Abstract
Here we describe a method to conditionally induce epithelial cell transformation by the use of the 4-Hydroxytamoxifen (4-OHT) inducible KalTA4-ERT2/UAS expression system1 in zebrafish larvae, and the subsequent live imaging of innate immune cell interaction with HRASG12V expressing skin cells. The KalTA4-ERT2/UAS system is both inducible and reversible which allows us to induce cell transformation with precise temporal/spatial resolution in vivo. This provides us with a unique opportunity to live image how individual preneoplastic cells interact with host tissues as soon as they emerge, then follow their progression as well as regression. Recent studies in zebrafish larvae have shown a trophic function of innate immunity in the earliest stages of tumorigenesis2,3. Our inducible system would allow us to live image the onset of cellular transformation and the subsequent host response, which may lead to important insights on the underlying mechanisms for the regulation of oncogenic trophic inflammatory responses. We also discuss how one might adapt our protocol to achieve temporal and spatial control of ectopic gene expression in any tissue of interest.
Keywords: Developmental Biology, Issue 96, zebrafish, live imaging, skin, conditional gene expression, KalTA4-ERT2/UAS, 4-Hydroxytamoxifen, HRASG12V, neutrophils
Introduction
The overgrowth of a preneoplastic cell progeny within an otherwise normal epithelial sheet strongly depends on an interaction with its microenvironment. The interaction with its host tissue is likely to be the major determinant of whether a preneoplastic cell is able to establish a clonal niche to further develop into a full blown cancer. Despite its importance in development, this initial phase of cancer progression is inaccessible in most commonly used model systems, in vivo.
The zebrafish, Danio rerio, is a well-established model organism for live imaging studies because of its transparency throughout development, the possibility for genetic manipulation, and accessibility for water-soluble drugs. Recent studies using a larval zebrafish model, overexpressing the human oncogene HRASG12V to transform epithelial cells, showed that host cell derived signals, in particular H2O2 lead to the recruitment of innate immune cells. Interestingly, the recruited immune cells, neutrophils and macrophages, were shown to have a trophic role in supporting preneoplastic cell proliferation2,3.
In order to understand the early events of tumor initiation and progression as well as the contribution of an inflammatory response, one needs to be able to image these events from the earliest time point onwards. Therefore it is necessary to control cell transformations in a temporal and tissue specific manner. We utilize the Gal4/UAS system that has been successfully adapted from Drosophila4 to conditionally express the human oncogene HRASG12V under the control of the skin specific promoter keratin 4 (krt4)5. The modified version of the transcriptional activator Gal4, namely KalTA46, enables expression of HRASG12V under the control of the Upstream Activating Sequence (UAS). To conditionally induce the transcriptional activator expression, KalTA4 has been fused to the mutant ligand-binding domain of the human estrogen receptor α (ERT2)7 which binds specifically 4-Hydroxytamoxifen (4-OHT)1. In the absence of 4-OHT the ERT2 is bound in the cytoplasm by heat-shock proteins. Upon 4-OHT binding the heat-shock proteins dissociate, allowing a nuclear translocation of KalTA4-ERT2 and subsequent activation of the UAS controlled gene of interest8 (Figure 1C, b). As this expression system depends on the presence of 4-OHT, the KalTA4 controlled expression of a gene of interest is reversible. In contrast to the Cre-ERT2/Lox system, which leads to an irreversible recombination event, the induction of transcriptional activation by KalTA4-ERT2 is reversible by addition or removal of 4-OHT.
The following protocol allows a conditional transformation of skin cells in zebrafish larvae and a subsequent monitoring of the interaction with innate immune cells by fluorescent protein expression. The basic methodologies employed in this protocol are similar to some commonly used techniques within the zebrafish community9–13.
The generation and combinatory use of transgenic lines together with the microinjection of transgenic constructs enables a transient approach to only transform single cells in a mosaic manner. The oxygen permeability of the embryonic and larval zebrafish skin raises the possibility for time-lapse live imaging studies by high-resolution microscopy from hours to days. Furthermore, its accessibility to water-soluble drugs will allow future mechanistic studies and the monitoring of cell biological events in vivo that could lead to a better understanding of the earliest stages of cancer development.
This protocol can be adapted to perform conditional gene expression in any tissue of interest in zebrafish larvae, in a mosaic manner. One can also optimize the microscope setting in order to live image deeper tissues other than skin.
