamic and quantitative Ca2+ measurements using improved cameleons,” Proc. Natl. Acad. Sci. U.S.A. 96(5), 2135–2140 (1999). 10.1073/pnas.96.5.2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S., Dunsby C., De Beule P. A. A., Owen D. M., Anand U., Lanigan P. M. P., Benninger R. K. P., Davis D. M., Neil M. A. A., Anand P., Benham C., Naylor A., French P. M. W., “Multifocal multiphoton excitation and time correlated single photon counting detection for 3-D fluorescence lifetime imaging,” Opt. Express 15(20), 12548–12561 (2007). 10.1364/OE.15.012548 [DOI] [PubMed] [Google Scholar]
- 21.Rinnenthal J. L., Börnchen C., Radbruch H., Andresen V., Mossakowski A., Siffrin V., Seelemann T., Spiecker H., Moll I., Herz J., Hauser A. E., Zipp F., Behne M. J., Niesner R., “Parallelized TCSPC for dynamic intravital fluorescence lifetime imaging: Quantifying neuronal dysfunction in neuroinflammation,” PLoS ONE 8(4), e60100 (2013). 10.1371/journal.pone.0060100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLoskey D., Birch D. J., Sanderson A., Suhling K., Welch E., Hicks P., “Multiplexed single‐photon counting. I. A time‐correlated fluorescence lifetime camera,” Rev. Sci. Instrum. 67(6), 2228–2237 (1996). 10.1063/1.1147041 [DOI] [Google Scholar]
- 23.Grant D. M., McGinty J., McGhee E. J., Bunney T. D., Owen D. M., Talbot C. B., Zhang W., Kumar S., Munro I., Lanigan P. M., Kennedy G. T., Dunsby C., Magee A. I., Courtney P., Katan M., Neil M. A., French P. M., “High speed optically sectioned fluorescence lifetime imaging permits study of live cell signaling events,” Opt. Express 15(24), 15656–15673 (2007). 10.1364/OE.15.015656 [DOI] [PubMed] [Google Scholar]
- 24.Padilla-Parra S., Audugé N., Coppey-Moisan M., Tramier M., “Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells,” Biophys. J. 95(6), 2976–2988 (2008). 10.1529/biophysj.108.131276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enari Y., Hayasaka K., Hokuue T., Inami K., Ohshima T., Sato N., Akatsu M., Kawakami S., Miyabayashi Y., Tokuda H., Yanase H., Shimoi H., Fujimori T., “Cross-talk of a multi-anode PMT and attainment of a TOF counter,” Nucl. Instrum. Methods Phys. Res. A 547(2-3), 490–503 (2005). 10.1016/j.nima.2005.03.159 [DOI] [Google Scholar]
- 26.Hanley Q. S., Subramaniam V., Arndt-Jovin D. J., Jovin T. M., “Fluorescence lifetime imaging: multi-point calibration, minimum resolvable differences, and artifact suppression,” Cytometry 43(4), 248–260 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Michalet X., Colyer R. A., Scalia G., Ingargiola A., Lin R., Millaud J. E., Weiss S., Siegmund O. H., Tremsin A. S., Vallerga J. V., Cheng A., Levi M., Aharoni D., Arisaka K., Villa F., Guerrieri F., Panzeri F., Rech I., Gulinatti A., Zappa F., Ghioni M., Cova S., “Development of new photon-counting detectors for single-molecule fluorescence microscopy,” Philos. Trans. R. Soc. Lond. B Biol. Sci. 368(1611), 20120035 (2013). 10.1098/rstb.2012.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charbon E., “Single-photon imaging in complementary metal oxide semiconductor processes,” Philos. Trans. A. Math Phys. Eng. Sci. 372(2012), 20130100 (2014). 10.1098/rsta.2013.0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colyer R. A., Scalia G., Rech I., Gulinatti A., Ghioni M., Cova S., Weiss S., Michalet X., “High-throughput FCS using an LCOS spatial light modulator and an 8 × 1 SPAD array,” Biomed. Opt. Express 1(5), 1408–1431 (2010). 10.1364/BOE.1.001408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonioli S., Miari L., Cuccato A., Crotti M., Rech I., Ghioni M., “8-channel acquisition system for time-correlated single-photon counting,” Rev. Sci. Instrum. 84(6), 064705 (2013). 10.1063/1.4811377 [DOI] [PubMed] [Google Scholar]