Protocol
The following experimental procedures were conducted in strict accordance with the Animals (Scientific Procedures) Act 1986.
1. Preparation of Plasmid DNA for Microinjection
Purify the plasmid pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP with a commercial plasmid DNA purification kit according to the manufacturer’s instructions and dilute the plasmid DNA to a final stock solution concentration of 100 ng/μl.
Linearize the transposase containing plasmid pT3TS/Tol214 by BamHI restriction digestion, following the manufacturer protocol. This step can be extended overnight. Precipitate the linearized DNA by a phenol/chloroform extraction, following the standard protocol.
In vitro transcribe the transposase mRNA from the linearized plasmid with a commercial transcription kit for large amounts of capped RNA according to the manufacturer’s instructions, dilute and aliquot to a final stock solution concentration of 100 ng/μl in nuclease-free water. Ensure quality of the purified RNA with a native 1-2% TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) agarose gel electrophoresis (80-120 V). Store the RNA at -80 °C.
2. Microinjection of Plasmid DNA for Transient Gene Expression
Prepare an agarose injection plate to hold the fertilized eggs during injection. Pour liquid 3% agarose into a 90 mm Petri dish. Let the agarose cool to 40-50 °C and add the wedged-shaped microinjection mold TU-1 on top of the liquid agarose. After hardening at room temperature, let the agarose rest at 4 °C for 30 min before removing the mold. Store microinjection molds at 4 °C and reuse.
Prepare an aliquot of the injection solution. Perform the following steps on ice. Pipette 1 μl of plasmid DNA (final concentration of 10 ng/μl), 2 μl of transposase mRNA (final concentration 20 ng/μl), 2 μl of 10 mg/μl Rhodamine B isothiocyanate-Dextran, 2 μl of 5x Danieau solution (290 mM NaCl, 3.5 mM KCl, 2 mM MgSO4, 3 mM Ca(NO3)2, 25 mM HEPES, pH 7.6) to a final volume of 10 μl in nuclease-free water.
Prepare needles from borosilicate glass capillaries of 1 mm diameter containing an inner filament by pulling the capillary in opposite directions with a Micropipette Puller. Use the following program: heat 530, pull 200, velocity 80 and time 150.
Fill one needle with 2 μl of the injection solution (kept on ice). Avoid bubbles by carefully withdrawing the microloader tip while releasing the solution into the capillary. Carefully break the tip of the needle with a forceps.
Regulate the drop size with a pneumatic pico pump. Measure the drop diameter/volume (V = 4/3πr3) and adjust the drop size with an ocular micrometer under a stereoscope in dimethylpolysiloxane. We recommend 100 μm, corresponding to 0.5 nl. This assures uniform and reproducible concentrations per each injected embryo. Titrate the injected concentration and adjust for each construct and plasmid preparation.
Place the zygotes (Figure 1A) obtained from a cross of Tg(UAS:eGFP-HRASG12V)io006 15 with Tg(lysC:dsRed2)nz50Tg 16 into the molds of the injection plate by use of a 3 ml pastette. Meanwhile, keep the tip of the pre prepared injection needle in dimethylpolysiloxane to avoid the solution from drying up and clotting.
Inject the premeasured drop (100 μm corresponding to 0.5 nl) under a stereoscope through the chorion into the blastodisc17, prior to the first cleavage (Figure 1A). Injection into the one-cell stage increases the potential of a uniform distribution of the plasmid DNA and RNA in the developing cell mass and the possibility for germline transmission. It is notable that the expression of constructs more often tends to be mosaic.
3. Conditional Skin Cell Transformation by 4-Hydroxytamoxifen (4-OHT)
Transfer the injected embryos into 0.3x Danieau solution and keep them at 28.5 °C in an incubator. Remove unfertilized eggs from the dish at sphere stage17. The co-injected Rhodamine B isothiocyanate-Dextran can be utilized as an injection-control under a fluorescent stereoscope.
Screen embryos at 24 hpf (hr post fertilization) for cardiac expression of eGFP. Continue to raise cmlc:eGFP positive zebrafish embryos until 72 hpf. Carefully monitor the water-quality (0.3x Danieau).
Remove the chorion of those embryos that have not hatched with forceps at 72 hpf. Carefully poke the chorion with 1 forceps and pull it apart by using the other. Select embryos for lysC:dsRed2 expression using a fluorescent stereoscope.
Add 10 μl of a 10 mM stock of (Z)-4-Hydroxytamoxifen to 20 ml of 0.3x Danieau solution (final concentration of 5 μM). Use the 4-OHT induction-solution for one standard Petri dish (90 mm) on a batch of 40-60 embryos.
Transfer the embryos into a new Petri dish containing 20 ml of freshly made induction-solution. 4-OHT has to be stored and handled in the dark. The first signs of expression may be detected 2 hr post induction commencement. The induction-step can be extended overnight, keeping in mind that initial host cell: preneoplastic cell interactions can already occur a few hours after treatment.
4. Mounting and Live Imaging of Zebrafish Larvae
Prepare a 60 mm Petri dish by introducing a hole with a diameter of 18-20 mm. Use high-vacuum silicone grease to cover and seal the hole on the outer bottom of the Petri dish with a 25 mm cover glass (Figure 1B).
Prepare 1% low melting point (LMP) agarose. Keep one 1.5 ml tube of LMP agarose at 37 °C in a heating block.
Preselect Tg(krt4:KalTA4-ERT2;UAS:eGFP-HRASG12V;lysC:dsRed2) larvae for green fluorescent skin cell and dsRed2 positive innate immune cell expression, under a fluorescent stereoscope. Anesthetize larvae by the addition of 1 ml of 0.4% tricaine18 to the 20 ml induction solution.
Transfer the preselected larvae with a pastette into the liquid LMP agarose. Let the larvae sink into the LMP agarose and transfer them onto the cover glass (Figure 1B). Orientate the embryos under a stereoscope. The larvae have to be placed directly onto the glass to reduce the working distance (up to 6 larvae).
Cover the LMP agarose with the tricaine containing induction solution after it has hardened (Figure 1B arrow).
Image the embedded larvae using an inverted fluorescent, laser-scanning or spinning disc confocal microscope. Use a 40X or 63X objective as objectives with long working distances enable focusing through the entire larvae.
Representative Results
Zygotes of a cross between Tg(UAS:eGFP-HRASG12V)io006 and Tg(lysC:dsRed2)nz50Tg were collected 10-15 min post fertilization (Figure 1A) to obtain 1 cell-stage embryos. After injection the embryos were raised until 3 dpf, before induction with 4-OHT and mounting for live imaging (Figure 1B). 2-6 hr post induction the mounted embryos were placed either under an inverted fluorescent microscope (Figure 2A) or an inverted confocal microscope (Figure 2B-C) and imaged for 2-3 hr with 1.5 min intervals. Preneoplastic superficial skin cells can be identified by their more rounded morphology compared to normal keratinocytes, as well as the green fluorescent emission of the membrane localized eGFP-HRASG12V. Areas of imaging were selected for co-localization of preneoplastic skin cells and innate immune cells (Figure 2A arrowhead). Due to the fast migration process of the neutrophils the time-lapse intervals were chosen to be between 1-1.5 min with Z stack step sizes of 0.7-1.7 μm to obtain maximum intensity projections (Figure 2, Video 1). There is a wide range of behaviors that can be observed while live imaging: neutrophils migrate next to and under preneoplastic cells (Figure 2C, Video 1), form direct contacts to cells (Figure 2B and C), remove remnants of preneoplastic cells that undergo apoptosis (Video 1), or actively engulf parts of preneoplastic cells (not shown). To capture the full spectrum of interactions, it is advisable to follow their behaviors by time-lapse live imaging.