- 31.Richardson J., Walker R., Grant L., Stoppa D., Borghetti F., Charbon E., Gersbach M., Henderson R. K., “A 32× 32 50ps resolution 10 bit time to digital converter array in 130nm CMOS for time correlated imaging,” in Custom Integrated Circuits Conference, 2009. CICC'09. IEEE(IEEE, 2009), pp. 77–80. [Google Scholar]
- 32.Li D. D.-U., Arlt J., Tyndall D., Walker R., Richardson J., Stoppa D., Charbon E., Henderson R. K., “Video-rate fluorescence lifetime imaging camera with CMOS single-photon avalanche diode arrays and high-speed imaging algorithm,” J. Biomed. Opt. 16, 096012 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Poland S. P., Krstajić N., Knight R. D., Henderson R. K., Ameer-Beg S. M., “Development of a doubly weighted Gerchberg-Saxton algorithm for use in multibeam imaging applications,” Opt. Lett. 39(8), 2431–2434 (2014). 10.1364/OL.39.002431 [DOI] [PubMed] [Google Scholar]
- 34.Spalding G. C., Courtial J., Leonardo R. D., Holographic Optical Tweezers, Structured Light and Its Applications, Andrews D. L., ed.(Academic Press, 2008) pp. 139–168 (2008). [Google Scholar]
- 35.Helal B. M., Straayer M., Wei G.-Y., Perrott M. H., “A low jitter 1.6 GHz multiplying DLL utilizing a scrambling time-to-digital converter and digital correlation,” in VLSI Circuits, 2007 IEEE Symposium on(IEEE, 2007), pp. 166–167 10.1109/VLSIC.2007.4342700 [DOI] [Google Scholar]
- 36.De Heyn V., Van der Plas G., Ryckaert J., Craninckx J., “A fast start-up 3GHz–10GHz digitally controlled oscillator for UWB impulse radio in 90nm CMOS,” in Solid State Circuits Conference, 2007. ESSCIRC 2007. 33rd European(IEEE, 2007), pp. 484–487. [Google Scholar]
- 37.Poland S. P., Coelho S., Krstajić N., Tyndall D., Walker R., Monypenny J., Li D. D., Henderson R., Ameer-Beg S., “Development of a fast TCSPC FLIM-FRET imaging system,” in SPIE BiOS(International Society for Optics and Photonics, 2013), pp. 85880X–85880X–85888. [Google Scholar]
- 38.Kalisz J., “Review of methods for time interval measurements with picosecond resolution,” Metrologia 41(1), 17–32 (2004). 10.1088/0026-1394/41/1/004 [DOI] [Google Scholar]
- 39.Barber P., Ameer-Beg S., Gilbey J., Carlin L., Keppler M., Ng T., Vojnovic B., “Multiphoton time-domain fluorescence lifetime imaging microscopy: practical application to protein–protein interactions using global analysis,” J. R. Soc. Interface 6(Suppl_1), S93–S105 (2009). 10.1098/rsif.2008.0451.focus [DOI] [Google Scholar]
- 40.Sharman K. K., Periasamy A., Ashworth H., Demas J. N., “Error analysis of the rapid lifetime determination method for double-exponential decays and new windowing schemes,” Anal. Chem. 71(5), 947–952 (1999). 10.1021/ac981050d [DOI] [PubMed] [Google Scholar]
- 41.Van Munster E., Gadella T. W., “φFLIM: a new method to avoid aliasing in frequency‐domain fluorescence lifetime imaging microscopy,” J Microsc. 213(1), 29–38 (2004). 10.1111/j.1365-2818.2004.01249.x [DOI] [PubMed] [Google Scholar]
- 42.van Munster E. B., Gadella T. W., Jr., “Suppression of photobleaching-induced artifacts in frequency-domain FLIM by permutation of the recording order,” Cytometry A 58(2), 185–194 (2004). 10.1002/cyto.a.20013 [DOI] [PubMed] [Google Scholar]