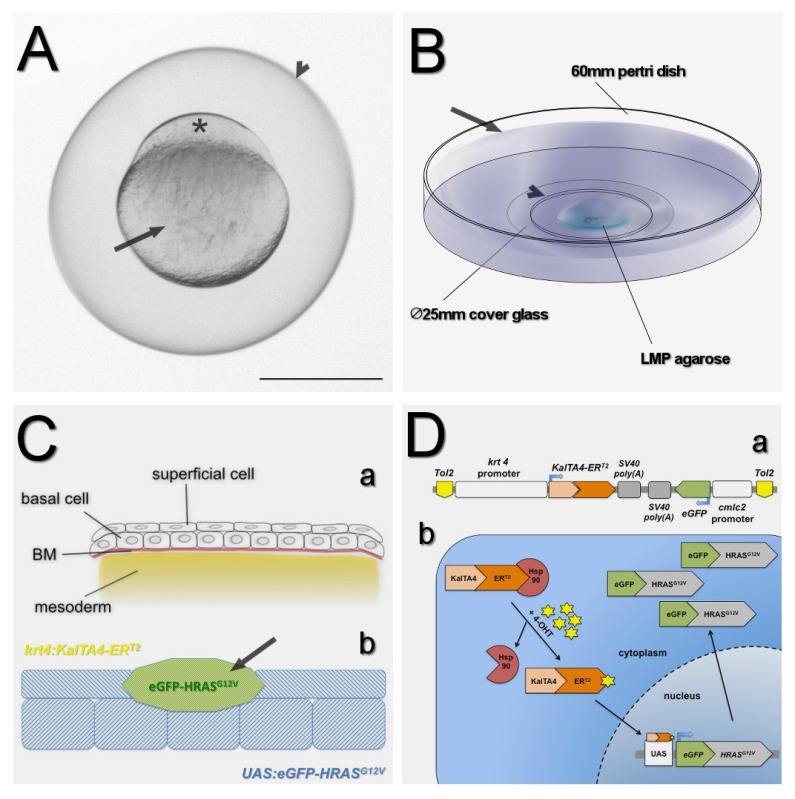 Figure 1: Schematic for conditional skin cell transformation in zebrafish larvae. (A) An image of a zygote of Tg(UAS:eGFP-HRASG12V;lysC:dsRed2) zebrafish at 10 min post fertilization (mpf). The fertilized egg is surrounded by the chorion (arrowhead). The site for injection is the blastodisc (asterisk), formed by cytoplasmic streams from the yolk (arrow) that defines the animal pole of the embryo. (B) Mounting of zebrafish larvae. 60 mm Petri dish with an 18-20 mm hole (arrowhead) that is sealed by a 25 mm cover glass. The zebrafish larvae are immobilized and mounted onto the cover glass by 1% LMP agarose. 0.3x Danieau solution (arrow) containing 5 μM 4-OHT is used to cover the LMP agarose. (C) 4-OHT induced transformations of superficial skin cells. (a) Schematic cross sectional profile of larval skin. The embryonic and larval skin is composed of two epithelial cell layers, a superficial cell layer and a basal cell layer of keratinocytes that are connected to the underlying basement membrane (BM). (b) Transient expression system to induce a preneoplastic cell clone amongst superficial skin cells. KalTA4-ERT2 is exclusively expressed in the outermost skin layer driven by the krt4 promoter (yellow). Mosaic expression of KalTA4-ERT2 activates UAS controlled (blue) eGFP-HRASG12V expression in superficial cells, leading to a clone of preneoplastic cells (green). (D) Schematic of the transient expression system. (a) Tol2 cassette of the pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP expression vector used transiently. (b) Conditional induction of the transcriptional activator expression. KalTA4 is fused to the mutant ligand-binding domain of the human estrogen receptor α (ERT2) which binds specifically 4-Hydroxytamoxifen (4-OHT). In the absence of 4-OHT the ERT2 is bound in the cytoplasm by heat-shock proteins (Hsp90). Upon 4-OHT binding Hsp90 dissociates, allowing a nuclear translocation of KalTA4-ERT2 and subsequent activation of the UAS controlled eGFP-HRASG12V expression. Scale bar A = 500 μm. Please click here to view a larger version of the figure.
Figure 1: Schematic for conditional skin cell transformation in zebrafish larvae. (A) An image of a zygote of Tg(UAS:eGFP-HRASG12V;lysC:dsRed2) zebrafish at 10 min post fertilization (mpf). The fertilized egg is surrounded by the chorion (arrowhead). The site for injection is the blastodisc (asterisk), formed by cytoplasmic streams from the yolk (arrow) that defines the animal pole of the embryo. (B) Mounting of zebrafish larvae. 60 mm Petri dish with an 18-20 mm hole (arrowhead) that is sealed by a 25 mm cover glass. The zebrafish larvae are immobilized and mounted onto the cover glass by 1% LMP agarose. 0.3x Danieau solution (arrow) containing 5 μM 4-OHT is used to cover the LMP agarose. (C) 4-OHT induced transformations of superficial skin cells. (a) Schematic cross sectional profile of larval skin. The embryonic and larval skin is composed of two epithelial cell layers, a superficial cell layer and a basal cell layer of keratinocytes that are connected to the underlying basement membrane (BM). (b) Transient expression system to induce a preneoplastic cell clone amongst superficial skin cells. KalTA4-ERT2 is exclusively expressed in the outermost skin layer driven by the krt4 promoter (yellow). Mosaic expression of KalTA4-ERT2 activates UAS controlled (blue) eGFP-HRASG12V expression in superficial cells, leading to a clone of preneoplastic cells (green). (D) Schematic of the transient expression system. (a) Tol2 cassette of the pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP expression vector used transiently. (b) Conditional induction of the transcriptional activator expression. KalTA4 is fused to the mutant ligand-binding domain of the human estrogen receptor α (ERT2) which binds specifically 4-Hydroxytamoxifen (4-OHT). In the absence of 4-OHT the ERT2 is bound in the cytoplasm by heat-shock proteins (Hsp90). Upon 4-OHT binding Hsp90 dissociates, allowing a nuclear translocation of KalTA4-ERT2 and subsequent activation of the UAS controlled eGFP-HRASG12V expression. Scale bar A = 500 μm. Please click here to view a larger version of the figure.
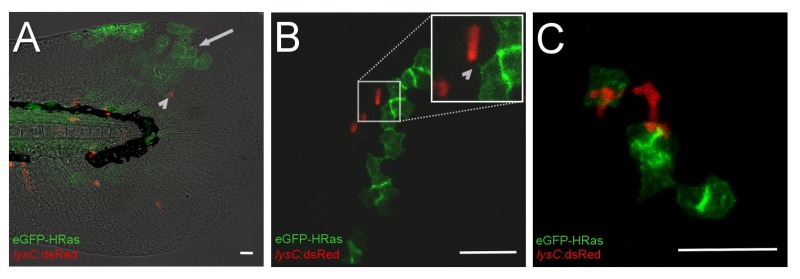 Figure 2: Live imaging of preneoplastic and innate immune cell interaction. (A-C) Images of a 4 dpf Tg(UAS:eGFP-HRASG12V;lysC:dsRed2) embryo, which has been injected with pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP and transposase mRNA at one cell stage. Images were taken after 6 hours of 4-OHT induction. (A) Lateral view of the tailfin region at 6 hours post treatment, showing patches of eGFP-HRASG12V expressing skin cells (arrow), and lysC:dsRed2+ neutrophils (arrowhead). (B-C) Representative images taken from a confocal time-lapse movie, showing neutrophil (red) interactions with eGFP-HRASG12V expressing skin cells (green). (B) A preneoplastic cell forms a short filopodial extension with the contacting neutrophil (arrowhead). (C) Neutrophils (red) is in close contact to a clone of preneoplastic skin cells (green). Scale bar in A = 100 μm; B/C = 50 μm. Please click here to view a larger version of the figure.
Figure 2: Live imaging of preneoplastic and innate immune cell interaction. (A-C) Images of a 4 dpf Tg(UAS:eGFP-HRASG12V;lysC:dsRed2) embryo, which has been injected with pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP and transposase mRNA at one cell stage. Images were taken after 6 hours of 4-OHT induction. (A) Lateral view of the tailfin region at 6 hours post treatment, showing patches of eGFP-HRASG12V expressing skin cells (arrow), and lysC:dsRed2+ neutrophils (arrowhead). (B-C) Representative images taken from a confocal time-lapse movie, showing neutrophil (red) interactions with eGFP-HRASG12V expressing skin cells (green). (B) A preneoplastic cell forms a short filopodial extension with the contacting neutrophil (arrowhead). (C) Neutrophils (red) is in close contact to a clone of preneoplastic skin cells (green). Scale bar in A = 100 μm; B/C = 50 μm. Please click here to view a larger version of the figure.
Video 1: Time-lapse live imaging of preneoplastic and innate immune cell interaction. Time-lapse of a 4 dpf Tg(UAS:eGFP-HRASG12V;lysC:dsRed2) embryo, injected with pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP and transposase mRNA at one cell stage. Confocal time-lapse movie from 6-9 hours after 4-OHT induction showing lysC:dsRed2+ neutrophil (red) interactions with eGFP-HRASG12V expressing skin cells (green). Neutrophils migrate next to and under preneoplastic cells and remove remnants of a preneoplastic cell that undergoes apoptosis. Time-lapse intervals of 1.5 min over 3 hr with Z stack step sizes of 1.3 μm.