- 43.Hanley Q. S., Lidke K. A., Heintzmann R., Arndt-Jovin D. J., Jovin T. M., “Fluorescence lifetime imaging in an optically sectioning programmable array microscope (PAM),” Cytometry A 67(2), 112–118 (2005). 10.1002/cyto.a.20177 [DOI] [PubMed] [Google Scholar]
- 44.vandeVen M., Ameloot M., Valeur B., Boens N., “Pitfalls and their remedies in time-resolved fluorescence spectroscopy and microscopy,” J. Fluoresc. 15(3), 377–413 (2005). 10.1007/s10895-005-2632-1 [DOI] [PubMed] [Google Scholar]
- 45.Gala K., Chandarlapaty S., “Molecular Pathways: HER3 Targeted Therapy,” Clin. Cancer Res. 20(6), 1410–1416 (2014). 10.1158/1078-0432.CCR-13-1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park Y., Jung H., Choi M., Chang W., Choi Y., Do I., Ahn J., Im Y., “Role of HER3 expression and PTEN loss in patients with HER2-overexpressing metastatic breast cancer (MBC) who received taxane plus trastuzumab treatment,” Br. J. Cancer 110(2), 384–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egan S. E., Giddings B. W., Brooks M. W., Buday L., Sizeland A. M., Weinberg R. A., “Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation,” Nature 363(6424), 45–51 (1993). 10.1038/363045a0 [DOI] [PubMed] [Google Scholar]
- 48.Olivier J. P., Raabe T., Henkemeyer M., Dickson B., Mbamalu G., Margolis B., Schlessinger J., Hafen E., Pawson T., “A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos,” Cell 73(1), 179–191 (1993). 10.1016/0092-8674(93)90170-U [DOI] [PubMed] [Google Scholar]
- 49.Wong L., Johnson G. R., “Epidermal growth factor induces coupling of protein-tyrosine phosphatase 1D to GRB2 via the COOH-terminal SH3 domain of GRB2,” J. Biol. Chem. 271(35), 20981–20984 (1996). 10.1074/jbc.271.35.20981 [DOI] [PubMed] [Google Scholar]
- 50.Li N., Lorinczi M., Ireton K., Elferink L. A., “Specific Grb2-mediated interactions regulate clathrin-dependent endocytosis of the cMet-tyrosine kinase,” J. Biol. Chem. 282(23), 16764–16775 (2007). 10.1074/jbc.M610835200 [DOI] [PubMed] [Google Scholar]
- 51.Carlier M.-F., Nioche P., Broutin-L’Hermite I., Boujemaa R., Le Clainche C., Egile C., Garbay C., Ducruix A., Sansonetti P., Pantaloni D., “GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex,” J. Biol. Chem. 275(29), 21946–21952 (2000). 10.1074/jbc.M000687200 [DOI] [PubMed] [Google Scholar]
- 52.Yamasaki S., Nishida K., Yoshida Y., Itoh M., Hibi M., Hirano T., “Gab1 is required for EGF receptor signaling and the transformation by activated ErbB2,” Oncogene 22(10), 1546–1556 (2003). 10.1038/sj.onc.1206284 [DOI] [PubMed] [Google Scholar]
- 53.Köllner M., Wolfrum J., “How many photons are necessary for fluorescence-lifetime measurements?” Chem. Phys. Lett. 200(1-2), 199–204 (1992). 10.1016/0009-2614(92)87068-Z [DOI] [Google Scholar]
- 54.Schneider P. C., Clegg R. M., “Rapid acquisition, analysis, and display of fluorescence lifetime-resolved images for real-time applications,” Rev. Sci. Instrum. 68(11), 4107–4119 (1997). 10.1063/1.1148354 [DOI] [Google Scholar]
- 55.Requejo-Isidro J., McGinty J., Munro I., Elson D. S., Galletly N. P., Lever M. J., Neil M. A., Stamp G. W., French P. M., Kellett P. A., Hares J. D., Dymoke-Bradshaw A. K., “High-speed wide-field time-gated endoscopic fluorescence-lifetime imaging,” Opt. Lett. 29(19), 2249–2251 (2004). 10.1364/OL.29.002249 [DOI] [PubMed] [Google Scholar]
- 56.Agronskaia A., Tertoolen L., Gerritsen H., “High frame rate fluorescence lifetime imaging,” J. Phys. D Appl. Phys. 36(14), 1655–1662 (2003). 10.1088/0022-3727/36/14/301 [DOI] [Google Scholar]