Discussion
During embryogenesis and larval development, the embryonic zebrafish skin is composed of two epithelial layers, the superficial layer called periderm and a layer of basal keratinocytes that are attached to the underlying basement membrane19 (Figure 1C, a). The protocol presented here describes a straightforward method to conditionally induce preneoplastic-cell transformation in the superficial skin layer of zebrafish larvae, and allows for subsequent interaction analysis with innate immune cells by live imaging. The basic methodologies in our protocol follow well-established zebrafish techniques9–13, and our protocol can be easily adapted and modified.
The krt4 promoter5 used here drives KalTA4-ERT2 expression specifically in the outermost skin layer (Figure 1C, b). However, the modular MultiSite Gateway cloning strategy, used to generate the krt4:KalTA4-ERT2construct (Figure 1D, a), provides the possibility to clone any promoter of interest in front of KalTA4-ERT2 in a single cloning step. An adaptation of the method presented herein is therefore possible for most tissues during embryogenesis and larval stages. The availability of promoter sequences is hereby the limiting factor. Furthermore, almost any gene of interest cloned behind a UAS can be conditionally overexpressed at different developmental stages by use of the spatial and temporally controlled KalTA4-ERT2 expression. Therefore, our protocol can be adapted to perform conditional gene expression in any tissue of interest in zebrafish larvae, in a mosaic manner. One can also optimize the microscope setting in order to live image deeper tissues such as liver or pancreas.
We have described one application using the protocol here, similarly, one can overexpress other inflammation modulators using this protocol to study the regulation of inflammatory responses, as one can easily live image neutrophil and macrophage behavioral changes following the induction and arrest in the expression of a candidate inflammatory modulator. Furthermore, the adapted protocol would also be beneficial for those who want to trace cell movement and behavior change during different stages of tissue morphogenesis and organ formation and, finally live image the interaction of innate immune cell interactions with other specific cell lineages that can be targeted by the inducible KalTA4-ERT2/UAS expression system.
We used the pDestTol2CG vector from the zebrafish Tol2kit20–23 that contains a cmlc2:eGFP-pA cassette (Figure 1D, a) which allows subsequent selection of embryos by green fluorescent positive hearts as a selection marker20 that simplifies the F0 screening process. A stable insertion of the transposon flanked gene of interest into the genome is facilitated by co-injection of the plasmid DNA together with transposase mRNA. The preparation of plasmid DNA and the transposase mRNA for microinjection follows standard protocols. However, a successful integration of the construct into the genome depends on the Tol2-based transposition14. This highlights the particular attention to be paid to work on ice under sterile conditions to avoid contaminations and degradations by RNases and DNases. Microinjection into one-cell stage embryos is a robust and well-established technique for gain and loss of function studies in zebrafish9,10. Although a well-trained person can easily inject into the yolk of over 1,000 embryos in 1 hr, the injection into the blastodisc (Figure 1A, asterisk) requires more experience and training. Critical during this procedure is the handling of the needle. The needle has to be very thin to penetrate the cell without damage and therefore has to be broken only on its very tip. It is important to regularly control and measure the drop size during the injection to guarantee a uniform injection into each embryo. Carefully handled, the needle can be used to rotate the embryo. This is often needed to find the blastodisc and the optimal penetration angle.
The concentration of DNA and transposase mRNA used for injection almost certainly influences the expression efficiency. Higher doses can lead to an increased proportion of cells expressing the transgene but with higher concentrations of DNA (>50 ng/μl) and transposase mRNA (>100 ng/μl) also the potential for toxic effects amongst injected embryos does arise. We favor the use of 10 ng/μl DNA and 20 ng/μl transposase mRNA for injection, which corresponds to 50 pg of plasmid DNA and 100 pg of RNA per injected embryo. 100% of embryos injected with these concentrations do develop normally, and between 40-70% show eGFP-HRASG12V expression, after 4-OHT induction, depending on the injection efficiency into the single cell. Amongst positive embryos we normally find approximately 30% that have 1-10 clones, 40% that have 10-50 clones and 30% having more than 50 clones. Due to our research aims, we favor the use of embryos with fewer clones expressing eGFP-HRASG12V. We select embryos with 10-50 clones for our transient experiments. For the generation of transgenic founder fish, we recommend to only select embryos with both the eGFP positive cardiac marker and a large number of eGFP-HRASG12V positive clones in their epidermis.
(Z)-4-Hydroxytamoxifen undergoes a cis-trans (E-Z) interconversion when exposed to light24. It was found that the cis isomer (E) is 100x less anti-estrogenic than its trans counterpart25,26. Exposure to light therefore has to be avoided. The 4-OHT stock solution dissolved in ethanol can be stored at -20 °C in the dark over a long time period. We recommend the use of a box to protect the Petri dishes from light during target gene induction within the incubator and transport. Although the area of imaging is very small and exposure of 4-OHT to UV light is reduced to a minimum, a constant exchange of induction solution every 2 hours while imaging over longer time periods is recommended to maintain maximum expression levels. 4-OHT effectively permeabilizes through the chorion and the outermost skin layers during zebrafish embryogenesis, therefore it can induce gene expression very rapidly. We can readily observe eGFP emission after 2 hr of induction and it has been reported that first signs of target-gene expression can be observed after 1 hour and plateau after 3 hr of treatment8. The differences might be due to the different promoter used in our study. As has been previously reported that 4-OHT treatment is dose dependent8,27, we chose to use the highest reported concentration of 5 μM to achieve robust and reproducible expression levels in our transient approach. This should guarantee that all cells carrying the transgene will induce target gene expression.
It has to be taken into account that the transient approach presented here could lead to varying expression levels due to the possibility of multiple and random genome insertion events. Furthermore, the use of the transposase mRNA in the Tol2 transgenic background of Tg(UAS:eGFP-HRASG12V)io006 can lead to potential transpositions of the UAS transgene that could result in gene silencing or disruption. However, as the method presented here represents a transient approach, the pre-selection of embryos subsequent to injection is mandatory. Many labs have found that UAS sequences tend to be silenced in the transgenic offspring, hence injecting the pTol2-krt4:KalTA4-ERT2;cmlc2:eGFP construct into Tg(UAS:eGFP-HRASG12V)io006 embryos might not be as efficient as injecting a UAS construct into Tg(krt4:KalTA4-ERT2;cmlc2:GFP) embryos. The use of the stable transgenic line Tg(krt4:KalTA4-ERT2) crossed with Tg(UAS:eGFP-HRASG12V)io006 will allow a precise monitoring of the on/off kinetics of the time-dosage dependent 4-OHT gene activation.
A critical step during the mounting of larvae is the transfer into the LMP agarose and onto the cover glass (Figure 1B). There is only a short time frame for liquid LMP agarose temperature that allows a safe transfer and orientation of the preselected larvae without harming the fish by either heat shock or hardening of the gel. The larvae should be orientated directly onto the cover glass to reduce the working distance for the subsequent microscopic analysis. As this can be a limiting factor during microscopy the choice of appropriate objectives is mandatory. Once mounted, the larva can be maintained from hours to days.
The conditionality of the model system presented herein allows for repeated transformation by 4-OHT activation of the transgene. The expression system depends on the presence of 4-OHT and therefore the KalTA4 controlled expression of a gene of interest is reversible (Figure 1D, b). In contrast to the Cre-ERT2/Lox system, which facilitates an irreversible genomic recombination event28, KalTA4-ERT2/UAS can be activated and deactivated upon addition or removal of 4-OHT and this potential for repeated induction is an advantage of the technique over the Cre-ERT2/Lox system. However, the requirement for constant levels of 4-OHT is a limitation if the experiment has to be prolonged from days to weeks.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was funded by a Wellcome Trust Sir Henry Dale Fellowship to Y. F. 100104/Z/12/Z.
We would like to thank Dr. Carl Tucker and the team of the fish facility at the Queen’s Medical Research Institute, University of Edinburgh. We like to thank Michael Millar of the immunohistology and imaging facility of the MRC Centre for Reproductive Medicine, University of Edinburgh for support.
We thank Prof. Dr. Matthias Hammerschmidt and Dr. Heiko Loehr of the Institute of Developmental Biology, University of Cologne for providing the p5E-krt4 entry vector. We also would like to thank Dr. Dirk Sieger of the Centre of Neuroregeneration, Univerity of Edinburgh for providing the pDestTol2CG destination vector.
References
- Kajita M, Sugimura K, et al. Filamin acts as a key regulator in epithelial defence against transformed cells. Nature communications. 2014;5:4428. doi: 10.1038/ncomms5428. [DOI] [PubMed] [Google Scholar]
- Feng Y, Santoriello C, Mione M, Hurlstone A, Martin P. Live imaging of innate immune cell sensing of transformed cells in zebrafish larvae: parallels between tumor initiation and wound inflammation. PLoS Biology. 2010;8(12):e1000562. doi: 10.1371/journal.pbio.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Renshaw S, Martin P. Live Imaging of Tumor Initiation in Zebrafish Larvae Reveals a Trophic Role for Leukocyte-Derived PGE 2. Current Biology. 2012;22(13):1253–1259. doi: 10.1016/j.cub.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, Leach SD. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5(2):97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Ju B, et al. fluorescent protein expression in germ-line transmitted transgenic zebrafish under a stratified epithelial promoter from keratin8. Developmental Dynamics an Official Publication of the American Association of Anatomists. 2002;223(2):204–215. doi: 10.1002/dvdy.10051. [DOI] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Köster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochemical and Biophysical Research Communications. 1997;237(3):752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Breau Ma, Sasai N, Xu Q, Briscoe J, Wilkinson DG. An inducible transgene expression system for zebrafish and chick. Development. 2013;140(10):2235–2243. doi: 10.1242/dev.091520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Sun Z. Microinjection of mRNA and Morpholino Antisense Oligonucleotides in Zebrafish Embryos. J. Vis. Exp. 2009. p. e1113. [DOI] [PMC free article] [PubMed]
- Rosen JN, Sweeney MF, Mably JD. Microinjection of Zebrafish Embryos to Analyze Gene Function. J. Vis. Exp. 2009. p. e1115. [DOI] [PMC free article] [PubMed]
- Eisenhoffer GT, Rosenblatt J. Live Imaging of Cell Extrusion from the Epidermis of Developing Zebrafish. J. Vis. Exp. 2011. p. e2689. [DOI] [PMC free article] [PubMed]
- Kague E, Weber C, Fisher S. Mosaic Zebrafish Transgenesis for Evaluating Enhancer Sequences. J. Vis. Exp. 2010. p. e1722. [DOI] [PMC free article] [PubMed]
- Balciuniene J, Gene Balciunas D. Trapping Using Gal4 in Zebrafish. J. Vis. Exp. 2013. p. e50113. [DOI] [PMC free article] [PubMed]
- Balciunas D, Wangensteen KJ, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genetics. 2006;2(11):e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoriello C, Gennaro E, et al. Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in zebrafish. PloS One. 2010;5(12):e15170. doi: 10.1371/journal.pone.0015170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Developmental Biology. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental Dynamics : an Official Publication of the American Association of Anatomists. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Westerfield M. THE ZEBRAFISH BOOK: A guide for the laboratory use of zebrafish (Danio rerio) 5th Edition. Eugene: University of Oregon Press; 2007. [Google Scholar]
- Le Guellec D, Morvan-Dubois G, Sire J-Y. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio) The International Journal of Developmental Biology. 2004;48(2-3):217–231. doi: 10.1387/ijdb.15272388. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental Dynamics : an Official Publication of the American Association of Anatomists. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Protocols and Product Manuals. 12537-023. Invitrogen life technologies; 2010. MultiSite Gateway Three- Fragment Vector Construction Kit. [Google Scholar]
- Petersen LK, Stowers RS. A Gateway MultiSite Recombination Cloning Toolkit) PLoS ONE. 2011;6(9):e10. doi: 10.1371/journal.pone.0024531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E, Bartling L, Hake S. From Gateway to MultiSite Gateway in one recombination event. BMC Molecular Biology. 2006;7:46. doi: 10.1186/1471-2199-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DW, Katzenellenbogen JA. Synthesis of the (E) and (Z) isomers of the antiestrogen tamoxifen and its metabolite, hydroxytamoxifen, in tritium-labeled form. The Journal of Organic Chemistry. 1982;47(12):2387–2393. [Google Scholar]
- Murphy C, Langan-Fahey S. Structure-function relationships of hydroxylated metabolites of tamoxifen that control the proliferation of estrogen-responsive T47D breast cancer cells in vitro. Molecular. 1990;38(5):737–743. [PubMed] [Google Scholar]
- Furr BJA, Jordan VC. The pharmacology and clinical uses of tamoxifen. Pharmacology & Therapeutics. 1984;25(2):127–205. doi: 10.1016/0163-7258(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Akerberg Aa, Stewart S, Stankunas K. Spatial and Temporal Control of Transgene Expression in Zebrafish. PloS One. 2014;9(3):e92217. doi: 10.1371/journal.pone.0092217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PloS One. 2009;4(2):e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]


